Abstract
A new species of nematode, Contracaecum pyripapillatum, is reported from the Australian pelican, Pelecanus conspicillatus. This species resembles Contracaecum multipapillatum which was found in the same host. These two species can be differentiated from one another based on the shape of the preanal papillae, being pyriform in C. pyripapillatum and circular in C. multipapillatum. Genetically, the two species differ in the sequences of first and second internal transcribed spacers (ITS-1 and ITS-2, respectively) of ribosomal DNA (rDNA). C. pyripapillatum and C. multipapillatum differed in the ITS-1 (443bp in both species) and ITS-2 (between 231 and 233bp) sequences by 3.4–3.8% and 6.0%, respectively. Based on previous allozyme and mtDNA datasets, genotypes of C. multipapillatum A, B, and C have been reported from Europe and USA. Therefore, C. multipapillatum from Australia has been designated as C. multipapillatum D. A morphological examination of these genotypes is necessary to determine whether they represent distinct species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Contracaecum multipapillatum was first described by Drasche (1882) as Ascaris multipapillata. Lucker (1941) redescribed the species and distinguished C. multipapillatum from the specimens misidentified as C. multipapillatum by Skrjabin (1916), naming the latter species C. ainterlabium. For many years, the species has been known as a parasite of stomachs of fish-eating birds, such as storks, pelicans, cormorants, darters and herons from north and south America (Lucker 1941; Courtney and Forrester 1974; Vidal-Martinez et al. 1994; Sepulveda et al. 1994, 1999; Navone et al. 2000; Kinsella et al. 2004). Larval stages of the species were reported from freshwater fishes and mullet (Huizinga 1967; Deardorff and Overstreet 1980). There was one report of infection by larval and adult stages of C. multipapillatum in a softshell turtle (Apalone ferox) in Florida (Foster et al. 1998). However, based on the reports of larvae from fishes, Deardorff and Overstreet (1980) stated that the species may extend to the Middle and Far East. In fact, some species reported subsequently are morphologically very similar to C. multipapillatum. For example, C. mexicanum from Pelecanus occidentalis in Mexico (Flores Barroeta 1957), C. philomultipapillatum from Bubulcus ibis ibis, Egretta thula thula and Egretta alba egretta in Argentina (Labriola and Suriano 1996) and Contracaecum bubakii from Phalacrocorax niger in Pakistan (Akram 1996) are morphologically very similar to C. multipapillatum and characters used to differentiate between species such as the number of precloacal or post-cloacal papillae are not of interspecific value (Labriola and Suriano 1996).
Recently, it has been shown that C. multipapillatum is a complex comprising at least three distinct species with distinct geographical distributions in Europe and North America (Nadler et al. 2000; Mattiucci et al. 2006; D’Amelio et al. 2007). However, these studies were based on molecular data alone and did not include any morphological or morphometric data. In their study of the phylogenetic relationships among selected species of ascaridoid nematodes based on allozyme analyses, Nadler et al. (2000) reported C. multipapillatum from Greece. Later, in another study based on the molecular data, D’Amelio et al. (2007) found that C. multipapillatum from the Nearctic region was genetically different from C. multipapillatum in Greece. Based on allozymes differences and mitochondrial oxidase cytochrome c oxidase subunit 2 gene sequence data, Mattiucci et al. (2006) suggested that C. multipapillatum comprised at least three genetic groups (designated A, B and C) from the Mediterranean area and the USA. Although these molecular analyses provided valuable information about the genetic identity of species from different continents, a detailed analysis of morphological differentiation within the species complex is required for nomenclatural designation (D’Amelio et al. 2007).
In the present study, specimens initially attributed to C. multipapillatum sensu lato, infecting the Australian pelican, Pelecanus conspicillatus, were found to comprise two distinct species. The aim was to describe these species and characterise them genetically based on the sequences of the first and second internal transcribed spacers of nuclear ribosomal DNA (ITS-1 and ITS-2, respectively) which are known to provide reliable genetic markers for the specific identification of anisakid nematodes (Zhu et al. 2000, 2001).
