Abstract
4E-BP, an eIF4E-binding protein, is well known as a cap-dependent translation inhibitor. Here, the 4E-BP homolog, Hl4E-BP, was isolated and identified from the hard tick Haemaphysalis longicornis. Hl4E-BP transcripts were ubiquitously expressed in the active stages, including the larvae, nymphs, and female adults, and the transcription levels were found to be higher in unfed than engorged ticks. In contrast, the expression levels of non-phosphorylated Hl4E-BP, which is a 13.4-kDa protein detected by the anti-recombinant Hl4E-BP antibody, were the highest in engorged ticks and significantly decreased progressively during the unfed starvation period of ticks. The functional role of Hl4E-BP as a metabolic brake was verified by histochemical observations on the lipid storage in midguts and fat bodies during the starvation period using ticks injected with dsHl4E-BP. The results indicate that Hl4E-BP is highly relevant to the lipid storage of ticks during the non-feeding starvation period. Our results suggest, for the first time, that Hl4E-BP may have a crucial role in the starvation resistance of ticks in an off-host condition via lipid metabolism control, although it was unclear whether Hl4E-BP might be involved in lipid synthesis regulation and/or lipid consumption inhibition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ticks are obligatory blood-sucking arthropods that can survive only by feeding on a vertebrate host. Their life is essentially composed of relatively short parasitic periods and long off-host periods without the intake of blood meals (Sonenshine 2005). Ticks spend part of their lives on a host animal; the feeding period is equivalent to less than 5 % of the lifespan (Umemiya-Shirafuji et al. 2010). This overtly prominent viability is important for understanding the biology and epidemiology of ticks and tick-borne pathogens. Midguts and fat bodies in unfed ticks in the off-host condition preserve digestive products (obtained when imbibing blood) as many of the nutrients (Sonenshine 2005; Williams et al. 1985). However, the molecular mechanisms underlying their metabolic features remain largely unknown.
Translation of protein synthesis in eukaryotes plays a critical role in development, differentiation, cell cycle progression, cell growth, and apoptosis (Gingras et al. 1999b, 2001). The eIF4E-binding protein, 4E-BP, is an important regulator of overall translation levels in cells (Gingras et al. 1999b, 2001; Lawrence and Abraham 1997). Recently, 4E-BP has been suggested to serve as a “metabolic brake” controlled by the target of rapamycin (TOR), which is activated under conditions of environmental stress to control the fat metabolism (Teleman et al. 2005; Hara et al. 1997). In mice, 4E-BP1 knockout mice showed markedly small white fat pads, and male mice display an increased metabolic rate (Tsukiyama-Kohara et al. 2001). In Drosophila, the fat levels of 4E-BP mutant flies decreased faster than those of wild flies in the starved condition (Teleman et al. 2005). It was also demonstrated that 4E-BP is a key molecule for lifespan extension under dietary restrictions by modulation of the mitochondrial function (Zid et al. 2009).
In this study, the 4E-BP homologue, Hl4E-BP, was identified and characterized from Haemaphysalis longicornis ticks. In particular, the functions of Hl4E-BP as a metabolic brake were verified by histochemical observations on the lipid storage in midgut epithelial cells and fat body trophocytes during non-feeding starvation period of ticks after RNA interference. The results indicate, for the first time, that tick 4E-BP affects the lipid storage levels and suggest that tick 4E-BP may be a key molecule contributing to the supply and preservation of survival energy of ticks during the long starvation period in the off-host condition.
Materials and methods
Ticks
Parthenogenetic H. longicornis ticks (Okayama strain) were maintained by feeding on the ears of Japanese white rabbits (Kyudo, Kumamoto, Japan) for many generations at our laboratory (Umemiya-Shirafuji et al. 2010; Kawano et al. 2011). In this study, the term “unfed” used regarding the state of ticks implies that ticks have not yet attacked new hosts after emerging in the subsequent life stage by hatching or molting.
