Abstract
Leucocytozoon, a widespread hemosporidian blood parasite that infects a broad group of avian families, has been studied in corvids (family: Corvidae) for over a century. Current taxonomic classification indicates that Leucocytozoon sakharoffi infects crows and related Corvus spp., while Leucocytozoon berestneffi infects magpies (Pica spp.) and blue jays (Cyanocitta sp.). This intrafamily host specificity was based on the experimental transmissibility of the parasites, as well as slight differences in their morphology and life cycle development. Genetic sequence data from Leucocytozoon spp. infecting corvids is scarce, and until the present study, sequence data has not been analyzed to confirm the current taxonomic distinctions. Here, we predict the phylogenetic relationships of Leucocytozoon cytochrome b lineages recovered from infected American Crows (Corvus brachyrhynchos), yellow-billed magpies (Pica nuttalli), and Steller’s jays (Cyanocitta stelleri) to explore the host specificity pattern of L. sakharoffi and L. berestneffi. Phylogenetic reconstruction revealed a single large clade containing nearly every lineage recovered from the three host species, while showing no evidence of the expected distinction between L. sakharoffi and L. berestneffi. In addition, five of the detected lineages were recovered from both crows and magpies. This absence of the previously described host specificity in corvid Leucocytozoon spp. suggests that L. sakharoffi and L. berestneffi be reexamined from a taxonomic perspective.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leucocytozoids are abundant blood parasites that exclusively infect birds. They are known to infect a wide range of avian host species across a wide diversity of habitats (Valkiūnas 2005). Many Leucocytozoon species are pathogenic to both wild and domestic birds, so understanding their diversity and pathogenicity could have vital conservation implications (Morii 1992; Hunter et al. 1997; Smith et al. 1998; Sehgal et al. 2006a, b; Bunbury et al. 2007). As with other avian blood parasites, leucocytozoids must utilize a vector and host to complete their life cycle; however, the specificity of the host-vector-parasite interaction of Leucocytozoon species remains the least studied of the three major avian hemosporidian parasites (Haemoproteus, Plasmodium, Leucocytozoon). Leucocytozoids are vectored by blood-sucking black flies (Diptera: Simuliidae), and with over 1800 recognized black fly species, Leucocytozoon has a large number of potential vectors (Crosskey and Howard 2004; Hellgren et al. 2008). An investigation of black fly blood meals revealed that individual species tend to target either mammals or birds and that ornithophilic species preferentially target large and abundant host species (Malmqvist et al. 2004). Based on these criteria, the authors conclude that corvids are an expected host for black flies.
Recent studies have suggested that corvids may be an important reservoir for avian blood parasites (Kim and Tsuda 2010; Leclerc et al. 2014). Corvids (family: Corvidae) are large, gregarious birds that are well adapted to life around humans (Johnston 2001). Increasing urbanization and sprawl favors birds that are able to exploit anthropogenic resources, thereby contributing to increased global populations (Marzluff et al. 2001; Marzluff and Neatherlin 2006). The expansion of cities has reinforced the importance of understanding the effects of urbanization on the ecology of wildlife-parasite interactions (Bradley and Altizer 2007). Knowledge of blood parasite-bird interactions in urban areas is characterized by contradictory findings on parasite prevalence, diversity, and host impact (reviewed in Delgado-V and French 2012). Urban-exploiting corvids represent an important host for further study of blood parasite ecology in an increasingly urbanized world.
Much of the current understanding of Leucocytozoon parasites infecting corvids is either outdated or incomplete. Over the past century, many studies have documented Leucocytozoon in corvids; however, these studies have relied solely upon blood smear or tissue sample examinations, detecting a range of prevalence from 2.7 to 36.4 % (Morgan and Waller 1941; Jones 1968; Murata 2002; Leclerc et al. 2014). In contrast, a recent study using molecular techniques detected both high prevalence (93.6 %) and a high diversity of Leucocytozoon parasites in corvids (Yoshimura et al. 2014). This and other studies revealed that leucocytozoids exhibit genetic diversity exceeding that observed in other blood parasites (i.e. Plasmodium and Haemoproteus spp.) (Martinsen et al. 2006, 2008; Sehgal et al. 2006a, b; Jenkins and Owens 2011). The detection of genetic diversity has led to the recent characterization of a cryptic species complex of the avian blood parasite Plasmodium homocircumflexum; in this case, the newly discovered species differs from its morphologically identical counterpart species in multiple ways, namely species-level genetic differences and increased pathogenicity in experimentally infected birds (Palinauskas et al. 2015). Mounting evidence suggests that Leucocytozoon could harbor undiscovered cryptic species as well (Bensch et al. 2004; de León and Nadler 2010; Lotta et al. 2013). A reexamination of Leucocytozoon in diurnal raptors, for example, yielded probable cryptic speciation, although morphological distinctions were later observed (Sehgal et al. 2006a; Valkiūnas et al. 2010). It is essential, therefore, to clarify the relationships among potential cryptic species infecting corvids.
