Abstract
Free-living Amoebae of Acanthamoeba genus include non-pathogenic and pathogenic strains that are currently classified in 18 different genotypes, T1–T18. In this study, a survey was carried out to evaluate the presence of Acanthamoeba strains in soil samples collected between 2012 and 2013 in Gran Canaria Island, Canary Islands, Spain. Samples were inoculated onto non-nutrient agar (NNA) plates and were checked for the presence of Acanthamoeba. Identification of Acanthamoeba strains was based on the morphology of the cyst and trophozoite forms. Subsequently, positive samples were cloned for their molecular characterization at the genotype level by sequencing the DF3 region located in the 18S rDNA gene of Acanthamoeba as previously described. Sequencing results revealed the presence of T2, T5 and T4 genotypes within the studied samples. To the best of our knowledge, this is the first report demonstrating the presence of Acanthamoeba in Gran Canaria Island and the first study at the genotype level in the Canary Islands.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Free-living amoebae (FLA) are widely distributed protozoa in the environment that colonise different habitats such as water, soil, dust and air sources (Marciano-Cabral and Cabral 2003; Siddiqui 2012; Lorenzo-Morales et al. 2013). Among FLA are some species which have been studied due to their implications on human health. These include mainly Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri and Sappinia diploidea (Visvesvara et al. 2007; Siddiqui 2012). Acanthamoeba pathogenic strains are the causative agents of a multifocal encephalitis called granulomatous amoebic encephalitis, a chronic central nervous system disease of immunocompromised hosts and various other system disease states including Acanthamoeba keratitis and pneumonitis (Lorenzo-Morales et al. 2005a). Furthermore, Acanthamoeba infection cases are often misdiagnosed due to lack of awareness and the expertise needed for diagnosis. Moreover, the most disturbing fact is that currently, there are no fully effective treatments against Acanthamoeba infections.
Moreover, Acanthamoebae are also known to serve as vehicles or vectors of many intracellular pathogenic microorganisms (Scheid and Schwarzenberger 2011, 2012). Instead of being digested, some of these so-called endocytobionts proliferate within the amoebae and also are protected against the actions of disinfectants and transported within the environment for their dispersion (Scheid and Schwarzenberger 2012).
The identification of Acanthamoeba at the genus level is based on distinctive features of trophozoites and cysts, especially the double walled cysts. Acanthamoeba sp. have been classified into three distinct morphological groups (I–III) (Pussard and Pons 1977). To date, evolutionary studies have led to the identification of 18 different genotypes (T1–T18) based on rRNA gene sequencing (Booton et al. 2005; Nuprasert et al. 2010; Qvarnstrom et al. 2013). Most studies have shown that 90 % of Acanthamoeba isolates that produce infections belong to the T4 genotype, although other genotypes have been reported as causative agents of amoebic infections in humans and other animals such as T1, T3, T5, T10, T11, T15, T17 and T18 (Lorenzo-Morales et al. 2013; Qvarnstrom et al. 2013).
Despite the fact that Acanthamoeba strains have been isolated from environmental sources worldwide, there are only a few studies regarding the isolation and genotyping of these amoebae in Spain (García A et al. 2011; Magnet et al. 2012, 2013). Moreover, there are no available genotype studies of Acanthamoebae from environmental sources in the Canary Islands and the only available studies were undertaken in Tenerife Island (Lorenzo-Morales et al. 2005a, b). Regarding clinical specimens, Acanthamoeba have been isolated from contact lens cases and lenses in Tenerife (Martín-Navarro et al. 2008) and also a case of a mixed keratitis was previously reported in the same island (Lorenzo-Morales et al. 2007a).
Therefore, the aim of this study was to determine the presence of these protozoa in soil samples in the island of Gran Canaria, Canary Islands, Spain. To the best of our knowledge, this is the first study of Acanthamoeba strains at the genotype level in the Canary Islands.
