Abstract
To determine if genetic diversity of Blastocystis hominis exists in China, 35 B. hominis isolates obtained from 19 asymptomatic infected individuals and 16 patients with gastrointestinal symptoms were genotyped by PCR using seven pairs of known sequenced-tagged site (STS) primers. Out of the 35 isolates, 29 were identified as one of the known genotypes, while five isolates showed two distinct genotypes, and one isolate was an unknown genotype as this was negative with all the STS primers. In this study, none of the isolates was classified as subtypes 4–7. Compared with the spectrum of human B. hominis subtypes obtained from five geographically different countries (Japan, Pakistan, Bangladesh, Germany, and Thailand), these results showed that subtype 1 was more a popular genotype (18/35) in China. In addition, two groups of the isolates from 19 asymptomatic infected individuals and those from 16 patients with intestinal symptoms were compared with the PCR-based subtype classification. The results suggest a possible relationship between subtype 1 and a pathogenic potential of this parasite.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Blastocystis hominis has been described as probably the most common intestinal parasites in humans (Clark 2000). Its prevalence of infection is higher in developing than in developed countries (Stenzel and Boreham 1996). Despite growing awareness of the presence of B. hominis in symptomatic patients with gastrointestinal disorders and in the absence of other causative factors, its pathogenic potential remains a controversy (Tan et al. 2002). Although studies have shown the extensive genetic variability present in Blastocystis isolates from humans and animals (Yoshikawa et al. 1996, 1998, 2000, 2003, 2004a,b; Böhm-Gloning et al. 1997; Clark 1997; Abe et al. 2003a–c; Noël et al. 2005), a definite correlation between genetically distinct genotypes and pathogenicity has not yet been confirmed due to the limited number of studies. The purpose of this study is to analyze genotypes of 35 B. hominis isolates obtained from the asymptomatic infected individuals and symptomatic patients in China by PCR with the seven pairs of sequenced-tagged site (STS) primers reported in Yoshikawa et al. (2004a) so as to assess the degree of heterogeneity of these organisms in China and their possible relationship with the parasite’s pathogenic role.
Materials and methods
Sample collection and culture of B. hominis isolates
A total of 35 B. hominis isolates (HC05-1 to HC05-35) were used in this study. Sixteen isolates (HC05-1 to HC05-10, HC05-21 to HC05-23, HC05-28, HC05-31, and HC05-35) were collected from patients with intestinal symptoms in the Affiliated Hospital of Gannan Medical College from July to August 2005, and 19 isolates (HC05-11 to HC05-20, HC05-24 to HC05-27, HC05-29 to HC05-30, and HC05-32 to HC05-34) were collected from symptom-free freshmen in Gannan Medical College who came to the Physical Examination Center of Affiliated Hospital of Gannan Medical College for entrance physical check-up in September 2005. Symptomatic patients had diarrhea and/or abdominal pain, but were negative for parasitic tests for Entamoeba histolytica, Giardia lamblia, Cryptosporidium sp., and helminth eggs. However, patients could not be examined for the presence of Campylobacter sp., Shigella sp., Yersinia sp., Cycolospora sp., Salmonella sp., enterotoxigenic Escherichia coli, and enteric viruses. The stool samples were collected either at the hospital or in the parasitology laboratory at Gannan Medical College. The microscopic diagnosis of B. hominis was confirmed by culturing in Locke–Egg (LE) medium (Clark and Diamond 2002) containing 10% newborn calf serum or in Jones’ medium (Jones 1946) supplemented with 10% newborn calf serum in the parasitology laboratory at Gannan Medical College. All isolates were cultured at 37°C in LE medium or in Jones’ medium and subcultured every 3 or 4 days.
Preparation of genomic DNA
B. hominis grown in LE medium or in Jones’ medium was isolated by centrifugation with lymphocytes separation medium, and then genomic DNA of B. hominis was extracted with Genomic DNA Purification Kit [TIANGEN BIOTCH (BEIJING)] according to the manufacturer’s protocol.
Genotyping by PCR with the STS primers
PCR was performed using seven pairs of STS primers (SB83, SB155, SB227, SB332, SB340, SB336, and SB337) to identify genotypes of B. hominis. The primers used in this study are those used by Yoshikawa et al. (2004a) (Table 1). The PCR conditions consisted of one cycle denaturing at 94°C for 3 min, 30 cycles including annealing at 59°C for 30 s, extending at 72°C for 60 s, denaturing at 94°C for 30 s, and additional cycle with a 5-min chain elongation at 72°C (GeneAmp PCR System 2700, Applied Biosystems, USA). The PCR products and molecular markers were electrophoresed in 1.5% agarose gel with Tris-acetate-EDTA electropheresis buffer. The size markers were 100 base-pair ladder [TIANGEN BIOTCH (BEIJING)]. The PCR amplification for each primer pair was repeated at least thrice. Bands were visualized by the imaging system (VILBER LOURMAT, France) after being stained with ethidium bromide.