The majority of Leucocytozoon species are specific only to the level of host family or order (Valkiūnas 2005). In contrast, there are two Leucocytozoon species specific to the corvid host family. Both described over a century ago, Leucocytozoon sakharoffi Sambon, 1908, was initially distinguished from Leucocytozoon berestneffi Sambon, 1908, based on how completely the host cell nucleus surrounds the parasite (Bennett and Peirce 1992). This initial morphological difference, however, did not bear out under further analysis. Subsequent studies revealed that the two parasite species could not be microscopically differentiated based on morphology of gametocytes or host cell (Ramisz 1962; Bennett and Peirce 1992; Valkiūnas 2005). However, other distinguishing features emerged to allow for the continued separation of the two corvid leucocytozoids: species differences in hepatic merogony, pre-patent period, and host transmissibility (Clark 1965; Khan and Fallis 1971; Valkiūnas 2005). Leucocytozoon might exhibit host specificity among the corvids: Leucocytozoon spp. found in American Crows and Common Ravens (Corvus brachyrhynchos, Corvus corax) were not experimentally transmitted to a blue jay (Cyanocitta cristata), and vice versa (Khan and Fallis 1971). In addition, the meronts described from the liver of C. cristata were determined to be identical in form and size to those found in magpies (Pica sp.) (Clark 1965; Valkiūnas 2005). Therefore, the original taxonomic description remains in place, with two Leucocytozoon parasites specific to the corvid family: L. sakharoffi infecting birds from the genus Corvus, and L. berestneffi restricted to corvids from the Cyanocitta and Pica genera (Valkiūnas 2005). This classification, however, is not universally accepted within the field of avian parasitology. A contradictory stance was taken by Bennett and Peirce (1992), who, after careful review of multiple historical studies, determined L. berestneffi to be a junior synonym of L. sakharoffi.
In this study, we used genetic data to clarify discrepancies in the literature regarding Leucocytozoon host specificity in corvids. Using molecular techniques and analyses, we investigated the specificity and diversity of Leucocytozoon in populations of American Crows (C. brachyrhynchos), yellow-billed magpies (Pica nuttalli), and Steller’s jays (Cyanocitta stelleri). We used a fragment of the cytochrome b gene (cyt b) from the mitochondrial DNA (mtDNA) to infer the phylogenetic relationship of Leucocytozoon sequences obtained from these populations. We hypothesized that distinct phylogenetic clustering of Leucocytozoon lineages according to host species would reflect the taxonomic separation of L. sakharoffi and L. berestneffi. Our study is the first to use Leucocytozoon cyt b haplotypes from corvid populations to investigate host specificity in this avian family.
Methods
Study area and sampling
Blood samples were collected from 198 American Crow nestlings from 82 nests in Davis, CA, in 2013 (n = 110 nestlings from 45 nests) and 2014 (n = 88 nestlings from 37 nests). The study site spanned the urban campus of the University of California, Davis, into the surrounding campus-owned agricultural areas (e.g., vineyards, pasture, and row crops: described in Townsend and Barker 2014). Nests were situated on lateral tree branches and accessed by boom lift. Birds were sampled 10–28 days after hatching. Adult crows were captured in Davis, CA, using Australian drop-in crow traps in January of 2014 (n = 16 crows) and 2015 (n = 44 crows). Adults were captured from a large, overwintering urban flock (>7000 birds), comprising both residents and migratory birds (Hinton et al. 2015). Blood samples were taken from the jugular vein of the crows using 27 gauge ½ inch needles and 1-mL syringes. Samples were placed immediately in Queen’s lysis buffer (Seutin et al. 1991) at an approximate rate of 50-μL blood to 1-mL buffer and kept at room temperature until extraction. All field work pertaining to American Crows was performed under protocols approved by the Institutional Animal Care and Use Committee of the University of California, Davis (Permit Number: 16897).
Yellow-billed magpie samples (n = 44) were collected from California’s Central Valley from 2004 to 2010 and were subsampled at random from a previous study (Ernest et al. 2010). Steller’s jay samples were graciously donated from specimens at the Museum of Vertebrate Zoology, University of California, Berkeley. All Steller’s jays were collected from El Dorado County, CA, in November 2001.
Parasite detection and sequencing
DNeasy Blood and Tissue extraction kits (Qiagen, Valencia, CA) were used to extract genomic DNA from blood samples from American Crows and yellow-billed magpies. DNA was recovered from liver samples of Steller’s jays using the same extraction technique. Successful DNA extraction from crow samples was confirmed through genotyping, as described in Townsend et al. (2009). DNA extraction success in magpie and jay samples was verified by amplification of the brain-derived neurotrophic factor (Sehgal and Lovette 2003). We screened for Leucocytozoon blood parasites using PCR amplification of a partial sequence of the parasite mtDNA cyt b gene. A nested PCR was used to detect Leucocytozoon spp. using the primers NF/NR3-FL/R2L as described in Hellgren et al. (2004), with a variation in the temperature profile of the second, nested PCR reaction. That is, we used a step-down approach, with annealing temperatures of 58, 56, 54, and 52 °C for 3 cycles each, and the remaining 23 cycles at 50 °C. Each PCR reaction was run in 25-μL volumes, accompanied by a positive control which was previously verified by sequencing and microscopy, and a negative control in which purified water was used in place of DNA template. The PCR amplicons were visualized on a 1.8 % agarose gel using ethidium bromide staining under ultraviolet light.
