Abstract
Expressed sequence tag (EST) pools represent partial profiles of the gene expressions of organisms. In an effort to construct a Clonorchis sinensis EST pool, 2,387 ESTs were collected from an adult C. sinensis cDNA library and assembled into 1,573 clusters. Of these clusters, 1,225 ESTs (51%) were singletons and 348 clusters consisted of more than two ESTs. There were 848 clusters (54%) that shared significant identity with previously reported proteins, and of these, 401 clusters were categorized into 11 major functional protein classes. Three cDNA clones of fructose-1,6-bisphosphate (FBP) aldolase were selected from the C. sinensis EST pool and analyzed for phylogenic clustering. FBP clones encoded a complete polypeptide, which shared significant identity to those of vertebrate and invertebrate animals and clustered with those of trematodes. We believe that the EST pool described can be confidently used as a platform in multigene researches on C. sinensis gene expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human clonorchiasis is caused by the Chinese liver fluke, Clonorchis sinensis, which is endemic in Korea, Japan, and Southern China (Crompton 1999). People become infected with C. sinensis, which inhabits biliary tracts, by eating raw or inadequately cooked freshwater fishes. The symptoms of human clonorchiasis include obstructive jaundice, a dull epigastric pain, biliary stones, and ascites.
In its larval phase, C. sinensis develops through several stages, i.e., miracidium, sporocyst, redia, cercaria, and metacercaria before reaching the egg-laying adult stage. We conducted this analysis of the gene expression profiles of developmental stages to better understand C. sinensis biology and host–parasite relationships. Huge amounts of genetic information on C. sinensis are required to discover novel gene products and promising candidates for new drugs and vaccine. Some C. sinensis genes have already been identified and characterized (Hong et al. 2000, 2001; Lee et al. 2003), but the numbers are relatively small. Expressed sequence tag (EST) analysis has been proven to be a rapid and efficient means of characterizing the massive sets of gene sequences that are expressed in a life-stage-specific manner in a wide variety of tissues and organisms (Adams et al. 1995).
Fructose-1,6-bisphosphate (FBP) aldolase is an enzyme that catalyzes the conversion of FBP to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate in the glycolytic pathway (Inoue et al. 1997). FBP aldolase is a bifunctional enzyme that is involved in catabolic (glycolysis) and anabolic (gluconeogenesis and Calvin cycle) pathways. The FBP aldolases comprise two classes. Class I aldolases form Schiff bases with their substrates and are subgrouped further into types A, B, and C by their tissue distributions (Kuba et al. 1997), whereas class II aldolases require a divalent cation (usually Zn2+ or Fe2+) for activation. Aldolases types A and C are found in muscle and brain, respectively, and prefer FBP to fructose-1-phosphate (F1P), whereas aldolase type B is found in liver and equally catalyzes these two substrates. Aldolase is a conserved enzyme among animals, which suggests that it is a suitable model enzyme for genetic evolutionary studies.
Materials and methods
Adult C. sinensis
The metacercariae of C. sinensis were collected from topmouth gudgeons, Pseudorasbora parva, by artificial digestion and were orally administered to rabbits, New Zealand White. Adult C. sinensis were recovered from rabbit bile ducts 7 months after infection and were used for total RNA extraction.
Construction of a cDNA library
Adult C. sinensis were homogenized in 4 M guanidium isothiocyanate containing α-mercaptoethanol and centrifuged at 10,000 rpm (Beckman, JA 20 rotor) for 10 min. The supernatants were added to 5.7 M CsCl solution and total RNA was purified by ultracentrifugation at 34,000 rpm (Beckman, SW-41 Ti rotor) for 18 h at 20°C. Poly (A)+ mRNA was then isolated from total RNA by oligo (dT) column chromatography. The adult C. sinensis cDNA library was then constructed in λ-ZAP II bacteriophage vector, according to the manufacturer’s instructions (Stratagene, La Jolla, CA, USA). In brief, double-stranded cDNA (ds-cDNA) was synthesized from poly (A)+ mRNA and size-fractionated by sepharose S-400 spin column chromatography. The first and second fractions, containing ds-cDNAs larger than 0.6–0.4 kbp, were pooled and concentrated by ethanol precipitation. The ds-cDNA (ca. 100 ng) so obtained was then ligated to 1 μg of λ-ZAP II bacteriophage vector arms and packaged in vitro. The cDNA library had 1.0×107 plaque forming units per 1 μg of bacteriophage DNA with 0.1% background.