Material and methods
Sample sites and culture of Acanthamoeba
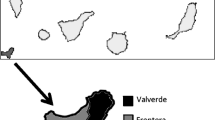
Twenty-four soil samples were collected across the island of Gran Canaria (located in the Atlantic Ocean about 150 km off the northwestern coast of Africa and about 1,350 km from Europe; coordinates 27°58′ N 15°36′ W, Fig. 1) from public access and recreational areas using sterile 15-ml tubes. Samples were dissolved in 20 ml of Neff’s saline and 150 μl of each sample were seeded onto 2 % non-nutrient agar plates and incubated at 25 °C. The cultures were monitored every 24 h, for up to 7 days. Positive isolates (identified under the inverted microscope) were cloned by dilution and transferred to the new culture plates for further molecular analyses as previously described (Lorenzo-Morales et al. 2006). Acanthamoeba isolated in this way were then transferred into axenic cultures by placing the amoebae into PYG medium (0.75 % Proteose peptone (w/v), 0.75 % yeast extract (w/v) and 1.5 % glucose (w/v)). A type strain of Acanthamoeba (Acanthamoeba castellanii Neff ATCC 30010) from the American Type Culture Collection (ATCC) was grown without shaking under the same conditions.
DNA extraction
DNA from cultures identified as positive for Acanthamoeba spp. by microscopy was extracted by placing 1–2 ml of Acanthamoeba cultures directly into the Maxwell® 16 tissue DNA purification kit sample cartridge (Promega, Madrid, Spain). Acanthamoeba genomic DNA was purified using the Maxwell® 16 instrument as described in the Maxwell® 16 DNA purification kits technical manual #TM284 (Promega, Madrid, Spain). DNA yield and purity were determined using the NanoDrop® 1000 spectrophotometer (Fisher Scientific, Madrid, Spain).
PCR and characterization at the genotype level
18S rDNA gene amplifications (DF3 region) were performed as previously described (Booton et al. 2005; Niyyati et al. 2009). PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and sequenced using a MegaBACE 1000 automatic sequencer (Healthcare Biosciences, Barcelona, Spain) in the University of La Laguna Sequencing Services (Servicio de Secuenciación SEGAI, University of La Laguna). The obtained sequences were aligned using Mega 5.0 software programme (Tamura et al. 2011). Genotype identification was based on sequence analysis of DF3 region as previously described by comparison to the available Acanthamoeba DNA sequences in GenBank database (Booton et al. 2005; Niyyati et al. 2009). A. castellanii Neff ATCC 30010 DNA was used as a positive control in the PCR reactions.
Phylogenetic analyses were carried out using maximum parsimony, minimum evolution and maximum likelihood optimality criteria, implemented in Mega 5.0 (Tamura et al. 2011). Transition/transversion ratios were estimated by maximum likelihood heuristic searches. Estimates of node support were obtained by performing 1,000 bootstrap replicates. Obtained sequences were compared to sequences available in GenBank database. Diagnostic Fragment 3 sequences for the new isolates are deposited in the GenBank database under the accession numbers: KF886742-KF886765.
Results and discussion
Fifteen of the 24 samples (62.5 %) included in this study were positive for Acanthamoeba based on morphological analysis and PCR results (Table 1). After sequencing of the obtained amplified products for the DF3 region as well as the phylogenetical analysis, Acanthamoeba strains belonging to genotype T2 (13.3 %), T4 (80 %) and T5 (6.7 %) were identified in the samples collected in the island of Gran Canaria (Fig. 2, Table 1).