PCR samples with parasite cyt b amplification were purified with ExoSAP (United States Biochemical Corporation, Cleveland, OH) according to the manufacturer’s directions. We identified lineages by sequencing the fragments bidirectionally using the BigDye version 1.1 sequencing kit (Applied Biosystems, Inc., Foster City, CA) on an ABI PRISM 3100™ automated sequencer (Applied Biosystems).
Phylogenetic analyses
All sequences were edited in Sequencher 4.9 (GeneCodes, Ann Arbor, MI). If the chromatograph of any sequence showed one or numerous double peaks, infection status was categorized as mixed. No attempt was made to parse out the individual sequences of mixed infections due to the possibility that more than two parasites were present in a sample; samples with mixed infections were scored as Leucocytozoon positive, but not included in any further genetic analyses. Remaining sequences were aligned by eye in MacClade 4.08a (Maddison and Maddison 2005). The sequence dataset was condensed to eliminate redundant sequences and then subject to a BLAST search to ensure that a parasite of the expected genus was accurately amplified. After trimming primers, a fragment length of 476 bp was obtained. Any unique sequence, differing by one or more nucleotide from other recorded sequences, was considered to be a distinct Leucocytozoon lineage. However, while each lineage is considered separately when determining diversity data and phylogenetic predictions, no assumption of species-level difference is implied by distinct lineages and should not be viewed as such.
The dataset used for phylogenetic reconstruction consisted of 28 Leucocytozoon lineages detected in corvids and 8 reference sequences of Leucocytozoon species downloaded from the MalAvi (Bensch et al. 2009) database or GenBank, each trimmed to 456 bp to ensure consistency of sequence length. The corvid lineages include our American Crow, yellow-billed magpie, and Steller’s jay samples from California, as well as nine Leucocytozoon lineages recovered from crow species (Corvus corone; Corvus macrorhynchos) in Japan (Yoshimura et al. 2014). A cyt b sequence from Leucocytozoon mathisi was downloaded from GenBank (DQ177252) and used as an outgroup. We analyzed the dataset in MrModeltest v3.7 (Posada and Crandall 1998) to determine which nucleotide substitution model was appropriate based on the Akaike Information Criterion (Huelsenbeck and Crandall 1997). Trees were constructed using a Bayesian inference performed in MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003). The GTR + G model was implemented, and two Markov chains were run simultaneously for 10 million generations; trees were sampled every 200 generations, resulting in 50,000 trees. After a burn-in period of 25 %, the remaining 37,500 trees were used to calculate posterior probabilities. To estimate bootstrap values, and to check for congruence of phylogenetic relationships across multiple approaches, a maximum likelihood (ML) approach was implemented in PAUP version 4 (Swofford 2002). A thorough bootstrap analysis using 1000 replicates was used to determine support of individual branches in the program RAxML (Stamatakis 2006). The Bayesian majority consensus tree and maximum likelihood phylogram were viewed and edited with FigTree 1.4 (Rambaut 2012) and Adobe Illustrator.
Results
Prevalence and diversity
Using PCR detection methods, the prevalence of Leucocytozoon parasites in American Crow samples over a 3-year period (n = 258) was 57.4 %. Infection prevalence remained relatively consistent across years, ranging from 50.0 to 62.7 %. Of the 44 yellow-billed magpie blood samples selected randomly from the population detailed in Ernest et al. (2010), 54.5 % (24) were infected with one or more Leucocytozoon lineages. One hundred percent of the Steller’s jays sampled (n = 20) harbored a Leucocytozoon lineage.
Partial cyt b mtDNA sequencing revealed 13 distinct Leucocytozoon lineages in the American Crow population (Table 1), 10 of which are a novel cyt b lineage not previously recorded in GenBank; three lineages have been recovered recently from black fly blood meals in Colorado (Murdock et al. 2015). Five lineages accounted for 89.1 % of single infections in the population. In the yellow-billed magpie population, we identified nine distinct cyt b lineages. Interestingly, five of these lineages are identical to a lineage also detected in the American Crow population. Of the four lineages discovered solely in magpies, three were previously unrecorded; one lineage (YBMag.4), however, had been isolated from a different corvid (Western Scrub Jay, Aphelocoma californica) in California (unpublished, GenBank no. KJ584594). We detected two novel Leucocytozoon lineages in the Steller’s jay samples. Neither of the lineages detected in Steller’s jays were identical to the lineages infecting American Crows or yellow-billed magpies. Mixed infections were recorded in 25.7 % of crow, 20.8 % of magpie, and 35 % of jay samples, respectively. All lineages have been deposited in GenBank under accession nos. KU842391–KU842409.
