Abstract
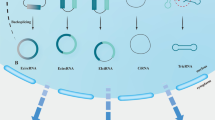
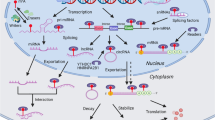
N6-methyladenosine (m6A), the richest post-transcriptional modification of RNA in eukaryotic cells, is dynamically installed/uninstalled by the RNA methylase complex (“writer”) and demethylase (“eraser”) and recognized by the m6A-binding protein (“reader”). M6A modification on RNA metabolism involves maturation, nuclear export, translation and splicing, thereby playing a critical role in cellular pathophysiology and disease processes. Circular RNAs (circRNAs) are a class of non-coding RNAs with a covalently closed loop structure. Due to its conserved and stable properties, circRNAs could participate in physiological and pathological processes through unique pathways. Despite the recent discovery of m6A and circRNAs remains in the initial stage, research has shown that m6A modifications are widespread in circRNAs and regulates circRNA metabolism, including biogenesis, cell localization, translation, and degradation. In this review, we describe the functional crosstalk between m6A and circRNAs, and illustrate their roles in cancer development. Moreover, we discuss the potential mechanisms and future research directions of m6A modification and circRNAs.



Similar content being viewed by others
Data availability
Not applicable.
References
Abdelmohsen K et al (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14(3):361–369
Alarcon CR et al (2015a) HNRNPA2B1 Is a mediator of m(6)A-Dependent nuclear RNA processing events. Cell 162(6):1299–1308
Alarcon CR et al (2015b) N6-methyladenosine marks primary microRNAs for processing. Nature 519(7544):482–485
Arguello AE, DeLiberto AN, Kleiner RE (2017) RNA chemical proteomics reveals the N(6)-Methyladenosine (m(6)A)-Regulated Protein-RNA Interactome. J Am Chem Soc 139(48):17249–17252
Aufiero S et al (2019) Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol 16(8):503–514
Barbagallo D et al (2019) CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers (basel) 11(2):194
Barbieri I et al (2017) Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552(7683):126–131
Bokar JA et al (1994) Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei internal mRNA methylation requires a multisubunit complex. J Biol Chem 269(26):17697–17704
Cai X et al (2018) HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett 415:11–19
Cai J et al (2022) CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol Ther Oncolytics 24:755–771
Capel B et al (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73(5):1019–1030
Chao CW et al (1998) The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 4(9):614–628
Chen LL (2020) The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21(8):475–490
Chen XY, Zhang J, Zhu JS (2019a) The role of m(6)A RNA methylation in human cancer. Mol Cancer 18(1):103
Chen X et al (2019b) Circular RNAs in immune responses and immune diseases. Theranostics 9(2):588–607
Chen RX et al (2019c) N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun 10(1):4695
Chen C et al (2021) N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics 11(9):4298–4315
Chen Y et al (2022) Activation of YAP1 by N6-methyladenosine-modified circCPSF6 drives malignancy in hepatocellular carcinoma. Cancer Res 82(4):599–614
Chi F, Cao Y, Chen Y (2021) Analysis and validation of circRNA-miRNA network in regulating m(6)A RNA methylation modulators reveals CircMAP2K4/miR-139-5p/YTHDF1 axis involving the proliferation of hepatocellular carcinoma. Front Oncol 11:560506
Chibwesha CJ, Stringer JSA (2019) Cervical cancer as a global concern: contributions of the dual epidemics of HPV and HIV. JAMA 322(16):1558–1560
Cocquerelle C et al (1993) Mis-splicing yields circular RNA molecules. FASEB J 7(1):155–160
Conn SJ et al (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160(6):1125–1134
Danan M et al (2012) Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res 40(7):3131–3142
Deng X et al (2018) RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res 28(5):507–517
Desrosiers R, Friderici K, Rottman F (1974) Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 71(10):3971–3975
Di Timoteo G et al (2020) Modulation of circRNA metabolism by m(6)A modification. Cell Rep 31(6):107641
Diaz-Diaz A et al (2020) Heat shock protein 90 chaperone regulates the E3 ubiquitin-ligase hakai protein stability. Cancers (basel) 12(1):215
Du WW et al (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44(6):2846–2858
Du A et al (2022) M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer 21(1):109
Du, H. et al., YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nature Communications, 2016. 7.
