Abstract
Recent studies showed that the huge diversity branching at or near the phylogenetic root of the fungal kingdom, mostly constituted by uncultured environmental clones, is actually characterized by intracellular predators/parasites of various eukaryotes. These form three related lineages: the Aphelidea, endoparasites of algae; the Rozellomycota, with Rozella species mainly endoparasites of water moulds, and Paramicrosporidium species endonuclear parasites of amoebae; and the Microsporidia, mainly endoparasites of animals. Increasing evidence suggests the emergence of Microsporidia from within Rozellomycota; however, their fungal or protistan nature is still unclear. Here, we report the molecular phylogeny based on the small subunit ribosomal RNA (SSU rDNA) gene, of an additional endoparasite of amoebae, corresponding to the old enigmatic chytrid Nucleophaga amoebae described in the nineteenth century. Our results show that Nucleophaga, possessing a morphotype intermediate between Rozella and Paramicrosporidium, emerges as a unique lineage within the Rozellomycota. The recovery and characterization of new members of Rozellomycota are of high value for the understanding of the early evolutionary history of the Fungi and related lineages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Previous molecular environmental surveys and phylogenetic analyses identified a large group of uncultured eukaryotes, basal to fungi within the large clade of the opisthokonts (animals, fungi and protistan relatives), interpreted as intermediate forms between protists and true fungi (Lara et al. 2010; Jones et al. 2011). The clade includes also Rozella spp., endoparasites mainly of other chytrids and chromistan oomycetes, and accordingly named Rozellida (Lara et al. 2010) or Rozellomycota (Corsaro et al. 2014), emerging together with the better-known Microsporidia, endoparasites of animals, on the most-basal branch in Fungi (James et al. 2006, 2013; Corsaro et al. 2014). Because some members of the rozellids appeared to be chytrid-like naked epibionts, Rozella was thought to be an atypical member and the group was renamed Cryptomycota (Jones et al. 2011). However, the original suggestion that the common morphotypes representing the group are epibionts primitively lacking a cell wall has been challenged (James and Berbee 2012; James et al. 2013; Corsaro et al. 2014; Karpov et al. 2014a). Indeed, epibiotic naked cells were likely the only observed stage out of many unobserved (Corsaro et al. 2014) or misinterpreted attached cysts (Karpov et al. 2014a), whereas Rozella (James and Berbee 2012; James et al. 2013) and Paramicrosporidium (Corsaro et al. 2014), are both endoparasites that have fungal-like cell walls. Moreover, the diversity at the base of the fungal tree was further increased by the inclusion of the Aphelids, endoparasites of green algae and chromistan algae, which also comprise several uncultured phylotypes (Karpov et al. 2013; Letcher et al. 2013).
There is compelling evidence (James et al. 2006, 2013; Karpov et al. 2013; Letcher et al. 2013; Corsaro et al. 2014) for a close relationship among Rozellomycota, Microsporidia and Aphelidea, with the Microsporidia emerging from modified endoparasitic rozellids (Corsaro et al. 2014). These groups emerge close to the Fungi along the clade holomycota (Fungi and protistan relatives), but the very few available data on characterized strains, in contrast with the huge diversity of uncultured sequences, is a limit to elucidating whether they belong to the Fungi or to the Choanozoa, resulting in contrasting systematic descriptions (Cavalier-Smith 2013; Corsaro et al. 2014; Karpov et al. 2014a).
We previously showed that microsporidia-like endonuclear parasites of free-living amoebae (Michel et al. 2000, 2009a) are actually highly modified Rozellomycota, likely the missing link with the Microsporidia, and we named them Paramicrosporidium (Corsaro et al. 2014). Indeed, despite its close morphological resemblance with true Microsporidia, Paramicrosporidium possesses ‘standard’ small subunit (SSU) and a typical eukaryotic rRNA gene structure, with the 5.8S separated from the large subunit (LSU) by the second internal transcribed spacer (ITS2). Such molecular traits are present in Rozella spp. and other chytrids but notoriously not in Microsporidia. Molecular phylogenies based on the SSU and the ribosomal DNA (rDNA) unit (SSU + 5.8S + LSU) clearly showed that Paramicrosporidium belongs to the Rozellomycota and is sister to the Microsporidia (Corsaro et al. 2014). Endonuclear true microsporidia infect fish and crustaceans and all belong to the family Enterocytozoonidae (Palenzuela et al. 2014). Otherwise, endonuclear symbionts of protists are prevalently prokaryotes.