Animals
Rabbits and mice were cared for in accordance with the guidelines approved by the Animal Care and Use Committee of Kagoshima University (Approval number A08010). These animals were maintained in a temperature- and humidity-regulated room under controlled lighting with free access to tap water and commercial regular chow throughout the experiments.
Identification of Hl4EBP cDNA
The full-length cDNA library was made using the vector-capping method as previously reported (Kato et al. 2005). A homolog of the 4E-BP, Hl4E-BP gene, was isolated from a hemocyte cDNA library previously constructed from H. longicornis (Umemiya-Shirafuji et al. 2010). The sequence of the plasmid containing the homolog was analyzed with the Applied Biosystems 3130x1 Genetic Analyzer (Applied Biosystems, California, USA). The amino acid sequence of Hl4E-BP was determined using GENETYX, version7 (Genetyx). The similarity of the deduced amino acid sequence in the protein database was examined using the BLASTp server (National Center for Biotechnology Information [NCBI], http://www.ncbi.nlm.nih.gov/Blast.cgi). The theoretical isoelectric point (pI) and molecular weight (MW) were determined using the Compute pI/Mw tool (http://au.expasy.org/tools/pi_tool.html). The alignment between Hl4EBP and other homologues was performed by CLUSTALW (http://www.genome.jp/tools/clustalw/).
Expression of recombinant Hl4E-BP protein and preparation of anti-recombinant Hl4E-BP sera
The open reading flame (ORF) of Hl4E-BP was amplified by PCR with the 5′-primer containing a recognition site for XhoI and the 3′-primer containing a recognition site for EcoRI (Table S1). The PCR products were purified using a GENECLEANKIT II kit (MP Biomedical, Ohio, USA) and inserted into the plasmid pRSET A vector (Invitrogen, California, USA), which had been cleaved at XhoI and EcoRI with DNA ligation kit ver. 2.1 (Takara, Shiga, Japan) according to the manufacturer’s protocol. The plasmid vector was then transformed into Escherichia coli (BL21 strain). To express the recombinant histidine-tagged Hl4E-BP protein, we added 1 mM isopropyl-β-d(−)-thiogalactopyranoside and cultured the mixture overnight at 20 °C. The E. coli lysate was centrifuged to obtain a pellet. The pellet was dissolved in a lysis buffer (50 mM NaH2SO4 and 300 mM NaCl, pH 8.0) with 1 mg/ml lysozyme followed by agitation for 1 h at 4 °C. After sonication and centrifugation, supernatant fluid containing recombinant Hl4E-BP was poured into a column (Poly-Prep® Chromatography Columns; Bio-Rad Laboratories, California, USA), and nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen, Venlo, Netherlands) were added. To induce binding of recombinant histidine-tagged Hl4E-BP protein to the agarose beads, supernatant fluid was agitated overnight at 4 °C, and recombinant Hl4E-BP was recovered from agarose beads with an elution buffer (50 mM NaH2SO4, 300 mM NaCl, and 20 mM imidazol, pH 8.0). The purified recombinant Hl4E-BP protein was dialyzed with TBS (40 mM Tris–HCl and 300 mM NaCl, pH 7.5). The solution containing 300 μg of recombinant Hl4E-BP was completely mixed with the same volume of Freund’s complete adjuvant or incomplete adjuvant (Sigma, MO, USA) and intraperitoneally injected into female mice (ddy, 6 weeks old). At day 14 after the first injection, two rounds of immunization were performed. The last immunization was performed similarly at day 28 with the same dose of recombinant Hl4E-BP in Freund’s incomplete adjuvant (Sigma). Finally, sera were collected at day 35.