Materials and methods
Parasites
Adult nematodes were collected from Australian pelicans, P. conspicillatus, from Victoria, Australia. Nematodes were washed extensively in physiological saline. A small piece of the mid-body of each individual nematode was removed with a scalpel for molecular study; the rest of the nematode was fixed in Berland’s fluid for 24h and then cleared in lactophenol for morphological examination. Some specimens were fixed in 70% ethanol for scanning electron microscopy (SEM).
Morphological examination
All specimens were studied morphologically by light microscopy. Drawings were made with the aid of a drawing tube and measurements were made directly with an eyepiece micrometre. Adult nematodes were identified to species based on the available keys and descriptions (Hartwich 1964, 1974). All measurements are given in millimetres, unless stated otherwise. Mean measurements are given, followed by the range in parentheses. Specimens have been deposited in South Australian Museum, Adelaide (SAM).
Scanning electron micrographs were obtained from anterior and posterior ends, dehydrated in ethanol, placed in analytical grade ethanol for at least 1h and then transferred into hexamethyldisilazane–ethanol solutions in which the amount of hexamethyldisilazane was gradually raised to 100%. After evaporation of the hexamethyldisilazane, specimens were placed on a stub, coated with gold and examined in a Philips 505 scanning electron microscope, at an accelerating voltage of 9.9–19.6kV and the images were captured digitally.
Molecular analyses
Genomic DNA from nematodes was isolated by sodium dodecyl sulphate–proteinase K treatment, column-purified (Wizard™ DNA Clean-Up, Promega) and eluted into 30µl of H2O. The ITS-1 was amplified by PCR with oligonucleotide primers SS1 (forward; 5'-GTTTCCGTAGGTGAACCTGCG-3') and NC13R (reverse; 5'-GCTGCGTTCTTCATCGAT-3') and the ITS-2 with oligonucleotide primers SS2 (forward; 5'-TTGCAGACACATTGAGCACT-3') and NC2 (reverse; 5'- TTAGTTTCTTTTCCTCCGCT -3'). The PCR (in 50µl) was performed in 10mM Tris–HCl, pH 8.4, 50mM KCl, 3mM MgCl2, 250µM each of dNTP, 50pmol of each primer and 1U Taq polymerase (Promega) in a thermocycler (Biometra) under the following cycling conditions: 94°C for 5min (initial denaturation), followed by 30 cycles of 94°C, 30s (denaturation), 55°C, 30s (annealing), 72°C, 30s (extension) and a final extension of 72°C for 5min. One microlitre (10–20ng) of genomic DNA was added to each PCR reaction. Samples with pelican DNA or without DNA were included in the PCR as negative controls; no amplicons were produced in the PCR from these negative control samples. An aliquot (5µl) of each amplicon was examined on a 1.5% w/v agarose gel, stained with ethidium bromide and photographed.
Single-strand conformation polymorphism (SSCP) analysis of amplicons was conducted to screen for nucleotide variation among samples. Five microlitre of individual amplicons were diluted in 5µl of H2O and mixed with 10-µl loading buffer (10mM NaOH, 95% formamide, 0.05% of both bromophenol blue and xylene cyanole). After denaturation at 96°C for 15min and snap cooling on a freeze block (−20°C), individual samples (12µl) were loaded into the wells of precast GMA™ S-2 × 25 gels (96 × 261mm; product no. 3548, Elchrom Scientific) and subjected to electrophoresis for 18 and 16h for ITS-1 and ITS-2 amplicons, respectively, at 72V and 7.2°C (constant) in a horizontal SEA2000™ apparatus (Elchrom Scientific) connected to a MultiTemp III (Pharmacia) cooling system. Following electrophoresis, gels were stained for 30min with ethidium bromide (0.5µg/ml), destained in H2O for at least 1h and then photographed (using 667 film, Polaroid) upon ultraviolet transillumination.