The first observations of intracellular organisms in various amoebae, flagellates and algae were reported in the late nineteenth century, including also the presence of intranuclear endosymbionts. Dangeard (1895) had reviewed the data available at that time on parasites of protists, noting that some endonuclear parasites found in amoebae were distinct from other known organisms, such as Holospora bacteria found in ciliates.
Dangeard distinguished two main amoeba parasites: the intracytoplasmic Sphaerita and the endonuclear Nucleophaga (Dangeard 1895). Because the latter appeared to be specific for the group Amoeba (=Thecamoeba) verrucosa (Amoebozoa, Thecamoebida), he proposed the name Nucleophaga amoebae. Later, Nucleophaga was reported also in other free-living amoebae like Amoeba (=Mayorella) vespertillio (Amoebozoa, Dermamoebida) as well as intestinal archamoebae like Iodamoeba and Entamoeba of vertebrates and insects (Mercier 1910; Kirby 1927; Lavier 1935; Brumpt and Lavier 1935). Dangeard argued that both Sphaerita and Nucleophaga grow by absorption, i.e. they are osmotrophic, and that they are closely related members belonging to the Chytridiales (Dangeard 1895), for a long time included in the heterogeneous group of ‘primitive fungi’ named Phycomycetes (Sparrow 1960). Indeed, beside the true fungal chytrid and zygomycotan lineages, the ‘Phycomycetes’ also comprise unrelated organisms belonging to the kingdom Chromista, such as oomycetes, hyphochytrids (Heterokonta) and plasmodiophorids (Rhizaria, Cercozoa). Similar parasites infecting flagellates and algae have biflagellate stages, e.g. Pseudosphaerita, and were later considered as closer to oomycetes. However, all these organisms remained unstudied and enigmatic ever since (Karling 1972), although sometimes still currently reported (e.g. Voge and Kessel 1958; Upton et al. 1991; Anderson et al. 1995).
Recently, we recovered Thecamoeba quadrilineata infected with endonuclear spore-forming parasites, called KTq-2, whose morphological features correspond to the description of N. amoebae (Michel et al. 2009b). Here, we show by SSU rDNA molecular phylogeny that Nucleophaga belongs to the Rozellomycota and present a new description of its morphology in light of new molecular phylogeny data. Overall, our data highlight the importance to identify culturable strains of these basal groups, which are also promising candidates for further genomic studies, to elucidate the early evolution of Fungi and relatives.
Materials and methods
Strains origin and culture
The amoeba host T. quadrilineata, harbouring the endonuclear parasite Nucleophaga, was recovered from moss meshwork in Austria (Michel et al. 2009b). Amoebae were cultivated on bacterized non-nutritive agar, and endoparasites were recovered by membrane filtration as described (Michel et al. 2009b; Corsaro et al. 2014).
The chitin/cellulose stain Calcofluor White (CFW, Sigma, 1 mg ml−l) was used to show the fungal-like cell wall of the endoparasite. Electron microscopy was performed as described (Michel et al. 2009b).
Phylogenetic analysis
The SSU rDNA was amplified by using the eukaryotic-specific primers SSU-I (5′-CGACTGGTTGATCCTGCCAGTAG-3) and SSU-II (5′-TCCTGATCCCTCCGCAGGTTCAC-3′), modified from Medlin et al. (1988) and sequenced by using the same primers and a set of internal primers. Sequences were aligned using MUSCLE and manually refined using BIOEDIT to exclude ambiguous sites, and Bayesian and maximum likelihood (GTR, Γ + I:4, 1000 bootstraps) phylogenetic trees were built as described (Corsaro et al. 2014). The list of taxa with their GenGank accession numbers is provided in the Supplementary Table 1.