18S rDNA DF3 linearized neighbour-joining tree obtained by using the Kimura two-parameter distance algorithm, produced in MEGA 5.0. The isolates obtained in the present study are identified in the tree (boxes). The type sequences were taken from GenBank and are presented under the following numbers: Acanthamoeba astronyxis strain PSHCW4 accession # KC493775, A. astronyxis strain CCAP 1534/1 #AF239293, A. castellanii strain CDC:0981:V006 accession #U07400, A. griffini S-7 ATCC 30731 accession #U07412, A. lenticulata isolate 12-SO #KC694184, A. lenticulata isolate 33195463 accession #KC438381, Acanthamoeba sp. isolate OSU09-002 accession # JQ669657, Acanthamoeba sp. genotype T2 Isolate OSU09-006 #JQ669661, Acanthamoeba palestinensis isolate TW-2 accession #KC694193, A. palestinensis strain CCAP 1547–1 #AF239296, Acanthamoeba sp. isolate BRO2-T16 accession #JX683392, Acanthamoeba sp. UWC9 accession #AF132134, Acanthamoeba sp. PM5 accession #JX494395, Acanthamoeba jacobsi AC304 accession #AY262364
Previous environmental studies in different regions have reported the presence of Acanthamoeba strains mainly in water and soil sources of many areas worldwide (Lorenzo-Morales et al. 2005a, 2005b, 2005c, 2006; De Jonckheere 2007; Shoff et al. 2008; Edagawa et al. 2009; Niyyati et al. 2009; García et al. 2011; Alves Dde et al. 2012; Magnet et al. 2012; Coşkun et al. 2013; Tanveer et al. 2013). In recent studies conducted in Spain, the percentage of positive samples for Acanthamoeba ranged between 2.8 and 99.1 %, T4 being the most common genotype (Magnet et al. 2013). Similarly, T4 was also reported as the most prevalent genotype in all other surveys conducted worldwide (Siddiqui 2012; Lorenzo-Morales et al. 2013). As well as the T4 genotype, other genotypes related to amoebic infections such as T5 and T11 have also been isolated in Spain (García et al. 2011; Lorenzo-Morales et al. 2011).
In the Canary Islands, studies on free-living amoebae and Acanthamoeba have only been carried out in the island of Tenerife, where Acanthamoeba was isolated from about 40 % percentage of samples from soil and water sources (Lorenzo-Morales et al. 2005a, b). No other islands from the archipelago have been surveyed for the presence of these potential pathogens, thus the importance of this study. Regarding clinical cases of Acanthamoeba, a mixed co-infection due to Acanthamoeba genotype T4 and Hartmannella has been reported in Tenerife island (Lorenzo-Morales et al. 2007a) and a study on the presence of Acanthamoeba in contact lens cases and lenses in Tenerife island described the isolation of genotypes T1, T3, T4 and T10, being T4 the most common genotype (78.6 %) within the studied samples (Martín-Navarro et al. 2008).
As it was mentioned before, Acanthamoeba strains could serve as vehicles and host for many pathogenic microorganisms such as virus, fungi, protozoa and bacteria as it has been previously reported (Scheid et al. 2010; Scheid and Schwarzenberger 2011, 2012). In the Canary Islands, the only available study on endocytobionts reported the presence of human pathogenic adenovirus within strains of Acanthamoeba that were isolated from water sources in the islands (Lorenzo-Morales et al. 2007b).
Gran Canaria Island was chosen because it is the second most populated island of the Canary Islands, a Spanish archipelago, with a population of 838,397, which constitutes approximately 40 % of the population of the archipelago. Therefore, soil sources were studied to investigate the presence of Acanthamoeba. The study results showed that a high percentage (62.5 %) of the collected samples were positive for Acanthamoeba spp. As expected, strains belonging to genotype T4 were the most common isolated ones; however, other strains belonging to T2 and T5 genotype were also identified in this survey. Interestingly, this is the first study at the genotype level in the Canary Islands and the first reports of the previously mentioned genotypes in this region. These data together with the reports worldwide about the high prevalence of T4 in clinical and environmental sources would give more detailed knowledge about the distribution of the different genotypes of Acanthamoeba worldwide and should raise awareness within clinicians and public health professionals in the Canary Islands and worldwide.