SDS-PAGE and Western blot analysis
Whole bodies of ticks in each developmental stage (larvae, nymphs, and female adults 1, 3, and 10 month(s) after hatching or molting, and larvae, nymphs, and female adults just after engorgement) were frozen and crashed with liquid nitrogen, dissolved in TBS containing a protease inhibitor (Complete Mini EDTA free; Roche, Basel, Switzerland) and 0.05 % TritonX-100, and then sonicated. The extracts were mixed with a 2× sample buffer (4 % SDS, 20 % glycerol, 125 M Tris, 1 % Bromophenol Blue, 10 % 2-mercaptoethanol) and boiled. After centrifugation (3,350 × g, 5 min at 4 °C), the supernatant was subjected to SDS-PAGE on 18 % acrylamide gel.
The separated proteins in gel after SDS-PAGE were transferred onto a polyvinylidene difluoride membrane for Western blot analysis. The membrane was blocked with 5 % skim milk in 0.1 % Tween 20–TBS and then incubated with mouse sera containing the first antibody diluted in 5 % skim milk in 0.05 % Tween 20–TBS. Anti-Hl4E-BP antibody and anti-HlActin antibody were used as the first antibody (Kawano et al. 2011). After washing, the membrane was incubated with peroxidase-conjugated goat anti-mouse IgG sera (Dako Denmark A/S, Glostrup, Denmark), and antibody binding signals were then detected using ECL plus a Western Blotting Detection System (GE Healthcare, Little Chalfont, UK).
To detect the phosphorylated 4E-BP protein, a nitrocellulose membrane was used for Western blot analysis. The membrane after transferring separated proteins by SDS-PAGE was incubated with anti-phospho-4E-BP1 rabbit monoclonal antibody (Thr37/46; Cell Signaling Technology, Massachusetts, USA) and then with peroxidase-conjugated goat anti-rabbit IgG sera (Dako Denmark A/S). The membrane was blocked with 2 % gelatin in 0.1 % Tween 20–TBS, and each antibody used for this Western blot was diluted in 2 % gelatin in 0.05 % Tween 20–TBS.
Isolation of total RNA and synthesis of cDNA
RNA was extracted from whole bodies of ticks in each developmental stage (1, 3, and 10 month(s) after hatching or molting and larvae, nymphs, and female adults just after engorgement) using TRI Reagent (Sigma) according to the manufacturer’s protocol. The purified RNA was dissolved in diethylpyrocarbonate-treated water and stored at −80 °C until use. Single-stranded cDNA was generated by reverse transcription using a Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics).
RNAi of unfed and starved ticks
The Hl4E-BP fragment was amplified by PCR from a recombinant plasmid DNA (described in the section “Expression of recombinant Hl4E-BP protein and preparation of anti-rHl4EBP serum”) using oligonucleotides, including T7 forward and T7 reverse primers, to attach to the T7 promoter recognition site on both the 5′ and 3′ ends (Table S1). As a negative control, cDNA of the firefly Luciferase (Luc) (Promega) gene was amplified by PCR using oligonucleotides, including T7 forward and T7 reverse primers. PCR products were purified with the GENECLEANKIT II kit (MP Biomedical). Purified DNA was used to synthesize the double-stranded RNA (dsRNA) with a T7 RiboMaxTM Express RNAi System (Promega) according to the manufacturer’s protocol. The quality of dsRNA was checked using 1.5 % agarose gel, and the concentration was quantified using a spectrometer. The dsRNA (4 μg/tick) of Hl4E-BP (dsHl4-EBP) or Luc (dsLuc) was injected through the fourth coxae into the hemocoel of unfed female adult ticks (1 month after molting) fixed on a glass slide with adhesive tape. Injection of dsRNA was carried out using 50-μl microcapillaries (Drummond Scientific Company, Pennsylvania, USA) drawn to fine-point needles by heating. Female ticks 1 month after molting were injected with dsRNA and left for at 25 °C in an incubator without any blood meals for at maximum 4 months. Their survival rates and gene silencing after dsRNA injection were monitored in addition to the observation of lipid consumption in midguts and fat bodies.
