Abstract
Fecal specimens were obtained from a total of 413 dairy calves from farms in Vermont, New York, Pennsylvania, Maryland, Virginia, North Carolina, and Florida. After removal of fecal debris by sieving and density gradient centrifugation, specimens were examined by fluorescence microscopy, polymerase chain reaction (PCR), and DNA sequencing analysis for the presence of microsporidia. Microscopic examination revealed no spores. PCR using generic primers for microsporidia revealed 70 positive calves. PCR was then conducted using specific primers for Enterocytozoon bieneusi, the most frequently found microsporidian in human infections. These primers revealed 13 positive calves from six farms in five states. DNA sequencing analysis of the 13 E. bieneusi-positive specimens confirmed the PCR results and indicated 96.8–99.8% similarity with E. bieneusi sequences in GenBank. This is the first report of E. bieneusi in cattle in North America.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fourteen species of microsporidia have been identified as opportunistic or emerging pathogens of humans. Enterocytozoon bieneusi, first identified in a male patient with acquired immunodeficiency syndrome (Desportes et al. 1985), is the species of microsporidia reported most frequently in humans with microsporidiosis (Reetz et al. 2002). Recent reports suggest that E. bieneusi is a potential zoonotic pathogen. Several genotypes of E. bieneusi have been identified in farm and companion animals in Europe, including pigs, cattle, cats, dogs, a llama, and chickens (Deplazes et al. 1996; Breitenmoser et al. 1999; Mathis et al.1999; Rinder et al. 2000; Dengjel et al. 2001; Reetz et al. 2002). Several genotypes were reported from swine at a slaughterhouse in Massachusetts (Buckholt et al. 2002). Additional genotypes have been reported from wildlife including beavers, muskrats, otters, foxes, and raccoons in the United States (Sulaiman et al. 2003), and in laboratory rhesus monkeys (Mansfield et al. 1998). The biological significance of these numerous genotypes, including any influence on the range of potential hosts, is unknown. However, because organisms found in animals and those found in humans differ by only a few base pairs in gene sequences, the potential for transmission from animals to humans is of concern.
Of 88 cattle examined from herds in Germany, E. bieneusi has been identified in eight calves (Rinder et al. 2000; Dengjel et al. 2001). Objects resembling microsporidian spores have been observed occasionally during routine microscopic examinations of feces from calves in our laboratory for many years, without confirmation or documentation of the observations. The present study was undertaken to specifically determine if microsporidia were present in calves on several farms in different geographic areas in North America using molecular methods to confirm the identity of positive specimens.
Materials and methods
Sources and collection of specimens
Feces were collected between June and October 2002 from a total of 413 calves. All calves were less than 42 days old, located on three dairy farms in Vermont and on two dairy farms each in New York, Pennsylvania, Maryland, Virginia, North Carolina, and Florida (Table 1). Most calves were purebred or mixed-breed Holstein stock. Eleven of the 15 farms were selected based on having approximately 20 or more calves that could be examined during a single visit. Calves were housed under a wide variety of conditions including individual hutches distant from adult cattle, individual wire enclosures within a single large shed, individual solid-wall stalls within a barn, and large pens with ten or more calves. Feces were collected directly from the rectum of each calf into plastic specimen cups that were immediately capped, labeled to identify the source, and placed on ice. Feces were transported to the USDA laboratory in Beltsville, Maryland on ice in insulated containers, and processed within 3 days of collection.
Cleaning of specimens from feces
Fifteen grams of feces from each of the 413 calves was suspended in 35–50 ml of deionized H2O and passed through a number 325-mesh wire screen. The sieved fecal suspension was further cleaned of fecal debris by centrifugation in capped 50-ml conical centrifuge tubes at 1,800 g for 10 min. Supernatant was decanted and the pellet resuspended in deionized H2O to 25 ml. The suspension was thoroughly mixed with an additional 25 ml of cesium chloride (1.4 g/ml) and centrifuged at 300 g for 20 min. Next, 4 ml of supernatant were aspirated from the top, pipetted into a 15-ml conical centrifuge tube, and deionized H2O was added to reach a final volume of 15 ml. The suspension was centrifuged at 1,800 g for 10 min, the supernatant decanted, and the pellet resuspended in 500 μl of deionized H2O to be examined by microscopic and molecular methods for the presence of microsporidia.