Duan HC et al (2017) ALKBH10B Is an RNA N(6)-Methyladenosine demethylase affecting arabidopsis floral transition. Plant Cell 29(12):2995–3011
Duan JL et al (2022) A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer 21(1):93
Dubin RA, Kazmi MA, Ostrer H (1995) Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167(1–2):245–248
Dudekulay DB et al (2016) CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42
Fan HN et al (2022) METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer 21(1):51
Fischer JW et al (2020) Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell 78(1):70–84
Goh YT et al (2020) METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res 48(16):9250–9261
Goodall GJ, Wickramasinghe VO (2021) RNA in cancer. Nat Rev Cancer 21(1):22–36
Grossman JG et al (2018) Recruitment of CCR2(+) tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology 7(9):e1470729
Guarnerio J et al (2016) Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165(2):289–302
Guo Y et al (2021) Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer 20(1):93
Han Y et al (2014) Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343(6176):1244–1248
Han D et al (2019) Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 566(7743):270–274
Hansen TB et al (2011) miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30(21):4414–4422
Hansen TB et al (2013a) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Hansen TB, Kjems J, Damgaard CK (2013b) Circular RNA and miR-7 in cancer. Cancer Res 73(18):5609–5612
Holdt LM et al (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7:12429
Horiuchi K et al (2013) Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 288(46):33292–33302
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280(5720):339–340
Hsu PJ et al (2017) Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27(9):1115–1127
Hsu PJ et al (2019) The RNA-binding protein FMRP facilitates the nuclear export of N (6)-methyladenosine-containing mRNAs. J Biol Chem 294(52):19889–19895
Hu Y et al (2020) Oocyte competence is maintained by m(6)A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell Death Differ 27(8):2468–2483
Huang Y et al (2015) Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res 43(1):373–384
Huang H et al (2018a) Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20(3):285–295
Huang C et al (2018b) A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32(9–10):639–644
Huang R et al (2020) N(6)-Methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry 88(5):392–404
Ianniello Z, Paiardini A, Fatica A (2019) N(6)-Methyladenosine (m(6)A): A promising new molecular target in acute myeloid leukemia. Front Oncol 9:251
Ivanov A et al (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10(2):170–177
Jacobs E, Mills JD, Janitz M (2012) The role of RNA structure in posttranscriptional regulation of gene expression. J Genet Genomics 39(10):535–543
Jeck WR et al (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2):141–157
Jia R et al (2019) Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov 5:45
Jiang X et al (2021) The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 6(1):74
Kelley DR et al (2014) Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol 15(12):537
Knuckles P, Buhler M (2018) Adenosine methylation as a molecular imprint defining the fate of RNA. FEBS Lett 592(17):2845–2859
Knuckles P et al (2018) Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev 32(5–6):415–429
Komar AA, Hatzoglou M (2011) Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10(2):229–240
Kristensen LS et al (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20(11):675–691
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20(12):1829–1842
Lasda E, Parker R (2016) Circular RNAs Co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS ONE 11(2):e0148407
Lee M, Kim B, Kim VN (2014) Emerging roles of RNA modification: m(6)A and U-tail. Cell 158(5):980–987
Lei M et al (2020) Translation and functional roles of circular RNAs in human cancer. Mol Cancer 19(1):30
Lesbirel S et al (2018) The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci Rep 8(1):13827
Li F et al (2014) Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res 24(12):1490–1492
Li Z et al (2015a) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–264
Li Y et al (2015b) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25(8):981–984
Li J et al (2018) Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget 9(3):3752–3764
Li X, Tian G, Wu J (2021a) Novel circGFRalpha1 promotes Self-renewal of female germline stem cells mediated by m(6)A writer mettl14. Front Cell Dev Biol 9:640402
Li B et al (2021b) circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun 12(1):295
Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28(20):2233–2247
Liu J et al (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10(2):93–95
Liu N et al (2015) N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518(7540):560–564