Observation of lipid storage
Midguts and fat bodies of female ticks injected with dsRNA were dissected out in a fixative (4 % paraformaldehyde including 2.5 % glutaraldehyde in PBS), immersed in a fixative with shaking at 4 °C overnight, and embedded in the Tissue-Tek® O.C.T. Compound (Sakura Finetek, Tokyo, Japan; Umemiya-Shirafuji et al. 2010). Frozen sections (4–5 μm thick) were cut on a cryostat (Leica CM 1850; Leica Microsystems, Wetzlar, Germany) and placed on MAS-coated glass slides (Matsunami Glass, Ind.) and air-dried. Lipid staining was performed with 60 % isopropanol for 1 min and oil red O (Wako Pure Chemical Industries, Osaka, Japan) for 15 min. Nucleus was stained with Mayer’s hematoxylin solution (Wako Pure Chemical Industries) for 5 min and then washed with distilled water. The section was covered with glycerol and a cover glass and observed under the microscope. ImageJ (http://rsbweb.nih.gov/ij/) was employed to calculate the oil red O positive area.
Statistical analyses
All statistical analyses were performed with the Student’s t-test. P < 0.05 values were considered significant.
DDBJ/EMBL/GenBank accession number
The nucleotide sequence data reported in this paper was deposited in the DDBJ/EMBL/GenBank nucleotide sequence database with the accession number AB685378.
Results
Identification of Hl4E-BP cDNA
The full length of Hl4E-BP cDNA consists of 1,424 nucleotides, including an ORF of 387 bp, from 112 to 498 bp (Fig. S1). The ORF of the cDNA sequence encodes a putative protein of 128 amino acid residues with a predicted molecular weight of 13.4 kDa. The isoelectric point is 9.73. The cDNA sequence contains a TOS (TOR signaling) site (Phe161–Asp104; Schalm and Blenis 2002), a RAIP motif (Arg51–Pro54; Tee and Proud 2002), a YXXXXLφ-binding motif (eIF4E-binding motif, Tyr90–Leu95; Mader et al. 1995), and four phosphorylation sites (Thr73–Pro74, Thr82–Pro83, Sre101–Pro102, and Thr106–Pro107; Mothe-Satney et al. 2000). The Hl4E-BP sequence is conserved with higher homology to ixodid ticks (I. scapularis, 83 %; GenBank ID: XP002404524.1) than to insects (Drosophila melanogaster, 50 % [GenBank ID: NP477295.1] and Culex quinquefasciatus, 45 % [GenBank ID: XP001846418.1]) and to vertebrates (Mus musculus, 50 % [GenBank ID: NP034254.1] and Osmerus mordax, 49 % [GenBank ID: ACO09797.1]) (Fig. 1).
Alignment of the deduced amino acid sequence of Hl4E-BP. The ixodid tick Ixodes scapularis (GeneBank ID: XP002404524.1), fruit fly Drosophila melanogaster (GeneBank ID: NP477295.1), mouse Mus musculus (GeneBank ID: NP034254.1), bony fish Osmerus mordax (GeneBank ID: ACO09797.1), and culicine mosquito Culex quinquefasciatus (GeneBank ID: XP001846418.1) are calculated in reference to Haemaphysalis longicornis. The common conserved three motifs, the RAIP motif, TOS (TOR signaling) motif, and eIF4E binding motif (YxxxxLφ), are indicated by dashed lines. The predicted four phosphorylated sites are marked by asterisks. The identical residues are highlighted in black. Similar residues are highlighted in gray. Amino acids are numbered on the right
Expression profiles of Hl4E-BP mRNA and protein
To detect Hl4E-BP expression in whole bodies of ticks, we performed RT-PCR and Western blot analysis. Hl4E-BP was ubiquitously expressed in the active stages, including the larvae, nymphs, and adults, and the transcription levels were higher in unfed ticks than in engorged ticks (Fig. 2a). In unfed and engorged ticks, non-phosphorylated Hl4E-BP protein with a molecular weight of approximately 13.4 kDa was detected by the anti-recombinant Hl4E-BP antibody (Fig. 2b). The expression of the non-phosphorylated Hl4E-BP protein was most prominent in engorged ticks and was significantly decreased with the extension of non-feeding periods (Fig. 2b). The alteration of the phosphorylation levels of Hl4E-BP was examined by Western blot using an anti-phospho-4E-BP1 monoclonal antibody (Fig. 2c). As a result, two different molecular weight bands were detected in engorged ticks, and one of their higher molecular weight bands was ubiquitous in unfed ticks, as shown by the anti-phospho-4E-BP1 monoclonal antibody.
