Abstract
A new myxosporean species, Myxobolus supamattayai n. sp., was isolated from wild mullet (Valamugil seheli) from the Andaman Sea, Thailand and described based on its morphology and molecular data. The myxosporean produced black plasmodia-like unique clinical sign on the skin with sporogonic stages and mature spores. Polysporous plasmodia, up to 2.5 mm in diameter, were found in epithelium tissue in the scale pocket. The spores measured 6.6 (6.2–7.0) μm in length, 6.5 (6.2–6.7) μm in width, smooth, and round board to ellipsoidal in valvular view. Spores were enclosed with intracapsular process which represents 5–7 and 11–12 in amount revealed in light microscopy and ultrastructure, respectively. The polar capsules were pyriform and of equal size, measuring 3.5 (3.4–3.6) μm in length and 2.0 (1.9–2.2) μm in width, with four to five turns of polar filament arranged perpendicularly to longitudinal axis of the polar capsule. In conclusion, this new species is entirely different from those previously described; however, this finding was assured by the partial sequence of SSU rRNA gene (1,666 bp) analysis that differed from all known species of Myxobolus Bütschli, 1882. The phylogenetic tree of the sequence data sets including those of freshwater and marine of Myxobolus spp. and the sister group (Henneguya spp.) was constructed to establish the relationship of this new species in Myxobolus clade and to explore its relations between their sister groups. Phylogenetic analysis indicated that a monophyletic group with Myxobolus spp. which infected mullet represents the newly formed species. These results suggested the presumably nearby evolution prospecting of Myxobolus species that were found in the same host.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Myxobolus of the phylum Myxozoa contains more than 792 species; some are the most important parasitic pathogens of economic fish such as Myxobolus cerebrasis, causative of the whirling diseases in salmon. Most members of Myxobolus are histozoic parasite of freshwater fish; however, about 30 species live in marine or estuarine water. Some of them have coelozoic plasmodia of far less compact structure than histozoic trophozite (Lom and Dyková 2006). Although there are numerous detailed descriptions of species of the genus Myxobolus from teleost of nearly all geographical areas, information about this genus in Southeast Asia, mainly Myxobolus as a parasite in economic marine fish such as Asian seabass, grouper, and mullet, remains scarce. Mullet species is one of the most important economic fish of Southeast Asia, particularly in southern Thailand. Culture of mullet has been increased in Thailand in recent years, which is performed in cage–polyculture systems along with Asian seabass, grouper, and red snapper. In addition, wild-caught mullet has been decreasing while the demand is very high. Coinciding with the growing economic value of this fish, its increasing parasite loads are likely to affect the aquaculture industry.
The taxonomic methodology for identification of myxosporean species have been principally based on spore morphology for the primary classification of the species character until the acceptance of molecular data with reference to phylogenetic tree in taxonomic myxosporea (Liu et al. 2010). When determining the validity of the identification of similar morphological myxosporea, especially from identical tissue of the taxonomically closely related host species (Székely et al. 2009), these classical morphological methods are very difficult to use. Although host and tissue specificity are of great help in differentiating morphologically similar spores, there are some myxosporea, such as Myxobolus rotundus, that are classified from 27 cyprinid fish species and from different sites in the fish body (Eiras et al. 2005; Molnár et al. 2009; Zhang et al. 2010). However, recent reports have described a specific parasite of common bream using molecular data and the actinosporean stage development in oligochaete, Tubifex tubifex (Molnár et al. 2009; Székely et al. 2009). The wide host range and multiple infection sites are still yet to be explored. Unfortunately, spores of many species of the genus Myxobolus vary from each other; many species descriptions of this genus that have long been discovered are not definite and many are inconsistent with their documentation (Dyková et al. 2002). The combination of morphology and molecular-based classification, with the consideration of host range and tissue specificity, provides a precise approach to distinguish valid species from identified taxa (Molnár et al. 2009). A list of parasites of the wild mullet species has been introduced including 15 species of the genus Myxobolus (Bahri and Marques 1996; Kent et al. 2001; Buhri et al. 2003), which can be found in various organs of their host fish. Only Myxobolus episquamalis and Myxobolus exiguus were observed in the scales of mullet (Pulsford and Matthews 1982; Egusa et al. 1990). However, most of the mullet parasite, Myxobolus spp., were described based on their morphology while six species have been studied for both morphology and phylogeny (Buhri et al. 2003). The recent classification model, designed for gaining more sufficient and reliable resolution, requires morphological information along with molecular analysis. The morphological data have been obtained from the fine structures of the genus Myxobolus appearances followed from typical myxozoa pattern including shape of spore, polar capsule, polar filament turns, and ultrastructural studies. The SSU rRNA gene sequences, typically 18S rDNA, have indicated a proof of molecular classification (Fiala 2006).