Representative samples were selected based on their SSCP profile as well as from individual host and sequenced. Amplicons were purified over mini-columns (Wizard™ PCR Prep, Promega, WI, USA), eluted in 30µl H2O and then subjected to automated sequencing (BigDye® chemistry, Applied Biosystems), in both directions, using the same primers as for the primary PCR. Sequences were aligned using the computer programme ClustalX (Chilton et al. 1995) and then adjusted manually. Polymorphic positions were designated using International Union of Pure and Applied Chemistry codes. Sequence differences (based on pairwise comparisons) were calculated using the formula D = 1 − (M / L; Chilton et al. 1995).
Results
Nematodes from Australian pelicans were initially identified as C. multipapillatum. However, a detailed morphological examination showed that males had either pyriform or circular preanal papilla, suggesting that they might belong to a distinct species. No morphological differences were found among females. Alignment of ITS-1 and ITS-2 sequences showed that these nematodes comprised two distinct genotypes, which along with the morphological difference, suggested the existence of two distinct species, one not previously described and named here Contracaecum pyripapillatum.
Morphology
C. pyripapillatum n. sp.
Description
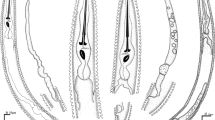
Labia large and fleshy, with interposed interlabia (Figs. 1a,b and 2a). Two large cephalic papillae on dorsal labium, one papilla on each subventral labium. Cephalic papillae located at mid-level of labium. Labia expanded antero-laterally. Margins of antero-lateral aspects of labia mildly rounded, without flanges. Slight depression visible anteriorly as well as on lateral aspects of each labium. Interlabia simple, triangular, wider at base. Interlabia approximately same height as labia. Excretory pore at base of lips. Collar prominent, interrupted laterally at base of lips, separated from body by prominent indention. Intestinal caecum three to four times longer than ventricular appendix (Fig. 1c).
C. pyripapillatum from P. conspicillatus: a dorsal labium (scale bar = 0.17 mm); b ventral interlabium with subventral labia (scale bar = 0.17 mm); c anterior end of nematode showing oesophagus, ventricular appendix and intestinal caecum (scale bar = 0.81 mm); d posterior end in male, ventral view, showing post-cloacal papillae (scale bar = 0.08 mm)
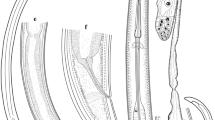
Scanning electron micrographs of C. pyripapillatum n. sp. (a to c) and C. multipapillatum D (d and e): a and e dorsal views of anterior end of nematodes showing dorsal labium with interlabia. Note simple tip of interlabia (scale bar = 0.1 mm); b pyriform precloacal papillae (scale bar = 0.05 mm); c tip of spicule (scale bar = 0.05 mm); d lateral view of anterior end of nematode showing subventral labium and two adjacent interlabia with simple tips. Arrow indicates lateral interruption in annulations of collar (scale bar = 0.1 mm)
Males (n = 4). Body length 29.1 (21.0–34.1), maximum width 0.94 (0.75–1.04). Diameter across lips 0.24 (0.18–0.27). Nerve ring 0.51 (0.43–0.59) from anterior end. Deirids at approximately same level as nerve ring, 0.52 (0.41–0.62) from anterior end. Oesophagus 5.23 (3.74–6.26) long, 18% (15–21%) of body length. Ventricular appendix 1.30 (1.11–1.43), 25% (22–30%) of oesophageal length. Intestinal caecum 4.43 (3.12–5.41) long, 85% (82–86%) of oesophageal length, 3.37 (2.82–3.92) times length of ventricular appendix. Spicules short, with small groove at tip, visible in scanning electron micrographs (Fig. 2c). Right spicule 2.12 (1.82–2.36) long, 8% (7–9%) of body length. Left spicule 2.06 (1.79–2.24) long, 8% (7–9%) body length. Post-cloacal papillae (Figs. 1d and 2b) consist of one pair of small simple papillae at level of cloaca, followed by pair of double papillae. Remaining post-cloacal ventral papillae simple, arranged in three rows; first row containing four papillae and others two papillae each; four to five single papillae distributed subventrally on post-cloacal area; similar in distribution to precloacal papillae. Precloacal papillae (Figs. 1d and 2b) simple, pyriform in shape with narrower margin lateral, forming two subventral lines extending anteriorly. Distance from cloaca to posterior end 0.17 (0.13–0.21), 0.6% (0.5–0.6%) of body length.