Results
Life cycle and morphology
Nucleophaga KTq-2 was found as a natural parasite of the free-living T. quadrilineata (Amoebozoa, Thecamoebida), showing endonuclear development ending in spore formation (Michel et al. 2009b). In coculture experiments, Nucleophaga successfully grows and produces spores also in Thecamoeba striata and Thecamoeba terricola and in some strains of the binucleate amoeba Sappinia. In this case, the two nuclei of Sappinia, closely attached in uninfected amoebae, become separated. Other amoebae were killed before spore formation (Michel et al. 2009b). Previous microscopical observations (Michel et al. 2009b) are herein re-interpreted (Fig. 1) taking in consideration the new molecular phylogeny data (see below).
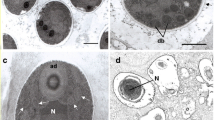
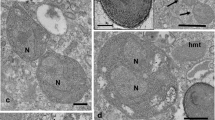
Morphology of Nucleophaga. a Light microscopy of two infected Thecamoeba trophozoites with intranuclear aggregates of spores. b Calcofluor White stain showing fungal-like cell wall. c Several spores showing a single nucleus (N) and a two-layererd cell wall (cw). The nuclear membrane (nm) of host amoeba remains intact along the entire parasite development (c–g). d Two early stages (P1, P2) feeding on the host karyosome (ka). Also visible are food vacuoles (fv) and exocytosis (ex) of digested remnants. e Several trophic stages filling the host karyoplasm. f Enlarged stage with rounded sporogenic area containing condensed semilunal material (double arrow). g Late enlarged trophic stage with irregular surfaces (arrows). h Detail of the irregular surface showing finger-like extensions (arrows). Scale bars 20 μm (a, b); 1 μm (c, h); 2 μm (d–g)
The spores of the parasite are engulfed by the amoebae through phagocytosis in food vacuoles and reach the amoebal nucleus (Michel et al. 2009b), where they grow forming aggregates visible by light microscopy (Fig. 1a). Spores are non-flagellated, with a single nucleus and different vacuoles, but apparently lack mitochondria, and possess a thick double-layered cell wall (Fig. 1c), positive to Calcofluor White (Fig. 1b). Once in the nucleus of the amoeba, the endoparasites enter into an apparently naked trophic stage (unicellular thallus, ∼1–2 μm), growing at the expense of the endosome (Fig. 1d–g). During growth, the early stages enlarge (up to 4.8 × 3.5 μm) and develop internal structures and vacuoles as well as an irregular surface in intimate contact with the host karyoplasm (Fig. 1g). This irregular surface was initially interpreted as ‘microvilli-like’ likely to promote osmotrophy efficiency. However, upon more accurate observations, the surface appears as finger-like extensions (Fig. 1h), suggesting that Nucleophaga would be able, at least in some stages of its life cycle, to perform phagocytosis. Parasite cells enlarge further (up to 5.5 μm) and enter a reproductive stage (unicellular sporangium), where a coat (initial cell wall) replaces the finger-like extensions. Within each sporangium, arise few rounded ‘sporogenic areas’ (about 1.4–2 μm), corresponding to nuclei of the new generation, with semilunar condensed nucleolar material (Fig. 1f), which finally develop into mature spores (sporangiospores, 1.5 × 2 μm). The nuclear membrane of the amoebae remains intact throughout the entire development of the parasite (Fig. 1d–g), and the amoebae maintain basic life functions, despite the disappearance of their nuclei.
Molecular phylogeny
To elucidate the phylogenetic position of N. amoebae, we used eukaryotic-specific primers to obtain near full SSU rDNA sequence of our strain KTq-2. BLAST search resulted in low-identity values with basidiomycotan, chytrid and uncultured fungal sequences, leading us to analyse fungal lineages and relatives (Supplementary Table 1).
Our phylogenetic analyses (Fig. 2) show Nucleariidae forming the basal clade of the holomycotans, followed by Aphelidea, Rozellomycota and the rest of the Fungi as separate lineages. Several major clades are recovered as well-supported holophyletic.