Phylogenetic and genetic distance analyses
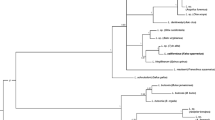
Both Bayesian inference (Fig. 1) and ML predictions (not shown, but bootstrap values imported to Bayesian tree) suggest that 26 of the 28 corvid lineages fall within a large clade with strong nodal support. However, the relationships between individual lineages within this clade remain mostly unresolved, displaying low nodal support. Furthermore, no pattern emerges that suggests a species-level distinction between Leucocytozoon spp. recovered from crows versus magpies and jays; lineages originating from each host species are interspersed throughout the large corvid clade. Also contained within this clade are two reference sequences (Leucocytozoon majoris; Leucocytozoon dubreuili).
Consensus tree displaying Leucocytozoon phylogenetic relationships as predicted by Bayesian inference, using GTR + G substitution model in MrBayes v3.1.2. Posterior probabilities >0.95 (above branch) and maximum likelihood bootstrap values (calculated in RAxML) >70 (below branch) are indicated. Lineages detected in the current study are indicated in bold. Sequences displaying GenBank accession numbers are either corvid lineages from a study in Japan (lineages named JapCor) or reference Leucocytozoon spp. downloaded from GenBank. L. mathisi is used as an outgroup. Node of clade which contains 26 of 28 corvid lineages likely predicting a single corvid Leucocytozoon sp. is marked with asterisk
Interestingly, according to our predicted phylogenies, two Leucocytozoon lineages recovered from both American Crow and yellow-billed magpie samples fall outside this large clade and appear to be most closely related to the Leucocytozoon fringillinarum reference sequence. A distance matrix (Table 2) confirms that these two corvid lineages (AmCrow.2.YBMag.5 and AmCrow.6.YBMag.6) are genetically similar to one another (displaying a distance of 2.2 %), while their average distance from all other corvid lineages is 7.9 %. The 17 other corvid Leucocytozoon lineages found in our study, which cluster together in the phylogeny, typically display genetic distances between 0 and 5.0 % from one another.
Discussion
Our results imply a lack of species-level host specificity previously believed to differentiate L. sakharoffi from L. berestneffi. While previous studies had relied mainly on morphological descriptions of blood stages, our study was one of the first to look at a genetic marker in corvid Leucocytozoon infections. The use of molecular techniques allowed further taxonomic inference where previously microscopic investigation dominated. Recent studies have revealed a hitherto undetected diversity of haplotypes among hemoproteid species, stressing the importance of molecular and phylogenetic analyses in species delineation (Perkins and Schall 2002; Beadell et al. 2004; Martinsen et al. 2008; Dimitrov et al. 2010; Oakgrove et al. 2014). Likewise, our molecular detection methods revealed both a high Leucocytozoon infection prevalence across corvid species (ranging from 54.5 to 100 %) and high lineage diversity; 19 distinct lineages (15 of which are novel) were detected.
Five Leucocytozoon lineages overlapped host species (i.e., these distinct haplotypes were detected in both American Crows and yellow-billed magpies). This finding suggests that crows and magpies—hosts currently thought to be parasitized by different Leucocytozoon species—could share lineages of Leucocytozoon. However, detection of a parasite lineage by PCR does not necessarily confirm infection in the individual. A positive PCR outcome can result either from successful sporozoite infection and replication (Valkiūnas et al. 2009), or failed infection where sporozoites are introduced but fail to develop, perhaps due to parasite-host incompatibility (Levin et al. 2013). Therefore, it is difficult to make definitive conclusions about Leucocytozoon host specificity. However, 11 of the 19 singly infected magpie samples (57.9 %) revealed lineages also recovered from crow samples. With a majority of the magpie lineages overlapping with crow lineages, it is difficult to imagine that failed infections accounted for all detections. Viable sporozoites typically exist in a bird’s bloodstream for 1–3 days, a short time frame for detections (Valkiūnas 2005). Therefore, we believe that the majority of our detections were from successful infections and that American Crows and yellow-billed magpies share at least some lineages of Leucocytozoon.