Female (n = 1). Body length 24.6, maximum width 0.89. Diameter across lips 0.2. Nerve ring 0.46 from anterior end. Deirids at approximately same level as nerve ring, 0.40 from anterior end. Oesophagus 4.11 long, 17% of body length. Ventricular appendix 1.09, 26% of oesophageal length. Intestinal caecum 3.43 long, 83% of oesophageal length, 3.15 times length of ventricular appendix. Vulva 11.05 from anterior end, 45% of body length. Distance from anus to posterior end 0.26, 1% of body length.
Type: holotype ♂ SAM AHC 34482, allotype ♀ SAM AHC 34483; paratypes, 3♂, SAM AHC 34484-5
Type host: P. conspicillatus (Aves, Pelicaniformes)
Localization in host: Stomach
Type locality: Melbourne, Victoria, Australia
C. multipapillatum (Drasche 1882)
Description
Labia large and fleshy, one dorsal and two subventral (Figs. 2d,e,a and 3b). Two large cephalic papillae on dorsal labium, one cephalic papilla on each subventral labium. Cephalic papillae located at mid-level of labium. Each labium expanded antero-laterally, almost in contact with adjacent labium. Margins of antero-lateral aspects of labia mildly rounded, without flanges. Slight depressions visible anteriorly as well as on lateral aspects of each labium. Interlabia simple, triangular, wider at base. Interlabia approximately same height as labia. Collar apparent, at base of lips, interrupted laterally, separated from body by prominent indentions. Intestinal caecum three to four times longer than ventricular appendix (Fig. 3c). Centrids arranged asymmetrically in mid-body region, right at half body length, left centrid placed more caudally at about two thirds of body length from anterior end.
C. multipapillatum D, from P. conspicillatus: a dorsal labium (scale bar = 0.17 mm); b ventral interlabium with subventral labia (scale bar = 0.17 mm); c posterior end in male, ventral view, showing post-cloacal papillae (scale bar = 0.08 mm); d anterior end of nematode showing oesophagus, ventricular appendix and intestinal caecum (scale bar = 0.65 mm)
Male (n = 1). Body length 26.0, width 0.85. Diameter across lips 0.23. Nerve ring 0.52 from anterior end. Deirids at approximately same level as nerve ring, 0.49 from anterior end. Oesophagus 5.64 long, 22% of body length. Ventricular appendix 1.46 long, 26% of oesophageal length. Intestinal caecum 5.2 long, 92% of oesophageal length and 3.56 times length of ventricular appendix. Spicules short, 7% of body length. Right spicule 1.82 and left spicule 1.76 long. Post-cloacal caudal papillae (Fig. 3d) consist of one pair of small simple papillae near level of cloaca, followed by pair of double papillae. Remaining post-cloacal ventral papillae simple, distributed in three rows; first row containing four papillae and the rest two papillae each; four to five single papillae distributed subventrally on post-cloacal area; similar in distribution to precloacal papillae. Precloacal papillae simple, forming two subventral lines extending anteriorly. Distance from cloaca to posterior end 0.10, 0.4% of body length.
Females (n = 10). Body length 30.2 (22.4–37.5), width 0.97 (0.81–1.19). Diameter across lips 0.25 (0.22–0.29). Nerve ring and deirids 0.52 (0.46–0.60) from anterior end. Oesophagus 5.87 (4.71–7.39) long, 19% (17–22%) of body length. Ventricular appendix 1.41 (0.98–2.03) long, 24% (21–27%) of oesophageal length. Intestinal caecum 5.00 (3.66–6.26) long, 83% (78–88%) of oesophageal length and 3.49 (3.08–3.94) times length of ventricular appendix. Vulva 13.1 (9.4–16.3) from anterior end, 43% (34–48%) of body length. Distance from anus to posterior end 0.32 (0.26–0.37), 1.1% (0.9–1.3%) of body length. Eggs 39 (24–49) × 49 (39–58)μm.