References
Alves Dde S, Moraes AS, Nitz N, de Oliveira MG, Hecht MM, Gurgel-Gonçalves R, Cuba CA (2012) Occurrence and characterization of Acanthamoeba similar to genotypes T4, T5 and T2/T6 isolated from environmental sources in Brasília, Federal District, Brazil. Exp Parasitol 131:239–244
Booton GC, Visvesvara GS, Byers TJ, Kelly DJ, Fuerst PA (2005) Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J Clin Microbiol 43:1689–1693
Coşkun KA, Ozçelik S, Tutar L, Elaldı N, Tutar Y (2013) Isolation and identification of free-living amoebae from tap water in Sivas, Turkey. Biomed Res Int 2013:675145. doi:10.1155/2013/675145
De Jonckheere JF (2007) Molecular identification of free-living amoebae of the Vahlkampfiidae and Acanthamoebidae isolated in Arizona (USA). Eur J Protistol 43:9–15
Edagawa A, Kimura A, Kawabuchi-Kurata T, Kusuhara Y, Karanis P (2009) Isolation and genotyping of potentially pathogenic Acanthamoeba and Naegleria sp. from tap water sources in Osaka, Japan. Parasitol Res 105:1109–1117
García A, Goñi P, Clavel A, Lobez S, Fernandez MT, Ormad MP (2011) Potentially pathogenic free-living amoebae (FLA) isolated in Spanish wastewater treatment plants. Environ Microbiol Rep 3:622–626
Lorenzo-Morales J, Monteverde-Miranda CA, Jiménez C, Tejedor ML, Valladares B, Ortega-Rivas A (2005a) Evaluation of Acanthamoeba isolates from environmental sources in Tenerife, Canary Islands, Spain. Ann Agric Environ Med 12:233–236
Lorenzo-Morales J, Ortega-Rivas A, Foronda P, Martínez E, Valladares B (2005b) Isolation and identification of pathogenic Acanthamoeba strains in Tenerife, Canary Islands, Spain from water sources. Parasitol Res 95:273–277
Lorenzo-Morales J, Lindo JF, Martinez E, Calder D, Figueruelo E, Valladares B, Ortega-Rivas A (2005c) Pathogenic Acanthamoeba strains from water sources in Jamaica, West Indies. Ann Trop Med Parasitol 99:751–758
Lorenzo-Morales J, Ortega-Rivas A, Martínez E, Khoubbane M, Artigas P, Periago MV, Foronda P, Abreu-Acosta N, Valladares B, Mas-Coma S (2006) Acanthamoeba isolates belonging to T1, T2, T3, T4 and T7 genotypes from environmental freshwater samples in the Nile Delta region, Egypt. Acta Trop 100:63–69
Lorenzo-Morales J, Martínez-Carretero E, Batista N, Alvarez-Marín J, Bahaya Y, Walochnik J, Valladares B (2007a) Early diagnosis of amoebic keratitis due to a mixed infection with Acanthamoeba and Hartmannella. Parasitol Res 102:167–169
Lorenzo-Morales J, Coronado-Alvarez N, Martínez-Carretero E, Maciver SK, Valladares B (2007b) Detection of four adenovirus serotypes within water-isolated strains of Acanthamoeba in the Canary Islands, Spain. Am J Trop Med Hyg 77:753–756
Lorenzo-Morales J, Morcillo-Laiz R, Martín-Navarro CM, López-Vélez R, López- Arencibia A, Arnalich-Montiel F, Maciver SK, Valladares B, Martínez- Carretero E (2011) Acanthamoeba keratitis due to genotype T11 in a rigid gas permeable contact lens wearer in Spain. Cont Lens Anterior Eye 34:83–86
Lorenzo-Morales J, Martín-Navarro CM, López-Arencibia A, Arnalich-Montiel F, Piñero JE, Valladares B (2013) Acanthamoeba keratitis: an emerging disease gathering importance worldwide? Trends Parasitol 29:181–187
Magnet A, Galván AL, Fenoy S, Izquierdo F, Rueda C, Fernandez Vadillo C, Pérez- Irezábal J, Bandyopadhyay K, Visvesvara GS, da Silva AJ, del Aguila C (2012) Molecular characterization of Acanthamoeba isolated in water treatment plants and comparison with clinical isolates. Parasitol Res 111:383–392
Magnet A, Fenoy S, Galván AL, Izquierdo F, Rueda C, Fernandez Vadillo C, Del Aguila C (2013) A year-long study of the presence of free living amoeba in Spain. Water Res 47:6966–6972
Marciano-Cabral F, Cabral G (2003) Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 16:273–307
