Abstract
Purpose
Colorectal cancer (CRC) is a severe health condition characterized by high mortalities. NudC domain containing 1 (NUDCD1) is abnormally upregulated in multiple tumors and is recognized as a cancer antigen. In CRC, NUDCD1 upregulation accelerates tumor progression by activating the IGF1R-ERK1/2 signaling pathway. Its specific regulatory mechanisms, however, remain unclear.
Methods
In the present study, we predicted the regulators of NUDCD1 and analyzed the expression profile of NUDCD1 in CRC tissues using the gene chip dataset. We also determined the regulation between miR-144, NUDCD1 and IGF1R-ERK1/2 signaling in vitro and in vivo. Then, the expression of miR-144 in CRC tissues was detected and its cell functions were verified in vitro.
Results
As predicted by bioinformatics, we found that NUDCD1 is a predicted target of miR-144 and confirmed that miR-144 directly binds to NUDCD1. In vitro and in vivo, miR-144 was determined to specifically regulate NUDCD1 expression and as such, can reduce the activity of the IGF1R-ERK1/2 signaling pathway. Moreover, miR-144 was significantly downregulated in CRC tissues; its levels were significantly negatively correlated with CRC primary range and lymph node metastasis. Cell function studies verified that miR-144 acts as a tumor suppressor, because it significantly inhibits the proliferation, metastasis, and invasion of CRC cells as well as inducing cell cycle arrest and apoptosis.
Conclusions
Our study demonstrates that miR-144 regulates IGF1R-ERK1/2 signaling via NUDCD1 to inhibit CRC cell proliferation and metastasis. The miR-144/NUDCD1/IGF1R-ERK1/2 signaling axis may be crucial in the progression of CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC), a common gastroenterological malignancy, is the second leading cause of cancer-related deaths worldwide (Bray et al. 2018). Recently, CRC cases have been rapidly increasing in China (Chen et al. 2016). Because it is asymptomatic in its early stages, CRC is typically diagnosed when it has progressed to a more advanced state. Metastatic CRC is even more deadly; patients with distant metastasis typically have low 5-year survival rates (Brenner et al. 2014). The underlying mechanisms of CRC remain unclear; its growth, invasion, and metastasis are the focus in oncology.
NudC domain-containing 1 (NUDCD1) is a 66 kDa cytoplasmic protein (Asselin-Mullen et al. 2017). NUDCD1 is involved in diverse biological processes, such as cell growth, apoptosis, and regulation of cell cycle (Han et al. 2018b; Shi et al. 2021; Song et al. 2019); its overexpression has been reported in multiple tumors (Song et al. 2019). Conversely, NUDCD1 silencing inhibits HeLa cell proliferation, metastasis, and invasion in vitro and in vivo (Wang et al. 2008). NUDCD1 can affect multiple tumor signaling pathways. For example, in NIH3T3 cells, NUDCD1 can hyper-activate the PI3K/AKT and ERK1/2 MAPK signaling pathways (Rao et al. 2014b). We previously demonstrated that NUDCD1 expression is increased in CRC tissues, while its downregulation inhibits CRC cell viability and proliferation and slows xenograft growth in nude mice by decreasing IGF1R-ERK1/2 signaling (Han et al. 2018b). The underlying effector molecules of NUDCD1, however, remain to be fully elucidated.
MicroRNA (miRNA) is a highly conserved gene family that inhibits transcription or triggers mRNA degradation by binding to its 3′-untranslated region (3′-UTR) (Goodall and Wickramasinghe 2021). The aberrant expression of miR-144 has been observed in several diseases and it is significantly downregulated in most tumors. For instance, low miR-144 expression has been observed in colorectal (Cheng et al. 2020; Ji et al. 2019) and other tumors (Cao et al. 2020; Zhou et al. 2020). miR-144 also functions as a tumor suppressor: it inhibits the invasion and metastasis of renal tumor cells by targeting the MAP3K8 signaling pathway (Liu et al. 2016) and activating a caspase-dependent apoptotic pathway (Ovcharenko et al. 2007). In CRC, cancer cell phenotypes induced by miR-144 overexpression are similar to those induced by NUDCD1 silencing (Han et al. 2018b). But, whether miR144 targets NUDCD1 and the link between them are unclear.