Expression profiles of Hl4E-BP in unfed and engorged ticks. The expression of Hl4E-BP in different developmental stages of ticks was analyzed by RT-PCR and Western blot. m month(s), En engorged ticks. a mRNA of Hl4E-BP. b Nonphosphorylated Hl4E-BP detected by the anti-recombinant 4E-BP antibody. c pHl4E-BP, phosphorylated and hypophosphorylated Hl4E-BPs, detected by the anti-phospho 4E-BP1 monoclonal antibody. H. longicornis actin is expressed constitutively and shown as a control
RNAi of Hl4E-BP and lipid storage of unfed ticks
To examine the function of Hl4E-BP as a key molecule contributing to the energy supply and/or preservation of survival energy of ticks during the long starvation period, lipid storage in midguts and fat bodies was histochemically examined using female adult ticks 2 and 4 months after injection with dsHl4E-BP or dsLuc. Silencing of Hl4E-BP was manifest in unfed and starved ticks 32–66 days after dsHl4E-BP injection, while the transcripts were barely detectable in ticks 66 days after dsHl4E-BP injection. Robust expression of Hl4E-BP transcripts was observed in ticks 120 days after dsRNA injection. These results strongly suggested that gene silencing of Hl4E-BP was sustained for at least 2 months after dsRNA injection. The survival rates during the period of non-feeding and starvation, at maximum, 120 days, were comparable in dsHl4E-BP-injected ticks and dsLuc-injected ticks (Fig. 3).
Expression profiles of Hl4E-BP in ticks after dsRNA injection and survival rates of ticks after dsRNA injection. a Gene silencing of Hl4E-BP was confirmed by RT-PCR. d Days after dsRNA injection. b Survival rates of ticks after dsRNA injection. The black line indicates the dsHl4E-BP-injected group, and the gray line indicates the dsLuc-injected control group
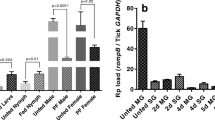
An abundant accumulation of lipid droplets was observed in the midguts and fat bodies of unfed female ticks immediately after dsHl4E-BP (Fig. S2) or dsLuc injection (data not shown). A similar accumulation level of lipid droplets was observed in the midguts and fat bodies of unfed ticks 2 and 4 months after dsLuc injection (Fig. 4b, d). However, the lipid accumulation levels in midguts and fat bodies of unfed ticks 2 and 4 months after dsHl4E-BP injection were dramatically decreased (Fig. 4a, c) and significantly lower than those after dsLuc injection (Fig. 5). The decrease of lipid storage in female ticks treated with dsHl4EBP was most prominent in ticks 4 months after dsRNA injection (Fig. 5).