Sequencing of ESTs
The cDNA library was converted into phagemid by mass in vivo excision using helper phage and transfected into E. coli SOLR strain. Individual bacterial colonies on LB/amp (50 mg/l) plates were selected randomly for plasmid DNA purification and sequencing. The cDNA sequence of each clone was read once from its 5′-end using T3 primer and an automatic sequencer (ABI, Applied Biosystems, Foster City, CA, USA).
Analysis of ESTs
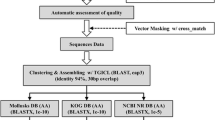
cDNA sequences were trimmed off vector and adaptor sequences, and the trimmed ESTs were assembled into cluster sequences using TIGR assembler version 2.0 (http://www.tigr.org). Homologous cluster sequences were searched for in the GenBank, NCBI database using Basic Local Alignment Search Tool X (BLAST X). Expectation values (e-values) of 1×10−5 were considered sufficient for a putative identification (Junqueira-de-Azevedo and Ho 2002).
Alignment of amino acid sequences
To evaluate EST quality, clusters of aldolases were collected from the C. sinensis EST pool and further analyzed to deduce polypeptides. In all organisms, glycolysis is a highly conserved metabolic pathway which produces energy at the substrate level. Animal aldolase sequences were retrieved from GenBank, NCBI and aligned with C. sinensis aldolases using Clustal X 1.81 (Thompson et al. 1997).
Phylogenetic tree inference
Phylogenetic trees were drawn by the neighbor-joining method using MEGA 2.1 (Kumar et al. 2001). Local bootstrap probability (LBP) was calculated from 1,000 replications using MEGA 2.1 (Hasegawa and Kishino 1994).
Results
Sequencing and annotation
After sequencing 3,000 clones randomly selected from the C. sinensis adult cDNA library, 2,387 ESTs were generated. These were analyzed to identify overlapping sequences and constructed into contigs encoding putative polypeptides and further assembled into 1,573 clusters. There were 1,225 (51.33%) clusters that represented singleton ESTs, 339 clusters assembled represented two to nine ESTs, and six clusters represented 10–19 ESTs. Three clusters comprised 23, 56, and 62 ESTs, respectively (Fig. 1). In length, the average size of clusters was 560 bp and the major fraction of the clusters ranged between 500 and 900 bp. The largest cluster was 2,114 bp in length (Fig. 2). Nucleotide sequencing data were deposited at the DDBJ/EMBO/GenBank nucleotide sequence database under accession numbers AT007520–AT009822.
BLAST searches revealed that 848 clusters (53.9% of the total) shared identity (e-value <10−5) with previously reported genes or proteins and were, thus, annotated to these proteins, while the remaining 725 clusters showed no significant similarity to any known gene. The EST number of a gene in an EST pool may represent the relative abundance of that gene’s expression. In adult C. sinensis, the most abundantly expressed genes were cysteine proteases, followed in descending order by mitochondrial genes, muscle proteins, alpha tubulins, and vitelline protein (Table 1). When grouping the annotated 848 clusters with functional features, 401 clusters were categorized into 11 major functional protein classes. The biggest group was composed of regulatory proteins and comprised kinases and phosphatases, and regulatory and signaling proteins, followed by enzymes involved in metabolic pathways and energy production. The third group comprised proteins constituting transcription and translation machineries, and the fourth group comprised structural proteins (Table 2). Among the annotated clusters, 75 had been previously identified in C. sinensis.
Nucleotide and amino acid sequences of C. sinensis aldolase cDNAs
The C. sinensis cDNA library contained three isoforms of fructose-1,6-bisphosphate aldolase, CsFbA-1, CsFbA-2, and CsFbA-3. CsFbA-1 have an open reading frame of 1,086 bp, encoding 362 amino acid residues, and the other two, CsFbA-2 and CsFbA-3, each have an open reading frame of encoding 363 amino acid residues. The deduced polypeptide of CsFbA-1 revealed sequence identities of 69 and 71% with CsFbA-2 and CsFbA-3, respectively. The sequence identity between CsFbA-2 and CsFbA-3 was 76% (Fig. 3).