If crows and magpies indeed share lineages, this finding would contradict the current classification of Leucocytozoon in corvids. At present, L. sakharoffi and L. berestneffi are considered host specific within corvids, according to their original taxonomic classification in 1908; L. sakharoffi infects the Corvus genus, while L. berestneffi infects magpies (Pica spp.) and jays (Cyanocitta spp.). This determination of host specificity is based upon the parasite’s (i) transmissibility amongst host species, (ii) length of pre-patent period of gametocyte development, and (iii) morphology of hepatic meronts (Clark 1965; Khan and Fallis 1971; Valkiūnas 2005). The current classification was reinforced when Khan and Fallis (1971) studied Leucocytozoon in American Crows (C. brachyrhynchos), common ravens (C. corax), and blue jays (Cyanocitta cristata), and found that Leucocytozoon initially found in Corvus spp. could not be experimentally transmitted to blue jays, and vice versa. Furthermore, the authors found that the minimum pre-patent period for infection is 92 h in jays (n = 3), while for crows (n = 2) and ravens (n = 1), it is at least 24 h longer. However, pre-patent period can vary widely within a Leucocytozoon sp. amongst its various hosts: e.g., the pre-patent period of L. sakharoffi in Corvus frugilegus is 192 h (Baker 1971), a sharp contrast to the 116 h recorded by Khan and Fallis. In addition to the supposed pre-patent period differences, the reliability of distinguishing L. sakharoffi and L. berestneffi by the morphology of hepatic meronts is tenuous. Valkiūnas (2005) catalogs these differences in his comprehensive review of avian hemosporidia, while noting that hepatic meronts found in one Eurasian magpie (Pica pica) were identical to those found in C. cristata. However, when describing hepatic meronts in yellow-billed magpies (one of our study species), Clark (1965) specifically emphasized that his findings actually reflect those observed in L. sakharoffi found in crows (Wingstrand 1947).
Despite the possible unreliability of two of the three distinguishing features in L. sakharoffi and L. berestneffi (pre-patent period; morphology of hepatic meronts), we cannot refute the experimental evidence of transmissibility of Leucocytozoon in crows and jays recorded by Khan and Fallis (1971). It should be noted, however, that a few details of the transmissibility study cast ambiguity on the results. First, the age of the bird receiving the parasite transmission varied by species: Raven and crow recipients were immature birds, while jay recipients were adults. Studies have shown that host age can affect both Leucocytozoon infection prevalence and immune response and sensitivity (Taft et al. 1994; Remple 2004; Forrester et al. 2001). Second, very few experimental details are included about the two black fly species (Simulium aureum and Prosimulium decemarticulatum) used to harbor and potentially transmit the infections. There is limited knowledge about the natural hosts of individual Leucocytozoon spp., and inconsistencies could be present regarding host-vector-host transmission pathways. Finally, their study did not include magpie species, leaving open the possibility that while Leucocytozoon host specificity in corvids exists, it could be inaccurate as currently classified.
A differing viewpoint contends that Leucocytozoon host specificity is lacking within corvids. A 1992 study by Bennett and Pierce presents a taxonomic review of leucocytozoids of the Corvidae family. The authors argue that early morphological descriptions, upon which the distinction of L. sakharoffi and L. berestneffi is based, were in fact comparing different stages of the parasite life cycle. This and other early studies (e.g., Coatney and West 1938; Wingstrand 1947) led the authors to conclude that L. berestneffi should be considered a synonym of L. sakharoffi. Also of note is a comparative study of the macrogametocytes of leucocytozoids occurring in Common Raven, Carrion Crow, and magpie, which did not reveal separation by host species (Ramisz 1962). In the course of our study, therefore, microscopic investigation of infected blood smears was de-emphasized; blood films collected for our study were incomplete and of inconsistent quality, which prevented reliable parasite detection capabilities, and we were unable to obtain tissue stage samples. However, for morphospecies comparisons, blood smears were not likely to have been informative, as the Leucocytozoon species we encountered are morphologically indistinguishable in the blood stages (Valkiūnas 2005). Instead, we relied solely on molecular techniques for parasite detection.
Our genetic distance and phylogenetic reconstruction analyses yielded two interesting trends. First, two lineages found in both crows and magpies were seemingly distinct from the other lineages, which could indicate they are in fact from a different Leucocytozoon sp. They were closely related to L. fringillinarum, a generalist parasite that has been recovered from over 200 host species (Valkiūnas 2005). Second, there was no genetic distance differentiation between our remaining 17 lineages. If a distinction between L. sakharoffi and L. berestneffi was evident, we would expect high distance values from crow lineages to both magpie and jay lineages but low distance to other crow lineages. Furthermore, we would expect low distance between magpie and jay lineages. These results, however, did not materialize (Table 2). For example, lineage AmCrow.1 (n = 21) displays distance values ranging from 2.3 to 7.1 % divergence from other lineages detected in crows while displaying distance values below 3.0 % from five of six lineages detected solely in magpies or jays; in fact, the lineage most genetically similar to AmCrow.1 was detected in Steller’s jays. Bayesian and ML inferences indicated that the majority of lineages detected in corvids forms a large and mostly unresolved clade. The results of our distance and phylogenetic analyses do not support the current nomenclature separating L. sakharoffi and L. berestneffi.