In the present study, Myxobolus supamattayai n. sp., a new species belonging to the genus Myxobolus, found in wild mullet (Valamugil seheli) from the Andaman Sea, Thailand was described. Light microscopic, ultrastructure study and phylogenetic analysis using SSU rDNA were performed to determine the evolutionary relationships of this particular species among genus Myxobolus to assess the reliability of spore morphology-based classification.
Materials and methods
Fish samples and sampling
The samples collected in 2007 and 2008 were 143 specimens of V. seheli ranging in weight from 30 to 120 g from two different areas on the coast of the Andaman Sea in Satun province (station 1—6°50′33″ N, 99°46′19″ E; station 2—6°47′03″ N, 99°49′44″ E), Thailand. These fish samples were caught using floating seine and beach seine, which are fishing tools of an artisanal fishery. According to the previous work from our group, the black cyst scale is the hallmark of host infected with interesting Myxobolus species; thus, the mullets exhibiting this symptom were separated to the restrictive tank. The infected and uninfected fish with or without symptoms were transported alive in aerated seawater to the Aquatic Animal Health Research Center, Prince of Songkla University, Thailand immediately after collection. Moribund fish were killed by overdose of clove oil, packed in a plastic bag, and kept in temperature-controlled freezer boxes during the transportation. Standard procedures of myxosporean examination, internal organs, body surface, and body fluid, were used to diagnose myxosporean infection in the collected fishes.
Morphology analysis
Morphological measurement of species description was obtained among 50 spores according to Lom and Arthur (1989) and Lom and Dyková (2006). Fresh spores and Giemsa-stained spores were analyzed to identify their morphology and photographed. Species descriptions were based on several Myxobolus populations from more than one host specimen. Spores were observed under an Olympus AX 70 microscope and photographed using Olympus DP 71 cool CCD camera with differential interference contrast (DIC) optics.
For ultrastructure studies by electron microscopy, fresh plasmodia were dissected from infected fish skin and fixed in 3% glutaraldehyde in cacodylate buffer (pH 7.4) at 4°C for 6 h. Samples were post-fixed in 1% (w/v) osmium tetroxide using the same buffer. Transmission electron microscopy samples were dehydrated through graded acetone series before embedding in Epon-812, ultrathin sectioned, and stained with uranyl acetate and lead citrate. They were examined under a JEOL transmission electron microscopy (JEM-123) at 80 kV. The images were digitally recorded for further analysis.
Genomic DNA extraction
The DNA extraction method was modified from Salim and Desser (2000). Briefly, spores were centrifuged and pelleted at 8,000×g for 10 min, washed twice with PBS, and resuspended in 200 μl of 2% (v/v) proteinase K in STE buffer (pH 8.0) containing 1% SDS. Spores were incubated overnight at 30°C. The spore suspension was boiled for 10 min and then plunged into liquid nitrogen; the same process was repeated three times. The suspension was examined using light microscopy to verify whether most spores released their sporoplasms. The DNA was subsequently extracted with phenol/chloroform/IAA (25:24:1, v/v/v) method, incubated overnight, and precipitated with 70% ethanol. The DNA pellet was obtained after centrifugation at 12,000×g for 15 min at 4°C, followed by washing with 70% ethanol, and was air dried and resuspended with 20–30 μl of double distilled water. The DNA samples were kept at −20°C until use.
PCR amplification and purification of the 18S rDNA
The 18S rDNA of the Myxobolus parasite was amplified with MX 5 (5′ CTGCGGAC GGCTCAGTAAATCAGT 3′) and MX 3 (5′ CCAGGACATCTTAGGGCATCACAGA 3′), which bind to the conserved regions in the 5′ and 3′ end, respectively (Andree et al. 1998). The PCR was performed in a 50-μl reaction containing 2 μl of template DNA (100–150 ng of genomic DNA), 100 pmol of each of forward and reverse primer, 250 mM dNTP, 5 μl of 10× PCR buffer, and 1 μl of 5 U/μl Taq DNA polymerase (Invitrogen). Amplifications were conducted with an MJ Research DNA Engine 100 (MJ Research) using the following cycling protocol: an initial denaturation step of 95°C for 5 min, 40 cycles of 95°C/1 min–60°C/1 min–72°C/1 min, and a final extension of 10 min at 72°C. The PCR products were electrophoresed on 1.5% agarose gel (Vivantis) in TAE buffer for approximately 30 min at 100 V. The expected band of the 18S rDNA fragments of Myxobolus were directly cut from the gel and further purified with QIAquick Gel Extraction kit (Qiagen). The purified DNA was immediately used as inserted DNA in cloning system.