Material examined: 1 male, 10 females (AHC 34486-7)
Host: P. conspicillatus
Localization in host: Stomach
Locality: Melbourne, Victoria, Australia
Genetic characterisation
C. pyripapillatum n. sp
A total of eight specimens was subjected to SSCP analysis. Based on the SSCP profiles and host individuals, ITS-1 and ITS-2 regions of four representative specimens were selected and sequenced (GenBank accession numbers: AM940062, AM940063, AM940064 and AM940065 for ITS-1 and AM940066, AM940067, AM940068 and AM940069 for ITS-2). The length of ITS-1 region was 443bp. Polymorphism (substitution of A or G) was detected in ITS-1 at alignment position 389 within specimen (Fig. 4). The sequence variation among specimens of C. pyripapillatum was 0.2%. The G + C content in ITS-1 was 42.9–43.1%. The ITS-2 region was 233bp in length and had a G + C content of 43.4%. No sequence polymorphism was detected in ITS-2.
Alignment of sequences of ITS-1 and ITS-2 sequences of C. multipapillatum D (accession numbers: AM940056, AM940057, AM940058, AM940059 for ITS-1, and AM940060 and AM940061 for ITS-2) and C. pyripapillatum n. sp. (AM940062, AM940063, AM940064 and AM940065 for ITS-1, and AM940066, AM940067, AM940068, and AM940069 for ITS-2). Numbers to the right of the alignment indicate the alignment position. Polymorphic sites were designated using IUPAC codes
C. multipapillatum D
A total of 16 specimens was subjected to SSCP analysis; four and two amplicons were selected for ITS-1 and ITS-2 regions, respectively, based on the SSCP profiles and from host individuals, and sequenced (GenBank accession numbers: AM940056, AM940057, AM940058, AM940059 for ITS-1 and AM940060 and AM940061 for ITS-2). The ITS-1 region was 443bp and its G + C content was 44.5–44.7%. Nucleotide polymorphism (substitution of C or T) in ITS-1 region was detected within one specimen at alignment position 104 (Fig. 4). The nucleotide variation in ITS-1 among specimens of C. multipapillatum was 0.2%. The ITS-2 region was 231bp in length and had a G + C content of 42.4%. No nucleotide polymorphism was detected in ITS-2. Based on the pairwise comparison of sequence differences, C. multipapillatum D and C. pyripapillatum differed by 3.4–3.8% and 6.0% in ITS-1 and ITS-2, respectively.
Discussion
Two species of Contracaecum from Australian pelicans included in the present study resembled C. multipapillatum morphologically. As redescribed by Lucker (1941), specimens described herein were identified as C. multipapillatum based on the size of spicules, arrangement of the post-cloacal papilla and the shape of lips. Some of these specimens were collected and described by Norman (2005). Although, Norman’s description of the post-cloacal papillae conforms with C. multipapillatum, he identified them as Contracaecum clelandi (Johnston and Mawson 1941). He described the first pair of post-cloacal caudal papillae as double, not simple, as originally described by Johnston and Mawson (1941) for C. clelandi. He noted that the large papilla in Johnston and Mawson’s (1941) description of C. clelandi was probably double. The description of this morphological character as well as measurements given by Norman (2005) indicate that his material is the same as that used in the present study and matches C. multipapillatum as redescribed by Lucker (1941). Johnston and Mawson’s (1941) description of C. clelandi is incomplete in several respects but is sufficient to permit the recognition of C. clelandi as a valid species. However, an examination of the type specimen deposited in SAM revealed that the type specimen is female.
The clearest distinction between C. multipapillatum and C. clelandi is that, in the former species, the first pair of post-cloacal papillae is double and the number of post-cloacal papillae and their distribution are more similar to those of Contracaecum spp. from marine mammals. Also, in C. multipapillatum, the papillae on the labium are located at about half the height of the labia, not close to its anterior end (as in C. clelandi) and there are significant differences in the distance of the cloaca and the anus from the posterior end in males and females, respectively. In our specimens, this distance was 0.1 and 0.26–0.37 for male and females, respectively, whereas it was 0.8 in Johnston and Mawson’s (1941) description of C. clelandi (sex of nematode not indicated).