Effects of Hl4E-BP RNAi on lipid storage in midguts and fat bodies of ticks during the long starvation period in the off-host condition. Lipid storage was detected by Oil red O staining. a Midgut of Hl4E-BP-knockdown adult female ticks 2 months after dsRNA injection; b midgut of dsLuc-injected ticks 2 months after dsRNA injection; c fat body of Hl4E-BP-knockdown ticks 4 months after dsRNA injection; d fat body of dsLuc-injected ticks 4 months after dsRNA injection. Arrowheads and arrows indicate the midgut epithelial cells and the fat body trophocytes, respectively
Effects of Hl4E-BP RNAi on lipid storage in midguts of ticks during the long starvation period in the off-host condition. Midguts were dissected from unfed adult female ticks 2 and 4 months after dsRNA injection. The black and gray bars show the percentages of Oil red O staining-positive area in the midgut from the dsHl4E-BP injected and dsLuc injected groups, respectively. *P < 0.05 by t-test
Discussion
4E-BP has been identified from a wide range of metazoan organisms, but there is no report on the characterization of 4E-BP in Chelicerata, including ticks, horseshoe crabs, scorpions, spiders, and mites, to this day. In the present study, the 4E-BP homolog, the Hl4E-BP gene, was isolated and identified from the hard tick H. longicornis. Three isoforms, 4E-BP1, 4E-BP2, and 4E-BP3, were identified from vertebrates (Homo sapiens, M. musculus, Rattus norvegicus, and Danio rerio; Rousseau et al. 1996; Poulin et al. 1998), while only one isoform was identified in invertebrate arthropods, such as D. melanogaster and Bombyx mori (Lasko 2000; Gu et al. 2011). In the present study, only one isoform was identified from H. longicornis ticks, but it is not clear whether or not other isoforms exist. The high sequence identity of amino acids of the Hl4E-BP gene was shared with D. melanogaster (50 %), M. musculus (50 %), and I. scapularis (83 %). The isolated Hl4E-BP gene has a conserved TOS site, a RAIP motif, an eIF4E-binding motif, and four phosphorylated sites similarly to other species (Schalm and Blenis 2002; Tee and Proud 2002; Mader et al. 1995; Mothe-Satney et al. 2000), suggesting the possibility that Hl4E-BP may have a binding affinity to eIF4E and phosphorylated regulation by TOR.
Hl4E-BP transcripts were ubiquitously expressed in the active stages, including the larvae, nymphs, and female adults, and the transcription levels were higher in unfed than engorged ticks (Fig. 2a). In contrast, the expression levels of non-phosphorylated Hl4E-BP, which is a 13.4-kDa protein detected by anti-recombinant Hl4E-BP antibody, were the highest in engorged ticks and significantly decreased progressively during the unfed starvation period of ticks (Fig. 2b). Two phosphorylated Hl4E-BPs were detected by the anti-phospho-4E-BP1 monoclonal antibody, and the lower phosphorylation level one (hypophosphorylated Hl4E-BP) was found in engorged ticks only (Fig. 2c). These results indicated that Hl4E-BP expression is usually regulated at transcription, translation, and phosphorylation, similarly to other organisms (Schalm and Blenis 2002; Tee and Proud 2002; Mader et al. 1995; Mothe-Satney et al. 2000; Rousseau et al. 1996; Poulin et al. 1998).
It is well known that non-phosphorylated 4E-BP shows high translation inhibitor activity via its binding with initiation factor eIF4E and hyperphosphorylated 4E-BP exhibits no translation suppression activity via its release from eIF4E (Gingras et al. 1999a; Ferguson et al. 2003; Kleijn et al. 1998; Raught and Gingras 1999). Therefore, it is inferred from the results of the phosphorylation status and amount of Hl4E-BP that a large amount of non-phosphorylated Hl4E-BP expressed in engorged ticks may contribute to enhance the metabolic activities for molting and/or reproduction and a moderate amount of non-phosphorylated Hl4E-BP expressed continuously in unfed ticks may play a role in energy supply for survival during the severe starvation period in the off-host condition. The biological role of the concomitant and robust expressions of non-phosphorylated Hl4E-BP and phosphorylated Hl4E-BP in engorged ticks remains unclear, but it may have functional relevance regarding the nutrient oversupply in ticks after very large blood meals.