DNA cloning of PCR products and sequencing
The 18S rDNA gene of Myxobolus species was cloned into pGEM®-T Easy Vector using the pGEM®-T Easy Vector Systems cloning kit (Promega) and transformed into Escherichia coli TOP10 cells according to the manufacturer’s instructions. The plasmids were purified from positive clones and selected upon the blue/white color screening method using the QIAprep Spin Miniprep Kit (Qiagen). Inserted parasitic genes in the plasmid was confirmed before sequencing by PCR using either the same primers as those used in the original amplification (MX 5 and MX 3) or the primers that flanked the insertion site (T7 and SP6). The purified plasmids containing insert were sequenced on a CEQ™ 8000 automatic sequencer (Beckman Coulter) using CEQ DTCS Dye Kit (Beckman Coulter) according to the manufacturer’s protocol. In addition, the internal part of 18S rDNA was sequenced using the internal forward (5′ CCAGTAGCGTATCTCAAAGTTGC 3′) and reverse (5′ CCCGTGTTGAGTCAAAT TAAGC 3′) primers which were designed based on conserved sequences within the SSU rDNA gene of this species.
Phylogenetic analysis
The 18S rDNA sequence of M. supamattayai n. sp. was compared to those of other species available in the GenBank using nucleotide BLAST protocol (Altschul et al. 1990). Taxa included in the alignments were based on similarities from BLAST results including congeners of the novel species and representatives from each of the major clades reported in previous studies (Fiala 2006). The malacasporean Tetracapsuloides bryosalmonae was used as an out-group species in phylogenetic analyses. In addition, 35 selected taxa of platysporinid rDNA sequence from the genus Myxobolus and related species, mostly Henneguya spp., used in the analysis were restricted to the same length of the sequence of those isolated from hosts living in all habitats: freshwater, brackish, and marine. CLUSTAL X algorithm (Thompson et al. 1994) was used for initial sequence alignment with default setting for gap opening/extension penalties, and regions of ambiguous sequence alignments were manually edited using the BioEdit sequence alignment editor (Hall 1999). We used neighbor-joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) methods to infer phylogenetic relationships among the Myxobolus spp. and its sister group. There were a total of 2,220 positions in the final dataset. Phylogenetic analyses of the NJ method and MP method were conducted using the MEGA V. 4.0 program (Kumar et al. 1993). For the NJ method, evolutionary distances were analyzed using the Kimura two-parameter (K2P) evolution sequence model (Kimura 1983). The evolutionary history was concluded using the NJ method. Statistical consensus trees inferred from 1,000 replications bootstrapping were taken to present the evolutionary history of the taxa analyzed. The MP tree was obtained using the Close-Neighbor-Interchange algorithm (Nei and Gojobori 2000) with search level 3 in which the initial trees were obtained with the random addition of sequence (100 replications). Parsimonious trees were shown with the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates). All alignment gaps/missing data were set as the complete deletion in both analysis methods. We used MEGA 4 for these analyses including the primary tree-based drawing of NJ tree.
The SeaView V 4 program (Gouy et al. 2010) was executed using the GTR model for ML analysis. The selecting models were evaluated by using jModelTest (Posada 2008). The BioNJ (Gascuel 1997) algorithm was selected for starting trees. Nearest-neighbor interchange and subtree pruning and regrafting were used for tree searching operation. The data sets were built with 500 bootstrap replications for the ML reconstruction.
Results
Morphology and clinical signs
Several cyst-like plasmodia, with diameter up to 2.5 mm, containing numerous sporogonic stages and mature spores were observed in the epithelium tissue above the scale. Twenty five of 143 specimens (17.48%) were infected with parasites classified, based on the spore morphology, as a myxosporean belonging to the genus Myxobolus Bütschli, 1882 (Lom and Dyková 2006). All characteristics of the genus Myxobolus were observed in the spores’ stage of these parasites found in macroscopical lesions localized in the epithelium scales pocket. Black cystic formations as parasite plasmodia were detected only in the target organ while no other tissues or organs contained visible parasites revealed by light microscopy. The majority of the parasitized fish showed more than 100 cyst-like structures in different locations, mostly in the pectoral to anal fin below the lateral line body area (Fig. 1a).
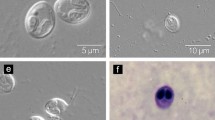
Myxobolus supamattayai n. sp. in mullet (Valamugil seheli) from the coast of the Andaman Sea. a Cysts of M. supamattayai n. sp. on the infecting mullet scales (arrows). b Pathology of M. supamattayai n. sp. on the scales (arrow). c Fresh mature spore (DIC photography). d Spore in Giemsa staining. e Fresh spore with ejected polar filament. All scale bars are 10 micrometers
Myxobolus supamattayai n. sp.