Two reference Leucocytozoon species (L. majoris; L. dubreuili) are contained within what we have speculated to be the probable Leucocytozoon corvid clade. The inclusion of these two generalist species with the corvid lineages is unexpected and warrants further assessment. One possible explanation for our findings is that the partial cyt b sequence was of insufficient phylogenetic robustness to accurately approximate the fine-scale species-level distinction we were testing. Previous studies have examined nuclear and/or apicoplast genes in addition to mitochondrial genes to gain a clearer picture of hemosporidian diversity (Bensch et al. 2004; Martinsen et al. 2008; Hellgren et al. 2014). As such, we attempted to amplify both the msp1 and ama-1 nuclear genes, and though each had been successfully amplified in recent studies on avian Plasmodium spp. (Hellgren et al. 2014; Lauron et al. 2014), this approach was unsuccessful with Leucocytozoon. Unfortunately, obtaining multiple genetic markers is a significant challenge in avian hemosporidian research (reviewed in Perkins 2014). However, Bensch et al. (2004) concludes that a 500-bp fragment of cyt b provides a phylogenetic signal as robust as the complete cyt b gene or the DHFR-TS nuclear gene. A second potential explanation for our results is that the evolutionarily recent speciation of L. sakharoffi and L. berestneffi is yet to be reflected in their respective cyt b sequence. However, a study on the relationships of corvid genera does not indicate that Corvus, Pica, and Cyanocitta spp. are recently diverged or even closely related within the corvid family (Ericson et al. 2005). Therefore, if these host-specific Leucocytozoon parasites have recently evolved, they are doing so independently of host speciation.
A final explanation is that L. sakharoffi and L. berestneffi, which are indistinguishable morphologically and undifferentiated genetically, are in fact part of one large corvid-parasite complex. Our data suggest that there is no definitive distinction in the Leucocytozoon species found in crows, magpies, and jays. It is clear, therefore, that further molecular studies are needed, ideally involving longer and/or additional genetic sequences from multiple genomes. In addition, a study attempting to replicate the transmissibility experiment of Khan and Fallis (1971) would be of immense value. A new experimental study should involve members of all three focal corvids at the center of the L. sakharoffi versus L. berestneffi classification issue and should ideally be a consistent age in order to minimize age-specific immune response differences. These further studies would help clarify the host-parasite specificity in the rapidly expanding corvid family.
References
Baker JR (1971) Sporogony and schizogony of Leucocytozoon sakharoffi in England. J Protozool 18:40
Beadell JS, Gering E, Austin J, Dumbacher JP, Peirce MA, Pratt TK, Atkinson CT, Fleischer RC (2004) Prevalence and differential host-specificity of two avian blood parasite genera in the Australo-Papuan region. Mol Ecol 13:3829–3844. doi:10.1111/j.1365-294X.2004.02363.x
Bennett GF, Peirce MA (1992) Leucocytozoids of seven old world passeriform families. J Nat Hist 26:693–707
Bensch S, Perez-Tres J, Waldenström J, Hellgren O (2004) Linkage between nuclear and mitochondrial dna sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution 58:1617–1621
Bensch S, Hellgren O, PÉrez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Res 9:1353–1358. doi:10.1111/j.1755-0998.2009.02692.x
Bradley CA, Altizer S (2007) Urbanization and the ecology of wildlife diseases. Trends Ecol Evol 22:95–102. doi:10.1016/j.tree.2006.11.001
Bunbury N, Barton E, Jones CG, Greenwood AG, Tyler KM, Bell DJ (2007) Avian blood parasites in an endangered columbid: Leucocytozoon marchouxi in the Mauritian Pink Pigeon Columba mayeri. Parasitology 134:797–804. doi:10.1017/S0031182006002149
Clark G (1965) Schizogony and gametocyte development of Leucocytoxoon berestneffi in the yellow-billed magpie, Pica nuttalli. J Protozool 12:584–589
Coatney GR, West E (1938) Some blood parasites from Nebraska birds. Am Midl Nat 19:601–612
Crosskey RW, Howard TM (2004) A revised taxonomic and geographical inventory of world blackflies (Diptera: Simuliidae). British Museum of Natural History, London
de León GPP, Nadler SA (2010) What we don’t recognize can hurt us: a plea for awareness about cryptic species. J Parasitol 96:453–464. doi:10.1645/GE-2260.1
Delgado-V CA, French K (2012) Parasite–bird interactions in urban areas: current evidence and emerging questions. Landsc Urban Plan 105:5–14. doi:10.1016/j.landurbplan.2011.12.019