In this study, we found that miR-144 interacts with NUDCD1, and its expression is dramatically downregulated in CRC tissues and cells. We further verified that, in CRC, low miR-144 expression leads to the abnormal upregulation of NUDCD1, which then promotes CRC progression by activating the IGF1R-ERK1/2 signaling pathway. We provide novel evidence that miR-144 acts as a tumor suppressor in CRC development; accordingly, the negative regulatory network between miR-144 and NUDCD1 may be a potential target for CRC interventions.
Materials and methods
Prediction of NUDCD1 regulators
Eleven databases from the miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/mirwalk2) online database were used to predict the upstream regulatory miRNAs of NUDCD1 (Dweep et al. 2011). Among them, if a miRNA was predicted by more than half, it was considered to have a potential NUDCD1-binding domain.
Microarray data
The expression profile of NUDCD1 was analyzed using the GDS4382 gene chip dataset (provided by Ahmed Khamas et al.) from GEO Datasets in National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/gds), which contains data from paired samples consisting of CRC tumor and adjacent non-cancerous tissues.
Cell culture and CRC patient samples
The normal colorectal epithelial cell line NCM460 was purchased from the Shanghai Chinese Academy of Sciences cell bank. HEK293T and the CRC cell lines SW620, HCT116, HT29, SW480, and DLD1 were purchased from ATCC. The cells were cultured in DMEM medium containing 10% FBS and incubated at 37 °C with 5% CO2.
Seventy pairs of cancerous and adjacent normal tissues (parallel tissues ≥ 5 cm from the borders of cancerous tissues) were collected from CRC patients (mean age: 56.8 + 15.2 years) undergoing gastrointestinal surgical resection. Prior to surgery, the patients did not receive chemotherapy. The specimens were frozen in liquid nitrogen. Patient age, gender, primary range (T grade), lymph node invasion (N grade), distal metastasis (M grade), and Duke grade were also collected. The study was approved by the Ethics Committee of Sichuan University, and informed consent was obtained from all patients.
Plasmid construction and transfection
agomiR-144 and agomiR-NC were designed and synthesized by RiboBio (Guangzhou, China). A NUDCD1 overexpression plasmid was constructed by cloning the coding sequence of NUDCD1 into the pcDNA3.1 plasmid (NUDCD1), untreated pcDNA3.1 plasmid was used as the control (pcDNA3.1-NC). We synthesized the 3'-UTR sequence of NUDCD1, which contained either the wild-type (the mutant sites were designed referring to the reference (Guo et al. 2015)) or mutant seed region of miR-144. The synthesized sequences were inserted into pSicheck-2 vectors (Promega, Madison, WI) to construct the luciferase reporter vectors pSicheck-NUDCD1-wt and pSicheck-NUDCD1-mut. The untreated pSicheck-2 vector, pSicheck-NC, was used as the control.
The plasmids and vectors were constructed using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China), and the cell transfections were performed using Transfection Reagent (Vazyme, Nanjing, China).
Dual-luciferase reporter assay
When HEK293T cells grew to 80% confluency, they were transfected with either pSicheck-NUDCD1-wt, pSicheck-NUDCD1-mut, or pSicheck-NC and simultaneously co-transfected with agomiR-144. After 48 h, the cells were harvested, and their fluorescence intensities were measured using a GloMax 20/20 luminometer (Promega Corporation, Madison, WI) and a dual-luciferase reporter assay kit (Promega, Madison, WI). Relative luciferase activity was compared to that of cells transfected with pSicheck-NC.