Molecular phylogeny of Nucleophaga. 18S rDNA phylogenetic tree of Fungi and relatives (holomycotans), rooted on holozoans (Animalia and part of Choanozoa). Major taxa of interest are indicated. Nucleophaga amoebae strain KTq-2 recovered in this study is in bold. Support values are for Bayesian posterior probability and Maximum Likelihood (1000 bootstraps). Filled and open circles represent values of 1/100 % and of at least 0.90/90 %, respectively
Our tree is congruent with some previous studies, where Rozellomycota emerged as the basal branch, sister to the remaining Fungi (James et al. 2006), forming a clade with the Microsporidia (James et al. 2006, 2013; Corsaro et al. 2014) and recovering aphelids as an outer basal lineage (Corsaro et al. 2014; Karpov et al. 2014a). However, other studies recovered aphelids and rozellids as a moderately supported clade, sister to the other Fungi (Letcher et al. 2013; Karpov et al. 2014b). Protein and rDNA multigene analyses on the few cultured members (i.e., Rozella and Amoeboaphelidium) allowed the recovery of the ARM clade (for Aphelids, Rozella and Microsporidia) (Karpov et al. 2013; Letcher et al. 2013), which is less stable when much more uncultured sequences are included (Karpov et al.; 2014a) (see Supplementary Figures in Karpov et al. 2013; Letcher et al. 2013). The overall topology of our tree is also recovered when including additional sequences and outgroups (Supplementary Figure 1).
In our analysis (Fig. 2), Nucleophaga KTq-2 emerges as a unique lineage within the highly supported holophyletic Rozellomycota. Rozella spp. form one of the first lineages and Paramicrosporidium spp. cluster with other environmental clones in a well-supported separate branch. Nucleophaga clusters moderately with some closely related (sequence identities ≥95 %) environmental sequences, from aquatic samples potentially rich in amoebae (see Supplementary Table 1 for source of environmental sequences), from which it is relatively distant (∼82 %). Nucleophaga shares a similar genetic distance with both Paramicrosporidium spp. (∼81 %) and Rozella spp. (∼80.5 %).
Our results demonstrate unambiguously the rozellid home for Nucleophaga. Because of the basal position of Rozellomycota, this confirms the early description by Dangeard (1895) of this organism as a ‘primitive fungal parasite’. Furthermore, our results also confirm the presence of more diversified morphotypes within Rozellomycota, which therefore include also various amoebal endoparasites, as previously hypothesized (Corsaro et al. 2014). More varied rozellids are thus expected to be found or rediscovered.
Discussion
Interestingly, Nucleophaga is similar to Paramicrosporidium (Michel et al. 2000, 2009a; Corsaro et al. 2014) in its infective stage, by having non-flagellated walled spores penetrating amoebae through host cell phagocytosis, but however differing by lack of structures resembling a polar filament or an anchoring disc. However, in the trophic stages, Nucleophaga appears more similar to thalli of Rozella. Albeit residing in the cytoplasm of their host cells, Rozella species grow as naked unicellular thalli (Held 1981), either mononucleate (Rozella allomycis) or multinucleate (Rozella polyphagi). The nucleolar material of Rozella is condensed peripherally in semilunar areas, recalling the nucleolar structure present in the ‘sporogenic areas’ of Nucleophaga. Also, Rozella possesses finger-like protoplasmic extensions (pseudopodia-like) possibly able to phagocytize host’s organelles (Powell 1984). As shown herein, finger-like extensions are present during a part of the trophic stage of Nucleophaga, but are never observed in Paramicrosporidium nor in Microsporidia. Finally, like other endobiotic chytrids, Rozella spp. produce sporangia (zoosporangia), where the entire thallus filling the host cell split by internal divisions to form naked zoospores (zoosporangia) or thick-walled resting spores. In Nucleophaga, the thallus converts to a sporangium, which is finally filled by aflagellate walled spores, more similar to those of Paramicrosporidium.