Dimitrov D, Zehtindjiev P, Bensch S (2010) Genetic diversity of avian blood parasites in SE Europe: cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Bulgaria. Acta Parasitol 55:201–209. doi:10.2478/s11686-010-0029-z
Ericson PGP, Jansén AL, Johansson US, Ekman J (2005) Inter-generic relationships of the crows, jays, magpies and allied groups (Aves: Corvidae) based on nucleotide sequence data. J Avian Biol 36:222–234. doi:10.1111/j.0908-8857.2001.03409.x
Ernest HB, Woods LW, Hoar BR (2010) Pathology associated with West Nile virus infections in the yellow-billed magpie (Pica nuttalli): a California endemic bird. J Wildl Dis 46:401–408
Forrester DJ, Foster GW, Morrison JL (2001) Leucocytozoon toddi and Haemoproteus tinnunculi (Protozoa: Haemosporina) in the Chimango caracara (Milvago chimango) in Southern Chile. Mem Inst Oswaldo Cruz 96:1023–1024
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90:797–802. doi:10.1645/GE-184R1
Hellgren O, Bensch S, Malmqvist B (2008) Bird hosts, blood parasites and their vectors - associations uncovered by molecular analyses of blackfly blood meals. Mol Ecol 17:1605–1613. doi:10.1111/j.1365-294X.2007.03680.x
Hellgren O, Atkinson CT, Bensch S, Albayrak T, Dimitrov D, Ewen JG, Marzal A (2014) Global phylogeography of the avian malaria pathogen Plasmodium relictum based on MSP1 allelic diversity. Ecography 38:001–009. doi:10.1111/ecog.01158
Hinton MG, Reisen WK, Wheeler SS, Townsend AK (2015) West Nile Virus activity in a winter roost of American crows (Corvus brachyrhynchos): is bird-to-bird transmission important in persistence and amplification? J Med Entomol 52:683–692, tjv040
Huelsenbeck JP, Crandall KA (1997) Phylogeny estimation and hypothesis testing using maximum likelihood. Annu Rev Ecol Syst 28:437–466
Hunter DB, Rohner C, Currie DC (1997) Mortality in fledgling great horned owls from black fly hematophaga and leucocytozoonosis. J Wildl Dis 33:486–491. doi:10.7589/0090-3558-33.3.486
Jenkins T, Owens IPF (2011) Biogeography of avian blood parasites (Leucocytozoon spp.) in two resident hosts across Europe: phylogeographic structuring or the abundance-occupancy relationship? Mol Ecol 20:3910–3920. doi:10.1111/j.1365-294X.2011.05221.x
Johnston RF (2001) The synanthropic birds of North America. In: Marzluff JM, Bowman R, Donnelly R (eds) Avian ecology and conservation in an urbanizing world. Kluwer, Norwell, pp 49–67
Jones J Jr (1968) Some parasites of the common crow, Corvus brachyrhynchos Brehm, from Ohio. Ohio J Sci 68:25–31
Khan RA, Fallis A (1971) Speciation, transmission, and schizogeny of Leucocytozoon in corvid birds. Can J Zool 49:1363–1367
Kim KS, Tsuda Y (2010) Seasonal changes in the feeding pattern of Culex pipiens pallens govern the transmission dynamics of multiple lineages of avian malaria parasites in Japanese wild bird community. Mol Ecol 19:5545–5554. doi:10.1111/j.1365-294X.2010.04897.x
Lauron EJ, Oakgrove KS, Tell LA, Biskar K, Roy SW, Sehgal RN (2014) Transcriptome sequencing and analysis of Plasmodium gallinaceum reveals polymorphisms and selection on the apical membrane antigen-1. Malar J 13:382. doi:10.1186/1475-2875-13-382
Leclerc A, Chavatte JM, Landau I, Snounou G, Petit T (2014) Morphological and molecular study of hemoparasites in wild corvids and evidence of sequence identity with Plasmodium DNA detected in captive black-footed penguins (Spheniscus demersus). J Zoo Wildl Med 45:577–588
Levin II, Zwiers P, Deem SL, Geest EA, Higashiguchi JM, Iezhova TA, Jiménez-Uzcátegui G, Kim DH, Morton JP, Perlut NG, Renfrew RB, Sari EH, Valkiunas G, Parker PG (2013) Multiple lineages of avian malaria parasites (Plasmodium) in the Galapagos Islands and evidence for arrival via migratory birds. Conserv Biol 27(6):1366–1377
Lotta IA, Matta NE, Torres RD, Sandino MM, Moncada LI (2013) Leucocytozoon fringillinarum and Leucocytozoon dubreuili in Turdus fuscater from a Colombian Páramo ecosystem. J Parasitol 99:359–362. doi:10.1645/GE-3156.1
Maddison DR, Maddison WP (2005) MacClade 4., v. 4.08 for OSX. Sinauer Associates, Sunderland, MA
Malmqvist B, Strasevicius D, Hellgren O, Adler PH, Bensch S (2004) Vertebrate host specificity of wild-caught blackflies revealed by mitochondrial DNA in blood. Proc Biol Sci 271:S152–S155. doi:10.1098/rsbl.2003.0120
Martinsen ES, Paperna I, Schall JJ (2006) Morphological versus molecular identification of avian Haemosporidia: an exploration of three species concepts. Parasitology 133:279–288. doi:10.1017/S0031182006000424
Martinsen ES, Perkins SL, Schall JJ (2008) A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol 47:261–273. doi:10.1016/j.ympev.2007.11.012
Marzluff JM, Neatherlin E (2006) Corvid response to human settlements and campgrounds: causes, consequences, and challenges for conservation. Biol Conserv 130:301–314. doi:10.1016/j.biocon.2005.12.026