Vegetative stages
More developed plasmodia contained late sporogonic stages and mostly mature spores. Black, polysporous spot to ellipsoidal-shaped histozotic cyst-like plasmodia measuring up to 2.5 mm in diameter were found in the epithelial tissue above the scales. The scales were destroyed as a plasmodium hole pocket (Fig. 1b). Missing scale and small ulcerated patches on the skin were occasionally found particularly in severely infected fish. Numerous plasmodia could be found in each infected fish; however, only one plasmodium per scale could be observed. In addition, this was not found in other organs.
Morphology of spores (based on 50 mature spores)
Spores with typical character of the genus Myxobolus were observed. The mature spores were round board to ellipsoidal in frontal view. Most of them are round with several edge markings (normally 5–7), whereas a few are variable in shape. Spore size was 6.6 (6.2–7.0) μm in length and 6.5 (6.2–6.7) μm in width. The two spherical polar capsules were equal in size and close together in the plane of sutural line at the apical end which measured 3.5 (3.4–3.6) μm in length and 2.0 (1.9–2.2) μm in width. Polar filaments formed four to five coils opening at the spores anterior nearby the sutural line. The coil arrangement was almost perpendicular to the sutural line. Polar filaments extruding from mature spores of fresh material were evaluated as 10.5 (10–11) μm in length (Fig. 1d). A small intercapsular appendix was located between the polar capsules at the anterior end of the spore and discernible vacuole-like body in the sporoplasm of the spore. Spore morphology observations under light microscopy and Giemsa staining are shown in Fig. 1c and e, respectively..
Type host
Blue spot mullet (Valamugil seheli)
Type locality
The coast of the Andaman Sea, Satun province, Thailand (6°47′03.68′′ N, 99°49′44.60′′ E)
Site of infection
Epithelial tissue located in the center of the scale
Prevalence
Up to 17.48% (25/143)
Type material
Slides with stained spore (syntype) and formalin-preserved infected host fish (V. seheli) have been deposited in the collection of the Princess Maha Chakri Sirindhorn Natural History Museum, Prince of Songkla University, Songkhla, Thailand (accession number PSUZC-20090216.01).
Etymology
The species name (M. supamattayai) is in homage to Assoc. Prof. Dr. Kidchakan Supamattaya, one of the pioneers in the study of aquatic animal disease in Thailand and the first director of this project (myxosporidia disease and biodiversity of parasite in economic marine fish of Thailand), who passed away in 2008.
Electron microscopy
Fine structure of mature spores presented the complete developing cells comprising two shell valves, mono-nucleated sporoplasm, and two polar capsules (Fig. 2a, b). The spore body was formed by two smooth shell valves, each one with two caudal projections for releasing the polar filament. The internal sutural line, the overlap of two spore valves, contained 11–12 intracapsular processes. The two equal polar capsules were pyriform, elongated, and a little converged at the apex of the spore. The polar capsules with a polar filament, basal straight central shaft, and coils of four to five turns were visible. Polar capsules were composed of thin electron-dense external wall along with thick and lighter inner wall. The apical channel for the polar filament discharge showed close contact with caudal projection in valves. The sporoplasmic cell was located in the posterior pole of the spore and the cytoplasm contained three distinct organelles; mononuclei were close to each other, low density electron-dense body, like a cell vacuole and sporoplasmosomes. These spherical-shaped vesicles were membrane bounded and contained an electron-dense and homogenous matrix. The nucleus was found in the sporoplasm cell, whereas other cells in mature spore could not show the basis of complete cell structure. A semi-schematic illustration of the spore, based on TEM observations, is shown in Fig. 3.
Transmission electron micrographs of the mature spore of Myxobolus supamattayai n. sp. a The detail of spore longitudinal ultrathin section showing the valves (V) with internal capsular process (arrow), the two polar capsules (P) and sporoplasm (Sp) with one nucleus and low density electron dense body (LB), like a vacuole. b The detail of spore sectioned transversally showing the valves (V) and sutural line (arrow), two polar capsules (P) with four to five polar filament turns
Semi-schematic illustration of a longitudinal view of a mature spore of the Myxobolus supamattayai n. sp., a parasite of blue spot mullet Valamugil seheli showing species-specific characters including spore size and shape, two equal polar capsules with four to five polar filament turns, uninucleated sporoplasm, vacuole with sporoplasmosome, and 11–12 intercapsular processes in sutural line
Molecular characterization and phylogenetic tree
Overall SSU rDNA sequences were determined for 12 samples (six samples from each sampling site). All the sequence samples showed 99.9% homology to the rDNA nucleotide sequences. The amplified and sequenced 18S rDNA gene of M. supamattayai n. sp. was 1,617 nucleotides in length excluding the region corresponding to MX 5–MX 3 primers while the G + C content was 43.0%. The sequence was deposited at the National Center for Biotechnology GenBank (accession number HQ166720). The 18S rDNA gene of this Myxobolus species isolated from mullets revealed the sequence identity with other parasite species in the same host: 88% with Myxobolus muelleri, 90% with M. episquamalis, 91% with Myxobolus bizerti, 88% with M. exiguus, 87% with Myxobolus ichkeulensis, and 88% with Myxobolus spinacurvatura. The nucleotide sequences of M. supamattayai n. sp. obviously differed, more than 25% of 1,666 base pairs, from those of all other Myxobolus species available in the GenBank.