Statistical analysis
Likelihood-ratio chi-square test was used to compare the frequency of genotypes of B. hominis isolates from symptomatic patients and asymptomatic infected individuals.
Results
Identification of genotype by PCR with the STS primers
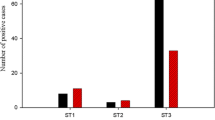
Genotypes of 35 isolates of B. hominis were obtained by PCR with the seven pairs of STS primers. The results are summarized in Table 2, and example of PCR amplification of several B. hominis isolates with the primers SB83, SB155, and SB227 is illustrated in Fig. 1. Twenty-nine isolates were identified as one of the known genotypes (Table 2, Fig. 1). Five isolates, HC05-5, HC05-6, HC05-7, HC05-8, and HC05-29, showed positive amplification with both SB83 and SB227 primers (Fig. 1a,c). Therefore, the five isolates were judged to be mixed isolates containing two distinct genotypes of subtype 1 and subtype 3. The remaining isolate HC05-10 unamplified with any STS primers was probably an unknown genotype. In this study, none of isolates was classified as subtypes 4–7. Our data showed that subtype 3 was the most common and subtype 1 was in the second rank.
Example of PCR amplification of several Blastocystis hominis isolates with the sequenced-tagged site (STS) primers SB83 (A), SB155 (B), SB227 (C). Five isolates, HC05-5 (lane 1), HC05-6 (lane 2), HC05-7 (lane 3), HC05-8 (lane 4), and HC05-29 (lane 14) are amplified with both SB83 (A) and SB227 (C), while other isolates are amplified with only one of the STS primers except for the isolate HC05-10 (lane 5), which is negative with all STS primers. M Molecular marker (100-bp ladder), 1 HC05-5, 2 HC05-6, 3 HC05-7, 4 HC05-8, 5 HC05-10, 6 HC05-11, 7 HC05-12, 8 HC05-13, 9 HC05-15, 10 HC05-19, 11 HC05-21, 12 HC05-23, 13 HC05-27, 14 HC05-29 {the first two letters of each isolates’ name indicate the origin [human (H)] and country [China (C)] from which the organism was isolated, and the numbers indicate the year [2005 (05)] in which the isolate was found. The number following the hyphen indicates the position of that particular isolate in that year’s series}
Genotype–phenotype association studies
To analyze the pathogenic potential of the distinct genotype(s) of B. hominis, two groups of isolates from symptomatic patients and from asymptomatic healthy individuals were compared in the subtype classification. Tentatively, the isolates recovered from 19 normal healthy individuals without any intestinal symptoms were categorized as the nonpathogenic isolates of B. hominis, while the isolates from 16 patients with diarrhea and/or abdominal pain without other pathogenic protozoans (E. histolytica, G. lamblia, and Cryptosporidium sp.) and helminths examined in this study were categorized as the pathogenic isolates. Based on the PCR amplification, subtype 1 is overrepresented in symptomatic patients, while subtype 3 in asymptomatic healthy individuals (Table 3). Because likelihood-ratio chi-square test shows significant differences between these groups (χ2=10.483, p=0.012), there was a possible correlation between subtype 1 and pathogenic potential of this parasite.
Discussion
In recent years, Yoshikawa et al. (1998, 2000, 2003) have developed seven pairs of STS primers derived from random amplified polymorphic DNAs (RAPDs). These primers can be used to identify the genotypes that correspond to phylogenetically different clades inferred from the small-subunit rRNA genes (SSU rDNA) (Arisue et al. 2003; Yoshikawa et al. 2003, 2004a). Moreover, this PCR-based technique has recently been reported to be a practical tool to classify and identify genotypes of B. hominis isolates from humans and animals, to detect some zoonotic genotypes from animal isolates, and to find unknown genotype(s) in clinical isolates of B. hominis (Abe et al. 2003a–c; Yoshikawa et al. 2003, 2004a,b). Admittedly, RAPD and the restriction fragment length polymorphism (RFLP) analysis of the SSU rDNA have been employed to show genomic polymorphism within species, but these specific primers have some advantages over RAPD and RFLP. The RAPD pattern cannot be compared with other reports accomplished in a different laboratory, and must be used to avoid contamination by other DNA (Yoshikawa et al. 1998). Similarly, because of the absence of reference strains and a standardized panel of restriction enzymes, RFLP profile cannot be effectively compared with other data obtained in a different laboratory. In addition, a recent sequence study of the SSU rDNAs of a wide range of Blastocystis isolates from humans and animals revealed that it could not be guaranteed that the same RFLP profile would always show the same sequence. Therefore, RFLP analysis of SSU rDNA is not a suitable tool of identifying or classifying the genotypes of B. hominis (Arisue et al. 2003; Yoshikawa et al. 2004a). In the present study, 35 B. hominis isolates obtained from 19 asymptomatic healthy individuals and 16 patients with diarrhea and/or abdominal pain were genotyped by PCR using known seven pairs of STS primers.