Multiple alignment of the amino acid sequences of the three C. sinensis FBP aldolases and eight FBP aldolases of vertebrates and invertebrates. Amino acids are represented by one-letter codes. Conserved amino acid residues of the FBP aldolases are indicated by an asterisk (*). A lysine residue (K229) forming a putative Shiff base is represented by a bold character on a black background. The aldolase isozymes are as follows: human aldolase A (P04075, Sakakibara et al. 1985), Xenopus laevis A (BAA19524, Hikasa et al. 1997), lamprey muscle type (M) (P53445, Zhang et al. 1995), Drosophila melanogaster Dm a (JX0233, Kai et al. 1992), Caenorhabditis elegans Ce-1 (P54216, Inoue et al. 1997), Onchocerca volvulus (AAD38430, McCarthy et al. 2002), E. multilocularis (CAC18550, Brehm et al. 2000), and S. mansoni (AAA57567, El-Dabaa et al. 1998)
Comparison of the primary structures of C. sinensis FBP aldolases
C. sinensis FBP aldolase sequences were compared with eight aldolases of vertebrate and invertebrate animals (Fig. 3). The deduced polypeptide sequences of CsFbA-1, CsFbA-2, and CsFbA-3 showed high degrees of identity with each other and with the conserved region of vertebrate and invertebrate aldolases. The sequence identity to human muscle-type FBP aldolase A shared 68.9% homology with CsFbA-1, 65.1% with CsFbA-2, and 67.9% with CsFbA-3. The primary structures of C. sinensis isoenzymes were typical class I aldolases, and they showed high homology with other aldolases. Among FBP aldolases, a lysine residue in the catalytic domain is highly conserved. This key lysine residue was conserved at 229 in CsFbA-1 and at 230 in CsFbA-2 and CsFbA-3 (Fig. 3). The conserved residues across animal FBP aldolases are Asp34, Lys41, Lys147, Arg149, His361, and a tyrosine residue at the C terminus (Kitajima et al. 1990). All these amino acid residues appeared at corresponding sites in the CsFbA-1, -2, and -3 polypeptides.
Phylogenetic analyses of FBP aldolases
A phylogenetic tree of three C. sinensis aldolases with 13 FBP aldolases of animal species showed that CsFbA-2 and CsFbA-3 clustered with Echinococcus multilocularis (LBP 0.54) and Schistosoma mansoni (LBP 0.87), respectively, and all formed a single clade (LBP 0.99). All aldolases of platyhelminthes in this tree, including CsFbA-1, formed one clade with an LBP value of 0.78 (Fig. 4).
Discussion
In this study, 2,387 ESTs from the cDNA library of C. sinensis were assembled to form 1,573 clusters. Two clusters representing the most abundant transcripts contained more than 50 ESTs accounting for 4.9% of the total ESTs analyzed (118 of 2,387 ESTs). One was composed of cysteine proteases and contained 62 ESTs, whilst the other was composed of mitochondrial genes and contained 56 ESTs, implying that the enzyme may support biliary epithelia destruction by adult C. sinensis and enable it to fight off host immune attack (Park et al. 1995; Nagano et al. 2004; Kang et al. 2004). The next most abundant gene transcripts were proteins constituting muscular tissues, such as limpet (1J758), protein KIAA1404, alpha-1 tubulin, tubulin alpha chain, and JF-2 (actin binding protein). These abundant expressions of muscle-related genes suggest the importance of muscular tissues and the major somatic structure that constitutes the oral and ventral suckers, subtegumental musculature, and myoepithelia of the long digestive tract, which enable adult flukes to abrade and feed on biliary epithelium (Kwon et al. 2005). Vitelline precursor protein was the third most abundantly expressed gene product. To perpetuate the species, adult C. sinensis produces daily large amounts of eggs, which incidentally contain yolk. Vitelline precursor proteins are produced in the vitelline glands, which are responsible in hardening the eggshell encasing the germ cell and surrounding yolk cells (Rice-Ficht et al. 1992; Tang et al. 2005).
Of the 848 putative homologous proteins, 401 proteins (25.5% of all total clusters) were categorizable into 11 major classes, as defined by a previous classification (Santos et al. 1999). A large number of these clusters (74.5% of total clusters) was not fully identified in the present study due to insufficient information. On the other hand, 75 clusters were closely related to proteins previously reported in C. sinensis and other animals. Therefore, most of the C. sinensis clusters described here are being reported for the first time.
Three types of FBP aldolase were found in the C. sinensis ESTs pool. They differed from each other with respect to amino acid number by one residue and in terms of sequential identity by 24–31%. Vertebrates and invertebrates possess three FBP aldolase isoenzymes (Shiokawa et al. 2002). The presence of these three isoenzymes in C. sinensis seems acceptable when it is considered that Drosophila melanogaster (Kai et al. 1992) and Caenorhabditis elegans (Inoue et al. 1997) possess more than two FBP aldolase isoenzymes.