Phylogenetic analysis was conducted using the NJ algorithm resulting in a single most parsimonious tree (Fig. 4), while the member species in each clade of the MP tree and ML tree remained the same. Some branches were swapped within clades, but this did not significantly change the apparent evolutionary relationship. The clades of Myxobolus species were divided into three main clades that were strongly supported by bootstrap values. However, the demise of Myxobolus clades distinctly interferes with the clade of Henneguya species. All the phylogenetic topology analysis trees demonstrated that the mullet parasite Myxobolus species containing seven species (M. supamattayai n. sp., M. muelleri, M. episquamalis, M. bizerti, M. exiguus, M. ichkeulensis, and M. spinacurvatura) correspond to a monophyletic group (clade A). This is separated into a distinct clade strongly supported by high bootstrap values. The representation of marine Myxobolus clade, infected mullet fish host, partitioned within the clade formed by the Henneguya species which was separated from other Myxobolus clades (B and C) was composed of 18 species which are involved in freshwater fish parasites.
Phylogenetic tree generated by neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) analyses of the 18S rDNA sequence of myxosporeans, root at Tetracapsuloides bryosalmonae. Numbers at nodes indicate bootstrap confidence levels presented as percentage for NJ, ML, and MP, respectively (NJ and MP = 1,000 repetitions and ML = 500 repetitions). GenBank accession numbers of each species are given in parentheses. The distance scale is shown beside the tree. Myxosporean identified in the present study is given in gray background. The dashed lines indicate the differences between the three trees, taking NJ as basic topology
Discussion
Morphological and ultrastructural taxonomic classification
The light morphology of the spores described in the present work corresponds to those of the genus Myxobolus, myxozoa parasite. Myxozoa species are very common parasites of fish. Many researchers demonstrated that more than one species of myxosporea parasite infect the same host fish. Although few studies on host specificity are reported, recent data indicated that most species are strictly host specific or only capable of developing in closely related fish host (Tajdari et al. 2005). According to previously described species, 15 species belonging to the Myxobolus genus have been demonstrated to infect mullet (Bahri and Marques 1996). Myxobolus species infecting mullet have been reported on various geological regions including the west coast of Tunisia, Mediterranean coast, in the Atlantic at Ria de Aveiro in northern Portugal, South Florida coast, Australia, and the Japanese coast (Maeno et al. 1990; Rothwell et al. 1997; Diamanka et al. 2008; Umur et al. 2010). All characteristics of this new species were also compared to confirm morphological similarities to the spores of different species of the genus Myxobolus described previously (see Table 1). Two species, M. episquamalis and M. exiguus, occurring on the host scales, which are the parasites infecting the epithelial tissue over the scales of species, are similar to that of M. supamattayai n. sp. However, M. episquamalis differ from our newfound species because of the less elongated shape of the spore and polar capsule, smaller overall dimensions, and the type and color of the plasmodia. Conversely, M. exiguus has a bigger spore size and longer polar filament length than those of M. supamattayai n. sp. Although spore shape of Myxobolus parvus (Bahri and Marques 1996) showed higher similarity to the spore of M. supamattayai n. sp., the first one has a bigger polar capsule. M. bizerti and M. ichkeulensis have the larger spore and polar capsule, more polar filament coils in the polar capsule, and longer polar filament length indeed than those observed from M. supamattayai n. sp. The shape of spore and polar capsule of M. spinacurvatura is larger than that found in M. supamattayai n. sp. even if it also occurs in the epithelial tissue over the scale (Lom and Dyková 1994). The ellipsoidal spores of Myxobolus branchialis have a longer space and in overall dimensions besides the polar capsule are bigger. Myxobolus cheni occurs in muscles and fins; in addition, the shape of the spores is larger and more ellipsoid. Myxobolus achmarovi, Myxobolus cephalus, and Myxobolus rohdei occur in the mesentery, gill filament, and kidney, respectively, and all these species have a larger spore and polar capsule than those of M. supamattayai n. sp. The morphological data of spores of Myxobolus mugcephalus parasitizing in gill arches was insufficient for comparison; however, it can be discriminated from M. supamattayai n. sp. by its smaller and more round board spore shape. M. muelleri, a high variable morphology detailed parasite, can be separated from M. supamattayai n. sp. by a number of considerable features. Spores seem much more oval than those of M. supamattayai n. sp., and a bigger polar capsule and six to eight coil turns to the polar filament in M. muelleri distinguish it from our species, which contains only four to five coils.