Finger-like pseudopodia (filopodia) are present in various lineages of the opisthokonts as well as in the Apusozoa and the Amoebozoa, from which opisthokonts seem to have emerged. In this eukaryote branch, filopodia are presumed to have evolved for feeding by phagocytosis (Cavalier-Smith 2013). Noteworthy, filopodia are absent in (more derived) Fungi, but they are typical for their closest relatives, the phagotrophic Nuclearoidea. Also, aphelids are able to phagocytize cell prey contents, and the finger-like structures of Nucleophaga may actually be filopodia, suggesting that it (at least temporally) may present, like Rozella, a residual phagocytosing ability. Interestingly, chytrid zoospores can possess very short pseudopodia, and various members, e.g. Amoebochytrium, Caulochytrium (Chytridiomycota inc. sed.), Catenaria and Paraphysoderma (Blastocladiomycota), still have amoeboid stages with filopodia (Sparrow 1960; Powell 1981; Hoffman et al. 2008; Gutman et al. 2009). However, in these cases, filopodia appear to be used to crawl, and phagocytosis was never reported.
Similarly, the likely primitive status and evolution of a naked trophont, shared by the basal holomycotans, should be studied further because naked thalli are also present in chytrids (Johnson et al. 2006) and even in endophytic higher fungi (Atsatt and Whiteside 2014).
Recent studies (Sebé-Pedrós et al. 2013) showed that opisthokonts share a common, but differentially conserved genetic and molecular toolkit of the actin-myosin system, with a loss of some of these components in the fungal lineage. Thus, deeper studies of this system in the basal holomycotans will be of great importance to reconstruct the evolutionary history of the transition from phagotrophy to osmotrophy, allowing for a new and better definition of the Fungi.
According to a widely accepted scenario, the Fungi have acquired a chitinous cell wall during most of their life cycle, including the trophic stage, changing thus from a phagotrophic to an absorptive feeding mode (Cavalier-Smith 1987). Filopodia should have been present in the ancestor and might gradually have been covered by a wall, being likely at the origins of fungal rhizoids observed in current chytrids. It may be expected that the very early fungi still retain a possibly limited phagocytosing ability during part of their life.
These various transitional steps are congruent with the phylogenetic position of available morphotypes of basal holomycotans: an ancestral free-living phagotrophic stage (Nuclearia), followed by a parasitic phagotrophic stage (aphelids), then by parasites in which phagotrophy appears to be still present but presumably in a residual form and gradually lost (Rozella, Nucleophaga), and ending with non-phagotrophic parasites (Paramicrosporidium, Microsporidia). The cell wall is finally retained also during the trophic stage (chytrids).
While it is widely accepted that nucleariids do not belong to the Fungi, the position of aphelids and rozellids is unclear. We previously proposed a rather conservative classification (Corsaro et al. 2014), waiting for further genomic studies to elucidate the status of these lineages, including also the peculiar Microsporidia (Corradi and Keeling 2014). However, Karpov et al. (2014a) recently proposed a new non-fungal higher taxon, Opisthosporidia, essentially based on aphelids, and keeping the erroneous name Cryptomycota for the rozellids. The term Cryptomycota is an unfortunate choice, as typical members like Rozella and now Nucleophaga, as well as the derived Microsporidia, are known since the nineteenth century as primitive fungi. These organisms are not ‘cryptic’, simply understudied. Furthermore, even if the non-fungal nature of this group would finally be confirmed by phylogenomics, the ending ‘-mycota’ would always be misleading, and the term ‘opisthosporidia’ would remain an unnecessary synonym for the choanozoan Rozellidea (Cavalier-Smith 2013), once emended to include also the Microsporidia.
While Paramicrosporidium was shown to represent the missing link between Rozellomycota and Microsporidia, we here present Nucleophaga, an intermediate step between Rozella and Paramicrosporidium, within the Rozellomycota. The characterization of additional members and further genomic studies are necessary to resolve the early evolutionary history of the Fungi and related lineages.
Taxonomy summary
Kingdom Fungi
Phylum Rozellomycota (Corsaro and Michel 2014)
Nucleophaga amoebae (Dangeard 1895)
MycoBank MB172878
Formal diagnosis
Unicellular, with aflagellated infective spores with chitinous wall; polar filament and mitochondria absent. Intranuclear parasites of amoebae as unwalled cells.