Microscopic examination
For microscopic examination of each fecal specimen, 15 μl of the suspended pellet was aspirated, pipetted onto a glass microscope slide coated with poly-l-lysine, and dried overnight. The dried preparation was fixed with methanol, dried, stained with Calcofluor white reagent (Becton Dickinson, Sparks, Md.), and examined with the aid of 40× and 100× oil immersion objectives on a Zeiss Axioskop fluorescence microscope at a wavelength of 470 nm.
DNA extraction
Using a QIAampTissue Kit (Qiagen, Valencia, Calif.) with a slightly modified protocol, total DNA was extracted from the suspensions cleaned of fecal debris. The protocol, described below, utilized reagents provided by the manufacturer and 50 μl of processed feces resuspended in 180 μl of ATL buffer thoroughly mixed by vortexing. To this suspension, 20 μl of proteinase K (20 mg/ml) was added, and the sample was mixed by vortexing. After incubating the mixture at 55°C overnight, 200 μl of AL buffer was added. The remaining protocol followed manufacturer's instructions with one exception. To increase the final DNA concentration in the elute, the nucleic acid was eluted in 100 μl of AE buffer.
PCR analysis
PCR analysis of small-subunit rRNA (SSU-rRNA) of E. bieneusi was performed with generic and specific primers. Generic primers designed to amplify DNA from all known human microsporidian species were used to screen all samples. All samples found positive with generic primers were subjected to a second PCR using E. bieneusi-specific primers.
The following generic primers Micro F (5′-CACCAGGTTGATTCTGCCTGA-3′) and Micro R (5′-CCTCTCCGGAACCAAACCCTG-3′) were used to generate an amplicon of 250 bp for E. bieneusi. These were based on primers previously reported (Fedorko et al. 1995, 2001; Fournier et al. 2002; Thurston-Enriquez et al. 2002). The PCR program was as follows: 95°C for 1 min, 40 cycles at 94°C for 45 s, 58°C for 20 s, and 72°C for 40 s, followed by a final extension at 72°C for 5 min.
Specific primers Eb F (5′-GCCTGACGTAGATGCTAGTC-3′) and Eb R (5′-ATGGTTCTCCAACTGAAACC-3′) were used to generate an amplified fragment of 1,294 bp (David et al. 1996). The PCR program was as follows: 94°C for 3 min, 35 cycles at 94°C for 75 s, 55°C for 1 min and 72°C for 2 min. Amplification was conducted using a 50 μl suspension of the following reagents: 10 mM Tris-HCl (pH 9), 50 mM KCl, 1.5 mM MgCl2, 0.2 mg/ml BSA, 1 μM of each primer, 0.2 mM of each deoxynucleotide triphosphate, and 2.5 U of Taq DNA polymerase (Qbiogene, Carlsbad, Calif.). PCR was performed with the aid of a PTC-200 thermocycler (Pertier Thermal Cycler, MJ Research, Waltham, Mass.). A negative control, consisting of a reaction mixture without the DNA template, was included in each experiment. As a positive control, DNA was extracted from E. bieneusi spores obtained from a human source. PCR products were analyzed on 1% agarose gel and visualized by ethidium bromide staining.
Sequence analysis
PCR products were purified using EXO-SAP enzyme (USB, Cleveland, Ohio). Purified products were sequenced using the same PCR primers in 10 μl reactions, Big Dye chemistries, and an ABI3100 sequencer analyzer (Applied Biosystems, Foster City, Calif.). Both strands of DNA were sequenced for each sample. Sequence chromatograms from each strand were aligned and inspected using Lasergene software (DNASTAR, Madison, Wis.). SSU-rRNA coding sequences from microsporidia detected in the present study were compared to SSU-rRNA sequences from GenBank.