Martín-Navarro CM, Lorenzo-Morales J, Cabrera-Serra MG, Rancel F, Coronado- Alvarez NM, Piñero JE, Valladares B (2008) The potential pathogenicity of chlorhexidine-sensitive Acanthamoeba strains isolated from contact lens cases from asymptomatic individuals in Tenerife, Canary Islands, Spain. J Med Microbiol 57:1399–1404
Niyyati M, Lorenzo-Morales J, Rezaie S, Rahimi F, Mohebali M, Maghsood AH, Motevalli-Haghi A, Martín-Navarro CM, Farnia S, Valladares B, Rezaeian M (2009) Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Exp Parasitol 121:242–245
Nuprasert W, Putaporntip C, Pariyakanok L, Jongwutiwes S (2010) Identification of a novel T17 genotype of Acanthamoeba from environmental isolates and T10 genotype causing keratitis in Thailand. J Clin Microbiol 48:4636–4640
Pussard M, Pons R (1977) Morphologie de la paroi kystique et taxonomie du genre Acanthamoeba. Proc Natl Acad Sci U S A 13:557–598
Qvarnstrom Y, Nerad TA, Visvesvara GS (2013) Characterization of a New Pathogenic Acanthamoeba Species, A. byersi n. sp., Isolated from a Human with Fatal Amoebic Encephalitis. J Eukaryot Microbiol 60:626–633
Scheid PL, Schwarzenberger R (2011) Free-living amoebae as vectors of Cryptosporidia. Parasitol Res 109:499–504
Scheid P, Schwarzenberger R (2012) Acanthamoeba spp. as vehicle and reservoir of adenoviruses. Parasitol Res 111:479–485
Scheid P, Hauröder B, Michel R (2010) Investigations of an extraordinary endocytobiont in Acanthamoeba sp.: development and replication. Parasitol Res 106:1371–1377
Shoff ME, Rogerson A, Kessler K, Schatz S, Seal DV (2008) Prevalence of Acanthamoeba and other naked amoebae in South Florida domestic water. J Water Health 6:99–104
Siddiqui R (2012) Khan NA (2012) Biology and pathogenesis of Acanthamoeba. Parasit Vectors 5:6. doi:10.1186/1756-3305-5-6. Review
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tanveer T, Hameed A, Muazzam AG, Jung SY, Gul A, Matin A (2013) Isolation and molecular characterization of potentially pathogenic Acanthamoeba genotypes from diverse water resources including household drinking water from Khyber Pakhtunkhwa, Pakistan. Parasitol Res 112:2925–2932
Visvesvara GS, Moura H, Schuster FL (2007) Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri and Sappinia diploidea. FEMS Immunol Med Microbio 50:1–26
Acknowledgments
This work was supported by the grants RICET (project no. RD12/0018/0012 of the programme of Redes Temáticas de Investigación Cooperativa, FIS), Spanish Ministry of Health, Madrid, Spain and the Project FIS PI10/01298 “Protozoosis emergentes por amebas de vida libre: aislamiento y caracterización molecular, identificación de cepas transportadoras de otros agentes patógenos y búsqueda de quimioterapias efectivas” from the Instituto de Salud Carlos III. ALA was funded by a grant “Ayudas del Programa de Formación de Personal Investigador, para la realización de Tesis Doctorales” from the Agencia Canaria de Investigación, Innovación y Sociedad de la Información from the Canary Islands Government. CMMN was supported by a postdoctoral grant from the Fundación Canaria Manuel Morales, La Palma, Canary Islands. MRB, CDT and AMCV were funded by CEI Canarias, Campus Atlántico Internacional. MRB was also supported by Becas Fundación Cajacanarias para Postgraduados. JLM was supported by the Ramón y Cajal Subprogramme from the Spanish Ministry of Economy and Competivity RYC-2011-08863. The authors are grateful to Jorge Rodríguez Rodríguez and Silvia Matos González for their help during the sampling procedure.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reyes-Batlle, M., Todd, C.D., Martín-Navarro, C.M. et al. Isolation and characterization of Acanthamoeba strains from soil samples in Gran Canaria, Canary Islands, Spain. Parasitol Res 113, 1383–1388 (2014). https://doi.org/10.1007/s00436-014-3778-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-014-3778-z