Liu N et al (2017) N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 45(10):6051–6063
Liu ZX et al (2018) Link between m6A modification and cancers. Front Bioeng Biotechnol 6:89
Liu R et al (2019) SEMA3C promotes cervical cancer growth and is associated with poor prognosis. Front Oncol 9:1035
Liu CX et al (2019) Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177(4):865–880
Liu J et al (2020a) N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367(6477):580–586
Liu N et al (2020b) Role of non-coding RNA in the pathogenesis of depression. Gene 735:144276
Liu Z et al (2021a) N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer 20(1):105
Liu B et al (2021b) Circular RNA circZbtb20 maintains ILC3 homeostasis and function via Alkbh5-dependent m(6)A demethylation of Nr4a1 mRNA. Cell Mol Immunol 18(6):1412–1424
Luxton HJ et al (2019) The oncogene Metadherin interacts with the known splicing proteins YTHDC1, sam68 and T-STAR and plays a novel role in alternative mRNA splicing. Cancers (basel) 11(9):1233
Ma JZ et al (2017) METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 65(2):529–543
Mackie GA (1998) Ribonuclease E is a 5’-end-dependent endonuclease. Nature 395(6703):720–723
Mauer J et al (2017) Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature 541(7637):371–375
Memczak S et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Mendel M et al (2018) Methylation of structured rna by the m(6)a writer mettl16 is essential for mouse embryonic development. Mol Cell 71(6):986–1000
Meng S et al (2017) CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 16(1):94
Meyer KD, Jaffrey SR (2017) Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol 33:319–342
Meyer KD et al (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149(7):1635–1646
Meyer KD et al (2015) 5’ UTR m(6)A promotes cap-independent translation. Cell 163(4):999–1010
Nicolet BP et al (2018) Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res 46(16):8168–8180
Nigro JM et al (1991) Scrambled exons. Cell 64(3):607–613
Noto JJ, Schmidt CA, Matera AG (2017) Engineering and expressing circular RNAs via tRNA splicing. RNA Biol 14(8):978–984
Park OH et al (2019) Endoribonucleolytic Cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell 74(3):494–507
Patil DP et al (2016) m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537(7620):369–373
Pendleton KE et al (2017) The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase intron retention. Cell 169(5):824–835
Ping XL et al (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24(2):177–189
Preusser C et al (2018) Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles 7(1):1424473
Rao X et al (2021) N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life 73(2):408–417
Rong B et al (2020) Ribosome 18S m(6)A methyltransferase METTL5 promotes translation initiation and breast cancer cell growth. Cell Rep 33(12):108544
Rong D et al (2021) m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids 26:637–648
Roost C et al (2015) Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc 137(5):2107–2115
Rossi F et al (2019) Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene 38(20):3843–3854
Roundtree, I.A. et al., YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife, 2017. 6.
Salzman J et al (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9(9):e1003777
Shen Y et al (2017) Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep 18(4):536–548
Shen Y et al (2021) G protein-coupled oestrogen receptor promotes cell growth of non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. J Cell Mol Med 25(1):284–296
Shi H et al (2017) YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res 27(3):315–328
Shulman Z, Stern-Ginossar N (2020) The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat Immunol 21(5):501–512
Sun HD et al (2018) Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep 8(1):10111
Sun T, Wu R, Ming L (2019) The role of m6A RNA methylation in cancer. Biomed Pharmacother 112:108613
Taketo K et al (2018) The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol 52(2):621–629
Tang C et al (2018) ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A 115(2):E325–E333
Tang C et al (2020) m(6)A-dependent biogenesis of circular RNAs in male germ cells. Cell Res 30(3):211–228
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17(5):272–283
van Tran N et al (2019) The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res 47(15):7719–7733
Visvanathan A et al (2018) Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37(4):522–533
Vo JN et al (2019) The landscape of circular RNA in cancer. Cell 176(4):869–888
Vu LP et al (2017) The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23(11):1369–1376
Wang X et al (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481):117–120
Wang X et al (2015) N(6)-methyladenosine modulates messenger rna translation Efficiency. Cell 161(6):1388–1399
Wang P, Doxtader KA, Nam Y (2016a) Structural basis for cooperative function of mettl3 and mettl14 methyltransferases. Mol Cell 63(2):306–317