Determination of minimum detection levels
Tests were conducted to determine the minimum level of detection for microsporidian spores in bovine feces by microscopic examination and PCR. Clean spores suspended in water were counted ten times in wells of three-well glass microscope slides and then thoroughly mixed with calf feces at the rate of 100 and 1,000 spores/g of feces. Ten, 15-g samples at each concentration of spores were cleaned of fecal debris using the same protocol as for farm derived specimens described above. After staining with Calcofluor white, each sample was examined by fluorescence microscopy to determine the number of samples in which spores could be detected. From two additional batches of 10, 15-g fecal samples spiked with either 100 or 1,000 spores/g of feces and cleaned of fecal debris, DNA was extracted and subjected to PCR using generic primers as described above for farm derived specimens.
Results
Detection levels in spiked specimens
At the spiking concentration of 100 spores/g of feces, spores were not observed in any of the ten specimens examined. At the spiking concentration of 1,000 spores/g, spores were detected in one of the ten specimens examined. Using generic PCR primers, at an original concentration of 100 spores/g of feces, none of the ten specimens was found positive; at 1,000 spores/g six of ten specimens were positive.
Detection of microsporidia from calves on farms
Of the 413 fecal specimens examined from calves on 15 farms in seven states (Table 1), spores of microsporidia were not detected by microscopic methods in any specimens after staining with Calcofluor white and examination by fluorescence microscopy.
Using generic PCR primers, 70 calves from 11 of 15 farms were found positive (Table 1). When specific PCR primers for E. bieneusi were used to further examine these 70 positive specimens, 13 calves from six farms were found positive (Table 1).
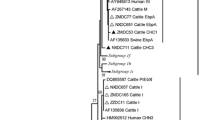
All PCR positive specimens were sequenced. The SSU-rRNA coding sequence of this microsporidian was compared to other SSU-rRNA sequences from GenBank. All SSU-rRNA gene sequences from E. bieneusi from the 13 calves were identical to one another. The sequence has been submitted to GenBank (accession no. AY257180). The sequence exhibited 96.8%–99.8% similarity with the E. bieneusi SSU-rRNA sequences reported by Hartskeerl et al. (1993), Zhu et al. (1993), DaSilva et al. (1996), Mathis et al. (1999), and Chalifoux et al. (2000) (GenBank L16868, L07123, AF024657, AF119100, and AF023245, respectively). The E. bieneusi SSU-rRNA gene from calves in the present study differed consistently from the human and rhesus monkey isolates at only two positions within the total 1,069 bp analyzed. At positions 193 and 388, calf isolates had a T and G whereas the primate isolates had G and A.
Using DNA sequence analysis for positive samples with generic primers (but not specific primers for E. bieneusi) unknown microsporidia species were identified, which were close to the insect microsporidia Orthosemella and Endoreticulatus.
Discussion
The present findings demonstrate for the first time the presence of microsporidia in cattle in North America. Over a widely dispersed geographic area from Vermont to Florida, calves on 11 of 15 farms tested positive with generic primers for microsporidia by PCR. Positive PCR specimens resulting from the use of generic primers represented microsporidia that could be parasites of invertebrates ingested with food or water. However, calves on six of these farms also tested positive with specific primers for E. bieneusi by PCR, and gene sequence data supported these findings. This reflects previous findings of E. bieneusi in European cattle (Rinder et al. 2000; Dengjel et al. 2001). These findings of E. bieneusi implicate calves as a potential source of this pathogen.