Results from this study showed that at least four genotypes of B. hominis were found in China. In the present study, the most dominant genotype was subtype 3, which is similar to that reported from four geographically distinct countries (Japan, Bangladesh, Pakistan, and Germany) by Yoshikawa et al. (2004a). However, the prevalence (18/35) of subtype 1 of B. hominis isolates obtained from China was higher than that (13/102) reported in subtype classification of B. hominis populations isolated from five geographically distinct countries by Yoshikawa et al.(2004a). Based on the recent molecular studies of B. hominis isolates, from humans and animals, subtypes 1–5 and 7 were zoonotic genotypes and subtype 6 is considered to be of animal origin because this genotype is only observed in the isolates from pigs and cattle (Abe et al. 2003a–c; Yoshikawa et al. 2003, 2004a,b). In this study, subtype 6 was not detected among 35 human isolates, further suggesting that this subtype is of animal origin. Because none of the isolates was classified as subtypes 4–7, and subtype 2 was only detected in two isolates from asymptomatic healthy individuals, the genotypes of subtype 2 and subtypes 4–7 appeared to be irrelevant with pathogenicity. Similar results were reported by Yoshikawa et al. (2004a).
In this study, one isolate, HC05-10, was negative with all STS primers in the PCR analysis. But its SSU rRNA genes were amplified using a pair of sense (SR1F) and anti-sense (SR1R) primers (Yoshikawa et al. 2000), and produced an approximately 1,800-bp product (data not showed). Because phylogenetic analysis of the SSU rRNA gene of isolate HC05-10 was not carried out, it is uncertain whether isolate HC05-10 was classified as an additional new clade or the clade inferred from the full SSU rRNA sequences of two isolates, HJ00-4 and HJ00-5, obtained from Japan (Yoshikawa et al. 2003). Because this isolate was collected from a patient with intestinal symptoms, further study might facilitate our understanding of its pathogenic potential.
The main controversy about B. hominis is its role in human disease. Because extensive genetic diversity has been demonstrated among B. hominis isolates from humans (Yoshikawa et al. 1996, 1998, 2000, 2003, 2004a; Clark 1997; Böhm-Gloning et al. 1997; Hoevers et al. 2000; Arisue et al. 2003; Noël et al. 2003), it has been postulated that certain demes or genetically distinct genotypes of B. hominis may exhibit pathogenicity. Because there is no suitable animal model available for B. hominis infection at present, the pathogenic potential of B. hominis cannot be demonstrated experimentally (Tan et al. 2002). Therefore, to reveal a possible correlation between certain genotypes and the pathogenic potential of this parasite, comparative studies of human B. hominis populations from geographically separate countries or of isolates from clinically symptomatic and asymptomatic patients were performed. Although there have been a few trials which have examined genotypes correlating with the pathogenic potential of this parasite, using RFLP analysis of SSU rDNA or PCR analysis with known STS primers, there was only a possible relationship or no distinct differences in genotypes between isolates from symptomatic and asymptomatic groups (Böhm-Gloning et al. 1997; Kaneda et al. 2001; Yoshikawa et al. 2004a). To this end, two populations of B. hominis isolated from the asymptomatic people and from symptomatic patients were also compared with the genotype classification. Because significant differences in frequency of subtypes between the two populations were observed when using likelihood-ratio chi-square test (Table 3) (χ2=10.483, p=0.012), it appeared that subtype 1 may be responsible for intestinal symptoms. As fecal specimens were not examined for bacteria and viruses causing intestinal symptoms and the sample sizes were too small in the present study, it might be unwise to directly correlate subtype 1 of B. hominis with intestinal symptoms. More studies are warranted to confirm this issue.