Latin diagnosis
Unicellulares; virulentae sporae sine flagellum cum pariete definito; filo polare et mitochondriis carentes. In nucleo amoebae parasiticae, cellulae sine pariete definito.
References
Anderson SA, Stewart A, Tolich Allen G (1995) Pseudosphaerita euglenae, a fungal parasite of Euglena spp. in the Mangere Oxidation Ponds, Auckland, New Zealand. N Z J Mar Freshw Res 29:371–379
Atsatt PR, Whiteside MD (2014) Novel symbiotic protoplasts formed by endophytic fungi explain their hidden existence, lifestyle switching, and diversity within the Plant Kingdom. PLoS ONE 9:e95266
Brumpt E, Lavier G (1935) Sur une nucléophaga parasite d’Endolimax nana. Ann Parasitol Hum Comp 13:439–444
Cavalier-Smith T (1987) The origin of fungi and pseudofungi. In: Rayner ADM, Braiser CM, Moore D (eds) Evolutionary biology of Fungi. Cambridge University Press, Cambridge, pp 339–353
Cavalier-Smith T (2013) Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa. Eur J Protistol 49:115–178
Corradi N, Keeling PJ (2014) Ecological genomics of the microsporidia. In: Martin F (ed) The ecological genomics of fungi. John Wiley & Sons, Inc, pp 261–278
Corsaro D, Walochnik J, Venditti D, Steinmann J, Müller K-H, Michel R (2014) Microsporidia-like parasites of amoebae belong to the early fungal lineage Rozellomycota. Parasitol Res 113:1909–1918
Dangeard P-A (1895) Mémoire sur les parasites du noyau et du protoplasme. Le Botaniste 4:199–248
Gutman J, Zarka A, Boussiba S (2009) The host-range of Paraphysoderma sedebokerensis, a chytrid that infects Haematococcus pluvialis. Eur J Phycol 44:509–514
Held AA (1981) Rozella and Rozellopsis: naked endoparasitic fungi which dress-up as their hosts. Bot Rev 47:451–515
Hoffman Y, Aflalo C, Zarka A, Gutman J, James TY, Boussiba S (2008) Isolation and characterization of a novel chytrid species (phylum Blastocladiomycota) parasitic on the green alga Haematococcus. Mycol Res 111:70–81
James TY, Berbee ML (2012) No jacket required—new fungal lineage defies dress code: recently described zoosporic fungi lack a cell wall during trophic phase. Bioessays 34:94–102
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822
James TY, Pelin A, Bonen L, Ahrendt S, Sain S, Corradi N, Stajich JE (2013) Shared signatures of parasitism and phylogenomics unite Cryptomycota and Microsporidia. Curr Biol 23:1548–1553
Johnson PTJ, Longcore JE, Stanton DE, Carnegie RB, Shields JD, Preu ER (2006) Chytrid infections of Daphnia pulicaria: development, ecology, pathology and phylogeny of Polycaryum laeve. Freshw Biol 51:634–648
Jones MDM, Forn I, Gadelha C, Egan MJ, Bass D, Massana R, Richards TA (2011) Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474:200–203
Karling JS (1972) The present status of Sphaerita, Pseudosphaerita, Morella and Nucleophaga. Bull Torrey Bot Club 99:223–228
Karpov SA, Mikhailov KV, Mirzaeva GS, Mirabdullaev IM, Mamkaeva KA, Titova NN, Aleoshin VV (2013) Obligately phagotrophic aphelids turned out to branch with the earliest-diverging fungi. Protist 164:195–205
Karpov SA, Mamkaeva MA, Aleoshin VV, Nassonova E, Lilje O, Gleason FH (2014a) Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Front Microbiol 5:112
Karpov SA, Mamkaeva MA, Benzerara K, Moreira D, López-García P (2014b) Molecular phylogeny and ultrastructure of Aphelidium aff. melosirae (Aphelida, Opisthosporidia). Protist 165:512–526
Kirby H (1927) Studies on some amoebae from the termite Mirotermes, with notes on some other Protozoa from the Termitidae. Q J Microsc Soc 71:189–222