Moreover, the ultrastructural morphology of the spores described in the present study also confirms that this parasite belongs to the genus Myxobolus Bütschli, 1882 according to previous studies and is the key for the determination of myxosporean genera (Lom and Dyková 2006). The ultrastructural data of mature spore of Myxobolus species infecting mullet remains scarce. However, fine structure of M. exiguus and M. episquamalis infecting the scales of grey mullet and flathead mullet has been described (Pulsford and Matthews 1982; Bahri and Marques 1996). M. supamattayai n. sp. is fundamentally similar to both Myxobolus species mentioned above which infected mullet and is similar throughout other myxosporeans, although there is a minor difference observed in size and structure. The presented mature spores and polar capsule were plugged with a stopper, which was covered by a cap-like structure at its apical end. These characters were similar to those recorded for the genus Myxobolus (Ali et al. 2007) and also from other myxosporean genera such as Sphaerospora (Supamattaya et al. 1991) and Zschokkella (Lom and Dyková 1996). The sporoplasm cell of genus Myxobolus is commonly characterized by a single binucleated sporoplasm (Pulsford and Matthews 1982; Bahri and Marques 1996). In contrast, this presented species showed only one nucleus in the mature spore of sporoplasm cell. However, uninucleated sporoplasm has been reported in some Myxobolus species such as Myxobolus stomum (Ali et al. 2003). Dense spherical inclusions and sporoplasmosomes were identified in the sporoplasm of the present species, which seem to be the common features of Myxobolus species and other myxosporeans (Abdel-Ghaffar et al. 2008).
Phylogenetic tree analysis by SSU rDNA approach
Species of the genus Myxobolus have been found in several clades over a broad analysis of the Myxozoa (Kent et al. 2001). Modern phylogenetic analysis classifications of myxosporeans have been described into two major clades: freshwater and marine species (Fiala 2006). SSU rDNA is widely used for studying phylogenetic relationships among taxa (Avise 2004). Moreover, it is the only molecular marker available for the broad range of Myxobolus species at present. Therefore, SSU rDNA sequences have been used as the main information source for reconstruction of Myxobolus evolutionary history. Obtained discrepancies between SSU rDNA gene phylogeny and taxonomy, based on the morphological data, may suggest that the molecular phylogeny is not congruent with the true phylogeny or may indicate that the characters of spore morphology are homoplasious. Consequently, molecular data of other genes, for instance, large subunit ribosomal DNA (LSU rDNA) or intergenic transcribed spacer (ITS), is required to confirm the myxosporean rDNA phylogeny (Fiala 2006). However, phylogenetic analyses of SSU rDNA suggest that pore morphology is of minor importance in phylogenetic relationships of myxosporean clades (Andree et al. 1999; Canning and Okamura 2004; Eszterbauer 2004; Holzer et al. 2004). Since different spore morphologies occur in the same clade even when there is another similar species in the other clade due to convergence, it seems that Myxobolus evolution may not depend on the change of spore shape. Moreover, separation of the minor clade of marine Myxobolus spp., infecting mullet, from other Myxobolus species in the Myxobolus clade was established in the present study.
Our comparative molecular genetic analysis included sequences from 36 species of expanded myxosporeans to the extent of SSU rDNA sequence availability. The intention of this analysis was to verify the differentiation of M. supamattayai n. sp., first from infected blue spot mullet Myxobolus species followed by the possibility in its closest relatives. Phylogenetic analyses formed one large clade that includes one of the out-group species, Tetracapsuloides bryosalmonae. This large clade was further divided into more sub-clades, one of which represented all of the mullet-infecting Myxobolus species, as marine myxozoa, including M. episquamalis, M. muelleri, M. spinacurvatura, M. ichkeulensis, M. bizerti, M. exiguus, and M. supamattayai n. sp.
In summary, M. supamattayai n. sp. does not correlate with any other Myxobolus spp. previously reported to infect mullet or another species among the genus Myxobolus. It tends to be differentiated from all known species of Myxobolus spp. by the presence of morphology, ultrastructure, and phylogenetic analysis.