References
Abe N, Wu Z, Yoshikawa H (2003a) Molecular characterization of Blastocystis isolates from birds by PCR with diagnostic primers and restriction fragment length polymorphism analysis of the small subunit ribosomal RNA gene. Parasitol Res 89:393–396
Abe N, Wu Z, Yoshikawa H (2003b) Molecular characterization of Blastocystis isolates from primates. Vet Parasitol 113:321–325
Abe N, Wu Z, Yoshikawa H (2003c) Zoonotic genotypes of Blastocystis hominis detected in cattle and pigs by PCR with diagnostic primers and restriction fragment length polymorphism analysis of the small subunit ribosomal RNA gene. Parasitol Res 90:124–128
Arisue N, Hashimoto T, Yoshikawa H (2003) Sequence heterogeneity of the small subunit ribosomal RNA genes among Blastocystis isolates. Parasitology 126:1–9
Böhm-Gloning B, Knobloch J, Walderich B (1997) Five subgroups of Blastocystis hominis isolates from symptomatic and asymptomatic patients revealed by restriction site analysis of PCR-amplified 16S-like rDNA. Trop Med Int Health 2:771–778
Clark CG (1997) Extensive genetic diversity in Blastocystis hominis. Mol Biochem Parasitol 87:79–83
Clark CG (2000) Cryptic genetic variation in parasitic protozoa. J Med Microbiol 49:489–916
Clark CG, Diamond LS (2002) Methods for cultivation of luminal parasitic protists of clinical importance. Clin Microbiol Rev 15:329–341
Hoevers J, Holman P, Logan K, Hommel M, Ashford R, Snowden K (2000) Restriction-fragment-length polymorphism analysis of small-subunit rRNA genes of Blastocystic hominis isolates from geographically diverse human hosts. Parasitol Res 86:57–61
Jones WR (1946) The experimental infection of rats with Entamoeba histolytica. Ann Trop Med Parasitol 40:130
Kaneda Y, Horiki N, Cheng XJ, Fujita Y, Maruyama M, Tachibana H (2001) Ribodemes of Blastocystis hominis isolated in Japan. Am J Trop Med Hyg 65:393–396
Noël C, Peyronnet C, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Sogin ML, Capron M, Viscogliosi E, Zenner L (2003) Phylogenetic analysis of Blastocystis isolates from different hosts based on the comparison of small-subunit rRNA gene sequences. Mol Biochem Parasitol 126:119–123
Noël C, Dufernez F, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Ho LC, Singh M, Wintjens R, Sogin ML, Capron M, Pierce R, Zenner L, Viscogliosi E (2005) Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species, and zoonosis. J Clin Microbiol 43:348–355
Stenzel DJ, Boreham PFL (1996) Blastocystis hominis revisited. Clin Microbiol Rev 9:563–584
Tan KSW, Singh M, Yap EH (2002) Recent advances in Blastocystis hominis research: hot spots in terra incognita. Int J Parasitol 32:789–804
Yoshikawa H, Nagano I, Yap EH, Singh M, Takahashi Y (1996) DNA polymorphism revealed by arbitrary primers polymerase chain reaction among Blastocystis strains isolated from humans, a chicken, and a reptile. J Eukaryot Microbiol 43:127–130
Yoshikawa H, Nagano I, Wu Z, Yap EH, Singh M, Takahashi Y (1998) Genomic polymorphism among Blastocystis strains and development of subtype-specific diagnostic primers. Mol Cell Probes 12:153–159
Yoshikawa H, Abe N, Iwasawa M, Kitano S, Nagano I, Wu Z, Takahashi Y (2000) Genomic analysis of Blastocystis hominis strains isolated from two long-term health care facilities. J Clin Microbiol 38:1324–1330
Yoshikawa H, Wu Z, Nagano I, Takahashi Y (2003) Molecular comparative studies among Blastocystis isolates obtained from humans and animals. J Parasitol 89:585–594
Yoshikawa H, Wu Z, Kimata I, Iseki M, Ali IK, Hossain MB, Zaman V, Haque R, Takahashi Y (2004a) Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol Res 92:22–29
Yoshikawa H, Abe N, Wu Z (2004b) PCR-based identification of zoonotic isolates of Blastocystis from mammals and birds. Microbiology 150:1147–1151
Acknowledgements
This work was supported by the Provincial Education Office of Jiangxi (Grant No. 251) and Medical Research Foundation of Gannan Medical College (No. 200538).
The experiments comply with the current regulations of People’s Republic of China.
The two authors, Shuilian Su and Riyong Lai, contributed equally to this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yan, Y., Su, S., Lai, R. et al. Genetic variability of Blastocystis hominis isolates in China. Parasitol Res 99, 597–601 (2006). https://doi.org/10.1007/s00436-006-0186-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-006-0186-z