Wang X et al (2016b) Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534(7608):575–578
Wang K et al (2017) Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett 394:1–12
Wang T et al (2020) The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer 19(1):88
Wang X et al (2021) Emerging roles of circular RNAs in systemic lupus erythematosus. Mol Ther Nucleic Acids 24:212–222
Warda AS et al (2017) Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep 18(11):2004–2014
Wei J et al (2018) Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell 71(6):973–985
Wei W et al (2021) Circ0008399 interaction with WTAP promotes assembly and activity of the m(6)A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res 81(24):6142–6156
Wen J et al (2018) Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell 69(6):1028–1038
Wiener D, Schwartz S (2021) The epitranscriptome beyond m(6)A. Nat Rev Genet 22(2):119–131
Wimalasena VK et al (2020) Using chemical epigenetics to target cancer. Mol Cell 78(6):1086–1095
Wu B et al (2018) Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun 9(1):420
Wu R et al (2019a) A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res 29(1):23–41
Wu Y et al (2019b) m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer 18(1):87
Wu X et al (2020) Circular RNA: A novel potential biomarker for skin diseases. Pharmacol Res 158:104841
Wu P et al (2021) N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis 12(4):298
Xiao W et al (2016) Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell 61(4):507–519
Xie F et al (2021) CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer 20(1):68
Xu C et al (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10(11):927–929
Xu J et al (2020) N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating beta-catenin signaling. Mol Cancer 19(1):163
Yang Y et al (2017a) Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27(5):626–641
Yang Z et al (2017b) MicroRNA-145 modulates N(6)-Methyladenosine levels by targeting the 3’-Untranslated mRNA region of the N(6)-Methyladenosine binding YTH domain family 2 protein. J Biol Chem 292(9):3614–3623
Yang D et al (2018a) N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res 46(8):3906–3920
Yang Y et al (2018b) Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 28(6):616–624
Yang X et al (2019) m(6)A promotes R-loop formation to facilitate transcription termination. Cell Res 29(12):1035–1038
Yang X et al (2020) METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer 19(1):46
Zaccara S, Jaffrey SR (2020) A Unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell 181(7):1582–1595
Zaccara S, Ries RJ, Jaffrey SR (2019) Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20(10):608–624
Zaphiropoulos PG (1996) Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 93(13):6536–6541
Zhang Y et al (2013) Circular intronic long noncoding RNAs. Mol Cell 51(6):792–806
Zhang XO et al (2014) Complementary sequence-mediated exon circularization. Cell 159(1):134–147
Zhang XO et al (2016) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26(9):1277–1287
Zhang C et al (2020) Circular RNA expression profile and m6A modification analysis in poorly differentiated adenocarcinoma of the stomach. Epigenomics 12(12):1027–1040
Zhang Y et al (2021) circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion. Sci Rep 11(1):24357
Zhao BS, Roundtree IA, He C (2018) Publisher correction: post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 19(12):808
Zhao J et al (2019) Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun 10(1):2300
Zheng G et al (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49(1):18–29
Zhou J et al (2015) Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526(7574):591–594
Zhou C et al (2017) Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep 20(9):2262–2276
Zhou S et al (2018) FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting beta-catenin through mRNA demethylation. Mol Carcinog 57(5):590–597
Zhou KI et al (2019) Regulation of Co-transcriptional Pre-mRNA Splicing by m(6)A through the low-complexity protein hnRNPG. Mol Cell 76(1):70–81
Zhou WY et al (2020) Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 19(1):172
Funding
This work was supported by National Natural Science Foundation of China (No. 82002889).
Author information
Authors and Affiliations
Contributions
Leyu Zhang consulted relevant literature and write the paper. Jingwen Liu and Xi Wang acted as the assists. Wei Zhao is responsible for the designing and funding. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that there is no conflict of interest.
Ethical approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Wang, X., Zhao, W. et al. Overview of m6A and circRNAs in human cancers. J Cancer Res Clin Oncol 149, 6769–6784 (2023). https://doi.org/10.1007/s00432-023-04610-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04610-8