C. pyripapillatum n. sp. is morphologically very similar to C. multipapillatum. The lengths of the oesophagus, ventricular appendix and intestinal caecum in C. multipapillatum are longer. The preanal ventral papillae of male C. pyripapillatum are pyriform in shape, whereas they are rounded in C. multipapillatum. The depression of the anterior margin of the labia is relatively deeper and the ratio of the height of the labium to its width was greater in the new species (Figs. 1a,b and 2a). Since the size of the oesophagus, ventricular appendix and intestinal caecum as well as their ratio are related to allometric growth (Huizinga 1967), they cannot be used reliably for differentiation. Also, different fixatives can affect the size and appearance of organs such as the lips (Fagerholm 1979). Therefore, at this stage, the only morphological character differentiating the two species is the shape of the precloacal papillae which are consistently pyriform in C. pyripapillatum (Figs. 1d and 2b).
Analysis of ITS-1 and ITS-2 sequence data supports the hypothesis that specimens similar to C. multipapillatum in the Australian pelican belong to two distinct species. In the ITS-1 region, 3.4–3.8% base pair nucleotide differences were found between these species. In contrast, the base pair nucleotide difference of 6.0% in ITS-2 region between C. multipapillatum and C. pyripapillatum was in the range of the base pair nucleotide difference within species complex of Contracaecum nematodes (0.0–6.8%; S. Shamsi unpublished data). Current evidence (S. Shamsi, unpublished data) based on ITS-1 and ITS-2 sequence data indicate that C. multipapillatum D and C. pyripapillatum are genetically more closely related than to other Contracaecum spp. However, based on the available data, it is not yet known whether genotypes A–C of C. multipapillatum sensu lato (described from the USA and Europe) are identical with the species found in Australia. To address this question, further comprehensive studies of specimens from different hosts and geographical locations are necessary.
References
Akram M (1996) Contracaecum bubakii, new species (Nematoda: Anisakidae) from little cormorant in Pakistan. Pak J Zool 28:131–132
Chilton NB, Gasser RB, Beveridge I (1995) Differences in a ribosomal DNA sequence of morphologically indistinguishable species within the Hypodontus macropi complex (Nematoda: Strongyloidea). Int J Parasitol 25:647–651
Courtney CH, Forrester DJ (1974) Helminth parasites of the brown pelican in Florida and Louisiana. Proc Helminthol Soc Wash 41:89–93
D’Amelio S, Barros NB, Ingrosso S, Fauquier DA, Russo R, Paggi L (2007) Genetic characterization of members of the genus Contracaecum (Nematoda: Anisakidae) from fish-eating birds from west-central Florida, USA, with evidence of new species. Parasitology 134:1041–1051
Deardorff TL, Overstreet RM (1980) Contracaecum multipapillatum (=C. robustum) from fishes and birds in the northern Gulf of Mexico. J Parasitol 66:853–856
Drasche R (1882) Helminthologische Notizen. Verh Zool-Bot Ges Wien 32:139–142
Fagerholm HP (1979) Nematode length and preservatives, with a method for determining the length of live specimens. J Parasitol 65:334–335
Flores Barroeta L (1957) Nematodos de aves y mamiferos. Rev Iber Parasitol 17:277–297
Foster GW, Kinsella JM, Moler PE, Johnson LM, Forrester DJ (1998) Parasites of Florida softshell turtles (Apalone ferox) from southeastern Florida. J Helminthol Soc Wash 65:62–64
Hartwich G (1964) Revision der vogelparasitischen nematoden mitteleuropas, II. Di Gattung Contracaecum Railliet & Henry, 1912. Mitteilungen aus dei Zoologisches Museum Berlin 40:15–53