FBP aldolase is a key enzyme in the glycolysis pathway and catalyzes a reversible aldol cleavage of FBP into two trioses, dihydroxyacetone phosphate and glyceraldehyde-3-phosphate. FBP aldolases form a Schiff base between substrate and a lysine residue and belong to class I aldolases. Class II aldolases use a divalent ion as a cofactor (El-Dabaa et al. 1998). Based on the possession of a putative Schiff’s-base-forming domain, three types of C. sinensis FBP aldolases are considered FBP aldolases. Class I FBP vertebrate aldolases are further subgrouped into three isozymes, i.e., A, B, and C. Aldolase type A, the muscle-type isozyme, has a preference for FBP rather than for fructose-1-phosphate . This isozyme generates chemical energy in muscular tissues, which lack a fructose to glycolytic intermediate metabolic pathway via F1P production (Horecher et al. 1972). Aldolase type B, the liver-type isozyme, exhibits substrate specificity suitable for metabolism in hepatocytes, in which fructose is metabolized to pyruvate via F1P, and aldolase type C, the brain-type isozyme, occurs in fetal tissues and adult nerve tissues and shows intermediate activities toward FBP and F1P.
Considerations of the structural arrangements of the amino acid groups of FBP aldolases often explain enzymatic activity differences. The amino acid arrangements in the C-terminal region of these enzymes largely determine their substrate specificities and catalytic activities. FBP aldolase type A is characterized by having a Lys230 residue at its 6-phosphate binding site and a His362 residue. These two residues are essential for high-level catalytic activity toward FBP. In FBP aldolase type B enzymes, asparagines and tyrosine residues substitute Lys230 and His362, respectively (Gamblin et al. 1991), whereas in polypeptides of C. sinensis FBP aldolases, a lysine residue is positioned at the 6-phosphate binding site and a histidine residue is located at 361 (CsFbA-1) and 362 (CsFbA-2 and -3), suggesting that these three putative isoenzymes could be assigned as FBP aldolase type A enzymes.
In this study, a large number of ESTs was collected from C. sinensis and an EST platform was formulated as an aid to future research at the multigene level.
References
Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O et al (1995) Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377(Suppl 6547):3–174
Brehm K, Jensen K, Frosch M (2000) mRNA trans-splicing in the human parasitic cestode Echinococcus multilocularis. J Biol Chem 275:38311–38318
Crompton DWT (1999) How much human helminthiasis is there in the world? J Parasitol 85:397–403
El-Dabaa E, Mei H, El-Sayed A, Karim AM, Eldesoky HM, Fahim FA, LoVerde PT, Saber MA (1998) Cloning and characterization of Schistosoma mansoni fructose-1,6-bisphosphate aldolase isoenzyme. J Parasitol 84:954–960
Gamblin S, Davies GJ, Grimes JM, Sackson RM, Lottlechild JA, Watson HC (1991) Activity and specificities of human aldolases. J Mol Biol 219:573–576
Hasegawa M, Kishino H (1994) Accuracies of the simple methods for estimating the bootstrap tree. Mol Biol Evol 11:142–145
Hikasa H, Hori K, Shiokawa K (1997) Structure of aldolase A (muscle-type) cDNA and its regulated expression in oocytes, embryos and adult tissues of Xenopus laevis. Biochim Biophys Acta 1354:189–203
Hong SJ, Seong KY, Sohn WM, Song KY (2000) Molecular cloning and immunological characterization of phosphoglycerate kinase from Clonorchis sinensis. Mol Biochem Parasitol 108:207–216
Hong SJ, Lee JY, Lee DH, Sohn WM, Cho SY (2001) Molecular cloning and characterization of a mu-class glutathione S-transferase from Clonorchis sinensis. Mol Biochem Parasitol 115:69–75
Horecher BL, Tsalas O, Lai CY (1972) Aldolases. In: Boyer PD (ed) Enzyme, vol. 7. Academic, New York, pp 158–213
Inoue T, Yatsuki H, Kusakabe T, Joh K, Takasaki Y, Nikoh N, Miyata T, Hori K (1997) Caenorhabditis elegans has two isozymic forms, CE-1 and CE-2, of fructose-1,6-bisphosphate aldolase which are encoded by different genes. Arch Biochem Biophys 339:226–234