The close relationship between 4E-BP and lipid metabolism has been reported (Teleman et al. 2005). In mammals, 4E-BP1-knockout mice have markedly small white fat pads, and male mice displayed an increase in metabolic rate (Tsukiyama-Kohara et al. 2001). In Drosophila insects, the fat levels of 4E-BP mutant flies decreased faster than those of control flies in the starved condition (Teleman et al. 2005), and 4E-BP regulated the expression of the mitochondrial gene on dietary restrictions (Zid et al. 2009). In ticks, it is widely accepted that both the epithelial cells of midguts and the trophocytes of fat bodies may function in the preservation and supply of survival energy for long starvation in the off-host condition (Sonenshine 2005; Koh et al. 1991). Therefore, the roles of Hl4E-BP in lipid storage in midgut and fat bodies in the unfed starvation period were investigated using ticks treated with dsHl4E-BP.
It was confirmed that gene silencing of Hl4E-BP was sustainable for at least 2 months (66 days, Fig. 3) after dsRNA injection. Abundant lipid storage in midguts and fat bodies of unfed ticks was dramatically decreased in ticks 2 and 4 months after dsHl4E-BP injection (Fig. 4a, c). The most prominent depletion of lipid storage was observed in ticks 4 months after dsHl4E-BP injection, when robust expression of Hl4E-BP transcripts was observed (120 days, Fig. 3). These results suggested that Hl4E-BP is highly relevant to lipid storage of ticks in the unfed starvation period. However, it was unclear in this study whether Hl4E-BP functions in lipid synthesis and/or lipid consumption inhibition. The robust transcription of Hl4E-BP in ticks 4 months after dsHl4E-BP injection might be interpreted in terms of the remarkably high levels of translation caused by the compensatory activity for the overconsumption of metabolic energy by knockdown of Hl4E-BP.
It was predicted that Hl4E-BP knockdown ticks might die faster than control ticks due to exhaustive energy depletion, but no significant differences were observed in mortality between dsHl4E-BP-treated and control groups during 4 months of unfed starvation. A likely explanation for this phenomenon is that the energy sources for ticks in the starvation period are lipids, carbohydrates, and protein.
The insulin/Akt/TOR signaling pathway regulates many organismal processes, including the lipid metabolism (Britton et al. 2002; Towler and Hardie 2007). In ticks, Akt (Umemiya-Shirafuji et al. 2012), TOR (Umemiya-Shirafuji, in preparation), GATA (Boldbaatar et al. 2010), and S6K (Boldbaatar et al. 2010), which are involved in this signaling pathway, have been already isolated and characterized using H. longicornis ticks in our laboratory. The biological roles of the lipid metabolism mediated by Hl4E-BP in ticks of the host-seeking starvation period should be elucidated in more detail from the standpoint of the insulin/Akt/TOR signaling pathway in the near future to achieve a better understanding of tick survival strategies towards the development of control measures of tick and tick-borne diseases.