Lara E, Moreira D, López-García P (2010) The environmental clade LKM11 and Rozella form the deepest branching clade of Fungi. Protist 161:116–121
Lavier G (1935) Sur une Nucleophaga parasite d'Entamoeba ranarum. Ann Parasitol Hum Comp 13:351–361
Letcher PM, Lopez S, Schmieder R, Lee PA, Behnke C, Powell MJ, McBride RC (2013) Characterization of Amoeboaphelidium protococcarum, an algal parasite new to the cryptomycota isolated from an outdoor algal pond used for the production of biofuel. PLoS ONE 8:e56232
Medlin L, Elwood HJ, Stickel S, Sogin ML (1988) The characterization of enzymatically amplified eukaryotic 16S-like rRNA coding regions. Gene 71:491–499
Mercier L (1910) Contribution à 1'étude de l'amibe de la blatte (Entamoeba blattae Bütschli). Arch Protistenkd 20:143–175
Michel R, Schmid EN, Böker T, Hager DG, Müller K-D, Hoffmann R, Seitz HM (2000) Vannella sp. harboring Microsporidia-like organisms isolated from the contact lens and inflamed eye of a female keratitis patient. Parasitol Res 86:514–520
Michel R, Müller K-D, Hauröder B (2009a) A novel microsporidian endoparasite replicating within the nucleus of Saccamoeba limax isolated from a pond. Endocytobios Cell Res 19:120–126
Michel R, Hauröder B, Zöller L (2009b) Isolation of the amoeba Thecamoeba quadrilineata harbouring intranuclear spore forming endoparasites considered as fungus-like organisms. Acta Protozool 48:41–49
Palenzuela O, Redondo MJ, Cali A, Takvorian PM, Alonso-Naveiro M, Alvarez-Pellitero P, Sitjà-Bobadilla A (2014) A new intranuclear microsporidium, Enterospora nucleophila n. sp., causing an emaciative syndrome in a piscine host (Sparus aurata), prompts the redescription of the family Enterocytozoonidae. Int J Parasitol 44:189–203
Powell MJ (1981) Zoospore structure of the mycoparasitic chytrid Caulochytrium protostelioides Olive. Am J Bot 68:1074–1089
Powell MJ (1984) Fine structure of the unwalled thallus of Rozella polyphagi in its host Polyphagus euglenae. Mycologia 76:1039–1048
Sebé-Pedrós A, Burkhardt P, Sánchez-Pons N, Fairclough SR, Lang BF, King N, Ruiz-Trillo I (2013) Insights into the origin of metazoan filopodia and microvilli. Mol Biol Evol 30:2013–2023
Sparrow FK Jr (1960) Aquatic phycomycetes, 2nd edn. The University of Michigan Press, Ann Arbor
Upton SJ, Brillhart DB, McAllister CT (1991) Two morphologically distinct types of Giardia sp. occur in cotton rats, Sigmodon hispidus. Tex J Sci 43:373–376
Voge M, Kessel JF (1958) Sphaerita in cysts of Entamoeba coli. J Parasitol 44:454–455
Acknowledgments
We thank R. Kurek for the assistance and previous microscopy data. This study was supported by internal fundings of each laboratory.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Maximum-Likelihood 18S rDNA tree of Fungi, other opisthokonts (Animalia and Choanozoa), Apusozoa and Amoebozoa, the latter used as root. Full tree topology is shown for the Rozellomycota, to demonstrate the relative position of the three cultured genus-level taxa (in bold) identified in this group. Other major groups are collapsed (see Suppl. Table 1). Branchs are shortened to 1/2 (Thecamonas, Dictyostelium) or 1/3 (Entamoeba). Bootstrap values >50 % are indicated (1000 replicates). (PDF 209 kb)
Supplementary Table 1
List of taxon names and GenBank accession numbers used in this study. (XLS 35 kb)
Rights and permissions
About this article
Cite this article
Corsaro, D., Walochnik, J., Venditti, D. et al. Rediscovery of Nucleophaga amoebae, a novel member of the Rozellomycota. Parasitol Res 113, 4491–4498 (2014). https://doi.org/10.1007/s00436-014-4138-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-014-4138-8