Junqueira-de-Azevedo Ide L, Ho PL (2002) A survey of gene expression and diversity in the venom glands of the pit viper snake Bothrops insularis through the generation of expressed sequence tags (ESTs). Gene 299:279–291
Kai T, Sugimoto Y, Kusakabe T, Zhang R, Koga K, Hori K (1992) Gene structure and multiple mRNA species of Drosophila melanogaster aldolase generating three isozymes with different enzymatic properties. J Biochem (Tokyo) 112:677–688
Kang TH, Yun DH, Lee EH, Chung YB, Bae YA, Chung JY, Kang I, Kim J, Cho SY, Kong Y (2004) A cathepsin F of adult Clonorchis sinensis and its phylogenetic conservation in trematodes. Parasitology 128:195–207
Kitajima Y, Takasaki Y, Takahashi I, Hori K (1990) Construction and properties of active chimeric enzymes between human aldolases A and B. J Biol Chem 265:17493–17498
Kuba M, Yatsuki H, Kusakabe T, Takasaki Y, Nikoh N, Miyata T, Yamaguchi T, Hori K (1997) Molecular evolution of amphioxus fructose-1,6-bisphosphate aldolase. Arch Biochem Biophys 348:329–336
Kumar S, Tamura K, Jakobson IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Kwon YD, Cho PY, Hong SJ (2005) Clonorchis sinensis: molecular cloning and localization of myosin regulatory light chain. Parasitol Res 97:21–26
Lee JS, Lee JW, Park SJ, Yong TS (2003) Analysis of the genes expressed in Clonorchis sinensis adults using the expressed sequence tag approach. Parasitol Res 91:283–289
McCarthy JS, Wieseman M, Tropea J, Kaslow D, Abraham D, Lustigman S, Tuan R, Guderian RH, Nutman TB (2002) Onchocerca volvulus glycolytic enzyme fructose-1,6-bisphosphate aldolase as a target for a protective immune response in humans. Infect Immun 70:851–858
Nagano I, Pei F, Wu Z, Wu J, Cui H, Boonmars T, Takahashi Y (2004) Molecular expression of a cysteine proteinase of Clonorchis sinensis and its application to an enzyme-linked immunosorbent assay for immunodiagnosis of clonorchiasis. Clin Diagn Lab Immunol 11:411–416
Park H, Ko MY, Paik MK, Soh CT, Seo JH, Im KI (1995) Cytotoxicity of a cysteine proteinase of adult Clonorchis sinensis. Korean J Parasitol 33:211–218
Rice-Ficht AC, Dusek KA, Kochevar GJ, Waite JH (1992) Eggshell precursor proteins of Fasciola hepatica, I. Structure and expression of vitelline protein B. Mol Biochem Parasitol 54:129–141
Santos TM, Johnston DA, Azevedo V, Ridgers IL, Martinez MF, Marotta GB, Santos RL, Fonseca SJ, Ortega JM, Rabelo EML, Saber M, Ahmed HM, Romeih MH, Franco GR, Rollinson D, Pena SDJ (1999) Analysis of the gene expression profile of Schistosoma mansoni cercariae using the expressed sequence tag approach. Mol Biochem Parasitol 103:79–97
Sakakibara M, Mukai T, Hori K (1985) Nucleotide sequence of a cDNA clone for human aldolase: a messenger RNA in the liver. Biochem Biophys Res Commun 131:413–420
Shiokawa K, Kajitawa E, Hara H, Yatsuki H, Hori K (2002) A developmental biological study of aldolase gene expression in Xenopus laevis. Cell Res 12:85–96
Tang Y, Cho PY, Kim BS, Hong SJ (2005) Molecular cloning and Characterization of vitelline precursor protein B1 from Clonorchis sinensis. J Parasitol 91:1374–1378
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Zhang R, Yatsuki H, Kusakabe T, Iwabe N, Miyata T, Imai T, Yoshida M, Hori K (1995) Structures of cDNAs encoding the muscle-type and non-muscle-type isozymes of lamprey fructose bisphosphate aldolases and the evolution of aldolase genes. J Biochem (Tokyo) 117:545–553
Acknowledgement
This work was supported by the Korea Research Foundation Grant (KRF-2002-041-E20086).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cho, P.Y., Lee, M.J., Kim, T.I. et al. Expressed sequence tag analysis of adult Clonorchis sinensis, the Chinese liver fluke. Parasitol Res 99, 602–608 (2006). https://doi.org/10.1007/s00436-006-0204-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-006-0204-1