Marzluff JM, McGowan KJ, Donnelly R, Knight RL (2001) Causes and consequences of expanding American Crow populations. In: Avian ecology and conservation in an urbanizing world. Springer US, pp 331–363
Morgan BB, Waller EF (1941) Some parasites of the eastern crow (Corvus brachyrhynchos brachyrhynchos Brehm). Bird-Banding 12:16–22
Morii T (1992) A review of Leucocytozoon caulleryi infection in chickens. J Protozool Res 2:128–133
Murata K (2002) Prevalence of blood parasites in Japanese wild birds. J Vet Med Sci 64:785–790. doi:10.1292/jvms.64.785
Murdock CC, Adler PH, Frank J, Perkins SL (2015) Molecular analyses on host-seeking black flies (Diptera: Simuliidae) reveal a diverse assemblage of Leucocytozoon (Apicomplexa: Haemospororida) parasites in an alpine ecosystem. Parasitol Vectors 8:1–7
Oakgrove KS, Harrigan RJ, Loiseau C, Guers S, Seppi B, Sehgal RNM (2014) Distribution, diversity and drivers of blood-borne parasite co-infections in Alaskan bird populations. Int J Parasitol 44:717–727. doi:10.1016/j.ijpara.2014.04.011
Palinauskas V, Žiegytė R, Ilgūnas M, Iezhova TA, Bernotienė R, Bolshakov C, Valkiūnas G (2015) Description of the first cryptic avian malaria parasite, Plasmodium homocircumflexum n. sp., with experimental data on its virulence and development in avian hosts and mosquitoes. Int J Parasitol 45:51–62. doi:10.1016/j.ijpara.2014.08.012
Perkins SL (2014) Malaria’s many mates: past, present, and future of the systematics of the order Haemosporida. J Parasitol 100:11–25
Perkins SL, Schall JJ (2002) A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol 88:972–978. doi:10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Rambaut A (2012) FigTree 1.4. Institute of Evolutionary Biology, University of Edinburgh
Ramisz A (1962) Protozoa of the genus Leucocytozoon Danilewski, 1890 in birds in the environs of Wroclaw. Acta Parasitol Pol 10:39–52
Remple JD (2004) Intracellular hematozoa of raptors: a review and update. J Avian Med Surg 18:75–88. doi:10.1647/2003-008
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sehgal RNM, Lovette IJ (2003) Molecular evolution of three avian neurotrophin genes: implications for proregion functional constraints. J Mol Evol 57:335–342
Sehgal RNM, Hull AC, Anderson NL, Valkiŭnas G, Markovets MJ, Kawamura S, Tell LA (2006a) Evidence for cryptic speciation of Leucocytozoon spp. (Haemosporida, Leucocytozoidae) in diurnal raptors. J Parasitol 92:375–379. doi:10.1645/GE-656R.1
Sehgal RNM, Valkiunas G, Iezhova TA, Smith TB (2006b) Blood parasites of chickens in Uganda and Cameroon with molecular descriptions of Leucocytozoon schoutedeni and Trypanosoma gallinarum. J Parasitol 92:1336–1343. doi:10.1645/GE-927R.1
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analyses. Can J Zool 69:82–90. doi:10.1139/z91-013
Smith RN, Cain SL, Anderson SH, Dunk JR, Williams S, Dunk JR, Williams ES (1998) Blackfly-induced mortality of nestling red-tailed hawks. Auk 115:368–375
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi:10.1093/bioinformatics/btl446
Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA
Taft SJ, Rosenfield RN, Bielefeldt J (1994) Avian hematozoa of adult and nestling Cooper’s Hawks (Accipiter cooperii) in Wisconsin. J Helminthol Soc Wash 61:146–148
Townsend AK, Barker CM (2014) Plastic and the nest entanglement of urban and agricultural crows. PLoS ONE 9:e88006
Townsend AK, Clark AB, McGowan KJ, Lovette IJ (2009) Reproductive partitioning and the assumptions of reproductive skew models in the cooperatively breeding American crow. Anim Behav 77:503–512
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC Press, Boca Raton
Valkiūnas G, Iezhova TA, Loiseau C, Sehgal RNM (2009) Nested cytochrome B polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. J Parasitol 95:1512–1515. doi:10.1645/GE-2105.1
Valkiūnas G, Sehgal RNM, Iezhova TA, Hull AC (2010) Identification of Leucocytozoon toddi group (Haemosporida: Leucocytozoidae), with remarks on the species taxonomy of leucocytozoids. J Parasitol 96:170–177. doi:10.1645/GE-2109.1
Wingstrand KG (1947) On some Haematozoa of Swedish birds with remarks on the schizogony of Leucocytozoon sakharoffi. Kungliga Svenska Vetenskapsakademiens Handlingar 24:31
Yoshimura A, Koketsu M, Bando H, Saiki E, Suzuki M, Watanabe Y, Kanuka H, Fukumoto S (2014) Phylogenetic comparison of avian haemosporidian parasites from resident and migratory birds in northern Japan. J Wildl Dis 50:235–242. doi:10.7589/2013-03-071
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Freund, D., Wheeler, S.S., Townsend, A.K. et al. Genetic sequence data reveals widespread sharing of Leucocytozoon lineages in corvids. Parasitol Res 115, 3557–3565 (2016). https://doi.org/10.1007/s00436-016-5121-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-016-5121-3