References
Abdel-Ghaffar F, El-Toukhy A, Al-Quraishy S, Al-Rasheid K, Abdel-Baki AS, Hegazy A, Bashter AR (2008) Five new myxosporean species (Myxozoa: Myxosporea) infecting the Nile tilapia Oreochromis niloticus in Bahr Shebin, Nile Tributary, Nile Delta, Egypt. Parasitol Res 103:1197–1205
Ali MA, Abdel-Baki AS, Sakran T, Entzeroth Abdel-Ghaffae F (2003) Light and electron microscopic studies of Myxobolus stomum n. sp. (Myxosporea: Myxosporea) infecting freshwater fishes of the River Nile, Egypt. Parasitol Res 88:9–15
Ali MA, Abdel-Baki AS, Sahran TH, Entzeroth R, Abdel-Ghaffae F (2007) Myxobolus lubati n. sp. (Myxosporea: Myxobolodae), a new parasite of Haffara seabream Rhabdosargus haffara (Forsskal, 1775), Red Sea, Egypt: a light and transmission electron microscopy. Parasitol Res 100:819–827
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tools. J Mol Biol 215:403–410
Andree KB, MacConnell E, Hedrick RP (1998) A nested polymerase chain reaction for the detection of genomic DNA of Myxobolus cerebralis in rainbow trout Oncorhynchus mykiss. Dis Aquat Organ 34:145–154
Andree KB, Szekely C, Molnár K, Gresoviac SJ, Hedrick RP (1999) Relationships among members of the genus Myxobolus (Myxozoa: Bilvalvulidae) based on small subunit ribosomal DNA sequences. J Parasitol 85:68–74
Avise JC (2004) Molecular marker, natural history, and evolution. Sinauer, Sunderland
Bahri S, Marques A (1996) Myxosporean parasites of the genus Myxobolus from Mugil cephalus in Ichkeul lagoon, Tunisia: description of two new species. Dis Aquat Organ 27:115–122
Buhri S, Andree KB, Hedrick RP (2003) Morphological and phylogenetic studies of marine Myxobolus spp. from mullet in Ichkeul Lake, Tunisia. J Eukaryot Microbiol 50:463–470
Canning EU, Okamura B (2004) Biodiversity and evolution of the Myxozoa. Adv Parasitol 56:44–131
Diamanka A, Fall M, Diebakate C, Faye N, Toguebaye BS (2008) Identification of Myxobolus episquamalis (Myxozoa, Myxobolidae) in flathead mullet Mugill cephalus (Pisces, Teleostei, Mugilidae) from the coast of Senegal (eastern tropical Atlantic Ocean). Acta Adriat 49:19–23
Dyková I, Fiala I, Nie P (2002) Myxobolus lentisuturalis sp. n. (Myxozoa: Myxobolidae), a new muscle-infecting species of the Prussian carp, Carassius gibelio from China. Folia Parasitol 49:253–258
Egusa S, Maeno Y, Sorimach M (1990) A new species of Myxozoa, Myxobolus episquamalis sp. n. infecting the scales of the mullet, Mugil cephalus L. Fish Pathol 25:87–91
Eiras JC, D’Souza J (2004) Myxobolus goensis n. sp. (Myxozoa, Myxosporea, Myxobolidae), a parasite of the gills of Mugil cephalus (Osteichthyes, Mugilidae) from Goa. India Parasite 11:243–248
Eiras JC, Molnár K, Lu YS (2005) Synopsis of the species of Myxobolus Butschli, 1882 (Myxozoa: Myxosporea: Myxobolidae). Syst Parasitol 61:1–46
Eiras JC, Abreu PC, Robaldo R, Júnior J (2007) Myxobolus platanus n. sp. (Myxosporea, Myxobolidae), a parasites of Mugil planus Günther, 1880 (Osteichthyes, Mugilidae) form Lagoa dos Patos, RS, Brazil. Arq Bras Med Vet Zootec 59:895–898
Eszterbauer E (2004) Genetic relationships among gill-infecting Myxobolus species (Myxosporea) of cyprinids: molecular evidence of importance of tissue specificity. Dis Aquat Org 58:35–40
Fiala I (2006) The phylogeny of Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. Int J Parasitol 36:1521–1534
Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14:685–695
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Holzer AS, Sommerville C, Wootten R (2004) Molecular relationships and phylogeny in a community of myxosporeans based on their 18S rDNA sequence. Int J Parasitol 34:1099–1111
Kent ML, Andree KB, Bartholomew JB, El-Matbouli M, Desser SS, Devlin RH, Feist SW, Hedrick RP, Hoffman RW, Khattra J, Hallett SL, Lester JG, Longshaw M, Palenzuela O, Siddall ME, Xiao C (2001) Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol 48:395–413
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kumar S, Tamura K, Nei M (1993) MEGA: molecular evolutionary genetics analysis. Pennsylvania State University Press, University Park
Lansberg JH, Lom J (1991) Taxonomy of the genera of the Myxobolus/Myxosoma group (Myxobolidae: Myxosporea), current listing of species and revision of synonyms. Syst Parasitol 18:165–186