The sequences obtained in the present study showed 96.8–99.8% similarity with the E. bieneusi SSU-rRNA reported by Hartskeerl et al. (1993), Zhu et al. (1993), DaSilva et al. (1996), Mathis et al. (1999) and Chalifoux et al. (2000). Many genotypes have been reported for E. bieneusi based on the characterization of the internal transcribed spacer of the rRNA gene. Isolates from approximately 79 humans, rhesus monkeys, pigs, cattle, cats, and a llama resulted in 14 different genotypes (Dengjel et al. 2001). Phylogenetic analysis of the sequences indicated no potential transmission barrier between isolates from humans and cats, pigs, and cattle (Dengjel et al. 2001). Isolates from 59 fur-bearing mammals, including beavers, muskrats, otters, foxes, and raccoons, identified 15 genotypes of E. bieneusi (Sulaiman et al. 2003). Some isolates from muskrats and raccoons formed two distinct groups, but others belonged to a major group consisting of all the previously described E. bieneusi genotypes from human and non-human sources. Results of the fur-bearing mammal study indicated that these animals, usually closely associated with surface waters, could also serve as potential sources of human-pathogenic E. bieneusi. But they could also serve as sources of infection for other animals, including cattle, either by contamination of water or by close contact. Nearly 30% of raccoons, animals frequently found in or near barns and animal feed storage areas, were found positive for E. bieneusi (Sulaiman et al. 2003). A similarly high percentage (30.5%) of infection with E. bieneusi was detected in apparently healthy pigs shipped from Pennsylvania, Massachusetts, New Hampshire, and Vermont to a slaughterhouse in Massachusetts, suggesting that swine could also be a significant source of environmental contamination with this pathogen (Buckholt et al. 2002).
Whether feed, water, or bedding were contaminated with spores from cattle within the herds, from other animals on the dairy farms, from humans caring for the calves, from wild animals, from birds, or from insect vectors, remains unknown. A farm with a high percentage of generic PRC positive specimens (NC-2) had a noticeably high fly population with many flies found in drinking water buckets. Of the 13 fecal specimens from this farm that were PCR positive using the generic primers, only two demonstrated similarity with E. bieneusi. The other 11 specimens demonstrated similarity with sequences in GenBank for microsporidia reported from insects. Fournier et al. (2002) detected an unknown microsporidia that was genetically similar to the insect microsporidian, Endoreticulatus in a swimming pool.
Because microscopic detection of microsporidian spores in calf feces spiked with 100 and 1,000 spores/g was only 0 and 10%, respectively, the lack of positive microscopic identification of spores in fecal specimens from the 413 calves in the present study suggests that if any calves from farms in this study were excreting spores they were likely at a concentration of less than 1,000 spores/g of feces. Utilizing PCR and generic primers, the detection of spores in calf feces spiked with 100 and 1,000 spores/g was 0 and 60%, respectively. Based on these findings, at a concentration of 1,000 spores/g of feces the actual number of positive calves would be underestimated by 40%. By this reckoning it is likely that the number of infected calves detected in the present study is an underestimation of the actual number of infected calves on the farms examined.
Our findings suggest that cattle could serve as potential reservoir hosts for E. bieneusi. Although the transmission of this pathogen from cattle or other animals to humans appears possible, and likely, based on the genetic identities of isolates from humans and animals, a specific link has not yet been demonstrated nor strongly suspected based on epidemiological evidence associated with any specific case.