References
Boldbaatar D, Battur B, Umemiya-Shirafuji R, Liao M, Tanaka T, Fujisaki K (2010) GATA transcription, translation, and regulation in Haemaphysalis longicornis tick: analysis of the cDNA and an essential role for vitellogenesis. Insect Biochem Mol Biol 40:49–57
Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA (2002) Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell 2:239–249
Ferguson G, Mothe-Satney I, Lawrence JC Jr (2003) Ser-64 and Ser-111 in PHAS-I are dispensable for insulin-stimulated dissociation from eIF4E. J Biol Chem 278:47459–47465
Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N (1999a) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Gene Dev 13:1422–1437
Gingras AC, Raught B, Sonenberg N (1999b) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68:913–963
Gingras AC, Raught B, Sonenberg N (2001) Regulation of translation initiation by FRAP/mTOR. Gene Dev 15:807–826
Gu SH, Young SC, Tsai WH, Lin JL, Lin PL (2011) Involvement of 4E-BP phosphorylation in embryonic development of the silkworm, Bombyx mori. J Insect Physiol 57:978–985
Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J (1997) Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem 272:26457–26463
Kato S, Ohtoko K, Ohtake H, Kimura T (2005) Vector-capping: a simple method for preparing a high-quality full-length cDNA library. DNA Res 12:53–62
Kawano S, Umemiya-Shirafuji R, Boldbaatar D, Matsuoka K, Tanaka T, Fujisaki K (2011) Cloning and characterization of the autophagy-related gene 6 from the hard tick, Haemaphysalis longicornis. Parasitol Res 109:1341–1349
Kleijn M, Scheper GC, Voorma HO, Thomas AA (1998) Regulation of translation initiation factors by signal transduction. Eur J Biochem 253:531–544
Koh K, Mori T, Shiraishi S, Uchida TA (1991) Ultrastructural changes of the midgut epithelial cells in feeding and molting nymphs of the tick Haemaphysalis longicornis. Int J Parasitol 21:23–36
Lasko P (2000) The Drosophila melanogaster genome: translation factors and RNA binding proteins. J Cell Biol 150:F51–F56
Lawrence JC Jr, Abraham RT (1997) PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci 22:345–349
Mader S, Lee H, Pause A, Sonenberg N (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 15:4990–4997
Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC Jr (2000) Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol 20:3558–3567
Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N (1998) 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem 273:14002–14007
Raught B, Gingras AC (1999) eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol 31:43–57
Rousseau D, Gingras AC, Pause A, Sonenberg N (1996) The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene 13:2415–2420
Schalm SS, Blenis J (2002) Identification of a conserved motif required for mTOR signaling. Curr Biol 12:632–639
Sonenshine DE (2005) The biology of tick vectors of human diseases. In: Goodman JL, Dennis DT, Sonenshine DE (eds) Tick-borne diseases of humans. ASM Press, Washington DC, pp 12–36
Tee AR, Proud CG (2002) Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol Cell Biol 22:1674–1683
Teleman AA, Chen YW, Cohen SM (2005) 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Dev Cell 9:271–281
Towler MC, Hardie DG (2007) AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res 100:328–341
Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, Harper ME, Tremblay ML, Sonenberg N (2001) Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med 7:1128–1132
Umemiya-Shirafuji R, Matsuo T, Liao M, Boldbaatar D, Battur B, Suzuki HI, Fujisaki K (2010) Increased expression of ATG genes during nonfeeding periods in the tick Haemaphysalis longicorni. Autophagy 6:473–481
Umemiya-Shirafuji R, Tanaka T, Boldbaatar D, Tanaka T, Fujisaki K (2012) Akt is an essential player in regulating cell/organ growth and development at nymphal and adult stages in the hard tick Haemaphysalis longicornis. Insect Biochem Mol Biol 42:164–173
Williams JP, Barker DM, Sauer JR et al (1985) Ultrastructural changes in the midgut epithelium of unfed lone star ticks with increasing age. Ann Entomol Soc Am 78:62–69
Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P (2009) 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139:149–160
Acknowledgements
We thank Dr. K. Kohara at the Transboundary Animal Diseases Research Center, Kagoshima University, for technical advice. This work was supported by a Grant-in-Aid for Scientific Research (A).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Nucleotide and deduced amino acid sequences of Hl4E-BP. M, a start codon metionine (DOCX 26 kb)
Fig. S2
Lipid storage observed by Oil red O staining in midguts and fat bodies of adult female ticks immediately after dsHl4E-BP injection. a Midgut; b fat bodies. The arrowheads and arrows indicate the midgut epithelial cells and the fat body trophocytes, respectively (DOCX 674 kb)
Table S1
‘Primers used in this study (DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Kume, A., Boldbaatar, D., Takazawa, Y. et al. RNAi of the translation inhibition gene 4E-BP identified from the hard tick, Haemaphysalis longicornis, affects lipid storage during the off-host starvation period of ticks. Parasitol Res 111, 889–896 (2012). https://doi.org/10.1007/s00436-012-2915-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-012-2915-9