Liu Y, Gu ZM, Luo LY (2010) Some additional data to the occurrence, morphology and validity of Myxobolus turpisrotundus Zhang, 2009 (Myxozoa: Myxosporea). Parasitol Res 107:67–73
Lom J, Arthur JR (1989) A guideline for the preparation of species descriptions in Myxosporea. J Fish Dis 12:151–156
Lom J, Dyková I (1994) Studies on protozoan parasites of Australian fishes III. Species of genus Myxobolus Bütschli, 1882. Eur J Protistol 30:431–439
Lom J, Dyková I (1996) Notes on the ultrastructure of two myxosporean (Myxozoa) species, Zschokkella pleomorpha and Ortholinea fluviatilis. Folia Parasitol 43:189–202
Lom J, Dyková I (2006) Myxozoan genera: definition and notes on taxonomy, life-cycle terminology and pathologenic species. Folia Parasitol 53:1–36
Maeno Y, Sorimach M, Ogawa K, Egusa S (1990) Myxobolus spinacarvatura n. sp. (Myxosporea: Bivalvulida) parasitic in deformed mullet, Mugil cephalus. Fish Pathol 25:37–41
Molnár K, Székely C, Hallett SL, Atkinson SD (2009) Some remarks on the occurrence, host-specificity and validity of Myxobolus rotundus Nemeczek, 1911 (Myxozoa: Myxosporea). Syst Parasitol 72:71–79
Nei M, Gojobori T (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Pulsford A, Matthews RA (1982) An ultrastructure study of Myxobolus exiguous Thelohan, 1895 (Myxosporea) from grey mullet, Creninguil labrosus (Risso). J Fish Dis 5:509–526
Rothwell JT, Virgonia JL, Callinan RB, Nicholls PJ, Langdon JS (1997) Occurrence of cutaneous infections of Myxobolus episquamalis (Myxozoa: Myxobolidae) in sea mullet, Mugill cephalus L, in Australia. Aust Vet J 75:349–352
Salim KY, Desser SS (2000) Description and phylogenetic systematics of Myxobolus spp. from cyprinids in Algonquin Park, Ontario. J Eukaryot Microbiol 47:309–318
Supamattaya K, Fischer-Scherl T, Hoffmann RW, Boonyaratpalin S (1991) Sphaerospora epinepheli n. sp. (Myxosporea: Sphaerosporidae) observed in grouper (Epinephelus malabaricus). J Eukaryot Microbiol 5:448–454
Székely C, Hallett SL, Atkinson SD, Molnár K (2009) Complete life cycle of Myxobolus rotundus (Myxosporea: Myxobolidae), a gill myxozoan of common bream Abramis brama. Dis Aquat Organ 85:147–155
Tajdari J, Matos E, Mendonça I, Azevedo C (2005) Ultrastructural morphology of Myxobolus testicularis sp. n., parasite of the testis of Hemiodopsis microlepis (Teleostei: Hemiodontidae) from the NE of Brazil. Acta Protozool 44:377–384
Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Umur Ş, Pekmezci GZ, Beyhan YE, Gürler AT, Açici M (2010) First record of Myxobolus muelleri (Myxosporea: Myxobolidae) in flathead grey mullet Mugil cephalus (Teleistei, Mugilidae) from Turkey. Ankara Üniv Vet Fak Derg 57:205–207
Zhang JY, Wang JG, Li AH, Gong XN (2010) Infection of Myxobolus turpisrotundus sp. n. in allogynogenetic gibel carp, Carassius auratus gibelio (Bloch), with revision of Myxobolus rotundus (s.l.) Nemeczek reported from C. auratus auratus (L.). J Fish Dis 33:625–638
Acknowledgments
This study was supported by the Thailand Research Fund through The Royal Golden Jubilee Ph.D. Program (RGJ-PHD) to Mr. Kittichon U-taynapun, the Center of Excellence on Agricultural Biotechnology, Science and Technology Postgraduate Education and Research Development Office, Commission on Higher Education, Ministry of Education (AG-BIO/PERDO-CHE). We would like to express our gratitude and sincere thanks to Dr. Raja Sudhakaran for revision the manuscript and the anonymous reviewers for their most valuable comments and suggestions. We thank the staff at Aquatic Animal Health Research Center, Department of Aquatic Science, Faculty of Natural Resources, Prince of Songkla University, Thailand as well as those at the Department of Biological Production and Environmental Science, Faculty of Agriculture, Miyazaki University, Japan for their kind help.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
U-taynapun, K., Penprapai, N., Bangrak, P. et al. Myxobolus supamattayai n. sp. (Myxosporea: Myxobolidae) from Thailand parasitizing the scale pellicle of wild mullet (Valamugil seheli). Parasitol Res 109, 81–91 (2011). https://doi.org/10.1007/s00436-010-2223-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-010-2223-1