Hartwich G (1974) Keys to genera of the Ascaridoidea. In: Chabaud AG, Anderson CRC, Willmott S (eds) CIH keys to the nematode parasites of vertebrates. vol. 2. Commonwealth Agricultural Bureaux, Farnham Royal, pp 1–15
Huizinga HW (1967) The life cycle of Contracaecum multipapillatum (Von Drasche, 1882) Lucker, 1941 (Nematoda: Heterocheilidae). J Parasitol 53:368–375
Johnston TH, Mawson PM (1941) Ascaroid nematodes from Australian birds. Transaction of Royal Society of South Australia 65:110–115
Kinsella JM, Spalding MG, Forrester DJ (2004) Parasitic helminths of the American white pelican, Pelecanus erythrorhynchos, from Florida, USA. Comparative Parasitology 71:29–36
Labriola J, Suriano DM (1996) Parasitic nematodes of birds from De Monte Pond, Buenos Aires, Argentina. Bol Chil Parasitol 51:59–65
Lucker JT (1941) A redescription of Contracaecum multipapillatum (von Drasche, 1882). J Parasitol 27:505–512
Mattiucci S, Olivero J, Paoletti M, Arrollo B, Nascetti G (2006). Genetic evidence for new species of Contracaecum (Nematoda, Anisakidae) parasites of the brown pelican, Pelecanus occidentalis, from Columbia: genetic relationships between congeners and larval identification. 11th International Congress of Parasitological Associations, 6th–11th August, Glasgow Scotland (Abstract)
Nadler SA, D’Amelio S, Fagerholm HP, Berland B, Paggi L (2000) Phylogenetic relationships among species of Contracaecum and Phocascaris (Nematoda, Ascaridoida) based on nuclear rDNA. Parasitology 121:455–463
Navone GT, Etchegoin JA, Cremonte F (2000) Contracaecum multipapillatum (Nematoda: Anisakidae) from Egretta alba (Aves: Ardeidae) and comments on other species of this genus in Argentina. J Parasitol 86:807–810
Norman R (2005) Parasitic diseases of the little penguin, Eudyptula minor with emphasis of the nematodes of the genus Contracaecum Raillliet and Heny, 1912 (Anisakidae). Ph.D. Thesis, Faculty of Veterinary Scinece, The University of Melbourne, 693 pp
Sepulveda MS, Spalding MG, Kinsella JM, Bjork RD, McLaughlin GS (1994) Helminths of the roseate spoonbill, Ajaiai ajaja, in southern Florida. J Helminthol Soc Wash 61:179–189
Sepulveda MS, Spalding MG, Kinsella JM, Forrester DJ (1999) Parasites of the great egret (Ardea albus) in Florida and a review of the helminths reported for the species. J Helminthol Soc Wash 66:7–13
Skrjabin KI (1916) Materials of helminthofauna of Paraguay. I. Nematodes. (Contribution to the study on helminthofauna of Paraguay. I. Nematodes). Zoologischeskii Vestnik 1:705–757 in Russian
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Vidal-Martinez VM, Osorio-Sarabia D, Overstreet RM (1994) Experimental infection of Contracaecum multipapillatum (Nematoda: Anisakinae) from Mexico in the domestic cat. J Parasitol 80:576–579
Zhu X, Gasser RB, Jacobs DE, Hung GC, Chilton NB (2000) Relationships among some ascaridoid nematodes based on ribosomal DNA sequence data. Parasitol Res 86:738–744
Zhu XQ, Gasser RB, Chilton NB, Jacobs DE (2001) Molecular approaches for studying ascaridoid nematodes with zoonotic potential, with an emphasis on Toxocara species. J Helminthol 75:101–108
Author information
Authors and Affiliations
Corresponding author
Additional information
Nucleotide sequence data reported in this paper are available in the GenBank database under the accession numbers AM940056–AM940069.
Rights and permissions
About this article
Cite this article
Shamsi, S., Gasser, R., Beveridge, I. et al. Contracaecum pyripapillatum n. sp. (Nematoda: Anisakidae) and a description of C. multipapillatum (von Drasche, 1882) from the Australian pelican, Pelecanus conspicillatus . Parasitol Res 103, 1031–1039 (2008). https://doi.org/10.1007/s00436-008-1088-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-008-1088-z