References
Breitenmoser AC, Mathis A, Burgi E, Weber R, Deplazes P (1999) High prevalence of Enterocytozoon bieneusi in swine with four genotypes that differ from those identified in humans. Parasitology 118:447–453
Buckholt MA, Lee JH, Tzipori S (2002) Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol 68:2595-2599
Chalifoux LV, Carville A, Pauley D, Thompson B, Lackner AA, Mansfield KG (2000) Enterocytozoon bieneusi as a cause of proliferative serositis in simian immunodeficiency virus-infected immunodeficient macaques (Macaca mulatta). Arch Pathol Lab Med 124:1480–1484
DaSilva AJ, Schwartz DA, Visvesvara GS, Moura H, Slemenda SB, Pieniazek NJ (1996) Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small subunit rRNA. J Clin Microbiol 34:986–987
David F, Schuitema AR, Sarfati C, Liguory O, Hartskeerl RA, Derouin F, Molina JM (1996) Detection and species identification of intestinal microsporidia by polymerase chain reaction in duodenal biopsies from human immunodeficiency virus-infected patients. J Infect Dis174:874–877
Dengjel B, Zahler M, Hermanns W, Heinritz K, Spillmann T, Thomschke A, Loscher T, Gothe R, Rinder H (2001) Zoonotic potential of Enterocytozoon bieneusi. J Clin Microbiol 39:4495–4499
Deplazes P, Mathis A, Muller C, Weber R (1996) Molecular epidemiology of Encephalitozoon cuniculi and first detection of Enterocytozoon bieneusi in faecal samples of pigs. J Eukaryot Microbiol 43:93S
Desportes I, Charpentier Y, Galian A, Bernard F, Cochand-Priollet B, Lavergne A, Ravisse P, Modigliani R (1985) Occurrence of a new microsporidian: Enterocytozoon bieneusi n. g., n. sp., in the enterocytes of a human patient with AIDS. J Protozool 32:250–254
Fedorko DP, Nelson NA, Cartwright CP (1995) Identification of microsporidia in stool specimens by using PCR and restriction endonucleases. J Clin Microbiol 33:1739–1741
Fedorko DP, Nelson NA, Didier ES, Bertucci D, Delgado RM, Hruszkewycz AM (2001) Speciation of human microsporidia by polymerase chain reaction single-strand conformation polymorphism. Am J Trop Med Hyg 65:397–401
Fournier S, Dubrou S, Liguory O, Gaussin F, Santillana-Hayat M, Sarfati C, Molina JM, Derouin F (2002) Detection of microsporidia, cryptosporidia and Giardia in swimming pools: a one-year prospective study. FEMS Immunol Med Microbiol 33:209–213
Hartskeerl RA, Schuitema AR, van Gool T, Terpstra WJ (1993) Genetic evidence for the occurrence of extra-intestinal Enterocytozoon bieneusi infections. Nucleic Acids Res 21:4150
Mansfield KG, Carville A, Hebert D, Chalifoux L, Shvetz D, Lin KC, Tzipori S, Lackner AA (1998) Localization of persistent Enterocytozoon bieneusi infection in normal rhesus macaques (Macaca mulatta) to the hepatobiliary tree. J Clin Microbiol 36:2336–2338
Mathis A, Breitenmoser AC, Deplazes P (1999) Detection of new Enterocytozoon genotypes in faecal samples of farm dogs and a cat. Parasite 6:189–193
Reetz J, Rinder H, Thomschke A, Manke H, Schwebs M, Bruderek A (2002) First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens). Int J Parasitol 32:785–787
Rinder H, Thomschke A, Dengjel B, Gothe R, Loscher T, Zahler M (2000) Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J Parasitol 86:185–188
Sulaiman I, Fayer R, Lal AA, Trout JM, Schaeffer F, Xiao L (2003) Molecular characterization of microsporidia indicates that fur-bearing mammals can be a source of human pathogenic Enterocytozoon bieneusi. Emerg Inf Dis (in press)
Thurston-Enriquez JA, Watt P, Dowd SE, Enriquez R, Pepper IL, Gerba CP (2002) Detection of protozoan parasites and microsporidia in irrigation waters used for crop production. J Food Protect 65:378–382
Zhu X, Wittner M, Tanowitz HB, Kotler D, Cali A, Weiss LM (1993) Small subunit rRNA sequence of Enterocytozoon bieneusi and its potential diagnostic role with use of the polymerase chain reaction. J Infect Dis 168:1570–1575
Acknowledgements
The authors thank Robert Palmer, Kristie Ludwig, and Courtney Anderson for technical services in support of this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fayer, R., Santín, M. & Trout, J.M. First detection of microsporidia in dairy calves in North America. Parasitol Res 90, 383–386 (2003). https://doi.org/10.1007/s00436-003-0870-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-003-0870-1