In vivo tumorigenesis assay
Male, SPF grade, 4-week-old BALB/c nude mice weighing 18–22 g were purchased from Chengdu Dossy Experimental Animals (Chengdu, China). They were allowed 1 week of habituation with food and water freely available. The experimental procedures were performed in accordance with the regulations of the Animal Ethics Committee of Sichuan University. 1 × 106 HCT116 cells were subcutaneously injected into the ribs of the mice, and those successfully inoculated were randomly assigned into one of two groups. Every 4 days, either agomiR-144 or agomiR-NC at 2 nmol/50 μL were injected into the tumors using Entranster ™-in vivo (Engreen, Beijing, China). Tumor sizes were determined every 4 days using the following formula: tumor volume = length × width2 × 0.5. The mice were sacrificed 28 days after the first intra-tumoral injection. The tumors from all mice were collected, weighed, and frozen in − 80 °C for subsequent testing.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cells or tissues in this study was extracted using the E.Z.N.A. Total DNA/RNA/Protein kit (Omega Bio-Tek, USA), the operations were conducted strictly according to the instructions. The quality of nucleic acids was confirmed to be good by gel electrophoresis. First-strand cDNA was synthesized from 2 µg total RNA using the PrimeScript™ RT reagent Kit (Takara, Japan). The qPCR reactions were performed using the SYBR Green Master Mix (Vazyme, Nanjing, China) on a CFX96TM detection system (BioRad, USA). U6 and GAPDH were used, respectively, as references for miRNA-144 and NUDCD1. The results were analyzed using the 2−∆∆CT method (Livak and Schmittgen 2001). The primers are listed in Table S1.
Western blotting
Proteins from cells or tissues in this study were extracted using the E.Z.N.A. DNA/RNA/Protein kit according to the instructions, and their concentrations were measured using the BCA protein concentration assay kit (Beyotime, Shanghai, China). The protein samples were mixed with SDS-PAGE loading buffer and boiled for 10 min. The proteins were separated using 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore, Ireland). The membranes were blocked with 5% skim milk at room temperature for 2 h, incubated with the primary antibody at 4 °C overnight, and incubated with the secondary antibody for 1 h at room temperature. The following primary antibodies were used: NUDCD1 (ab126902, abcam, USA), Bcl-2 (ab182858, abcam, USA), E-cadherin (ab133597, abcam, USA), N-cadherin (ab207608, abcam, USA), cyclin D1 (ab16663, abcam, USA), caspase3 (ab32351, abcam, USA), IGF1R (ab59348, abcam, USA), p-IGF1R (ab39398, abcam, USA), ERK1/2 (ab184699, abcam, USA), p-ERK1/2 (ab223500, abcam, USA), and GAPDH (ab9485, abcam, USA). After incubation, the membrane was placed in ECL luminescent liquid (Beyotime, Shanghai, China) and exposed to a gel imaging system for signal capture. The bands were analyzed using Image Lab Software v3.0 and normalized to GAPDH.
Online bioinformatic analysis
OncomiR (http://www.oncomir.org/) was used to explore miR-144 dysregulation in CRC (Wong et al. 2018). Kaplan–Meier survival analysis for miR-144 across different cancer types was performed using the Kaplan–Meier Plotter (https://kmplot.com/analysis/) (Nagy et al. 2018). All of above survival analysis was based on the data of miR-144 expression in cancer patients from online database. FunRich version 3.1.4 (http://www.funrich.org/) was used for miR-144 functional enrichment analysis (Fonseka et al. 2020). Finally, protein–protein interaction (PPI) networks were explored using the Search Tool for the Retrieval of Interacting Genes/Proteins database (https://cn.string-db.org/) (Szklarczyk and Jensen 2015).
Cell proliferation assay
CRC cells transfected with agomiR-144 or agomiR-NC were seeded in 96-well plates at 5 × 103 cells/well. Each group had three replicate wells. The cells were left to adhere for 6 h. After the non-adherent cells were removed, we quantified the number of viable cells at 0, 24, 48, and 72 h using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega, Madison, WI).
Cell migration and invasion assay
After 48 h of transfection, CRC cells transfected with agomiR-144 or agomiR-NC were cultured in serum-free DMEM medium for 12 h to remove serum effects. The cells were digested with trypsin without EDTA (Beyotime, Shanghai, China), resuspended using serum-free DMEM medium, and inoculated into Transwell chambers (Merck Millipore, Germany) with or without Matrigel at 2 × 105 cells/well. The chambers were placed into 24-well plates, and 500 μL of 10% FBS DMEM medium was added into each well to cover the lower surface of the chambers. The 24-well plate was incubated at 37 °C in 5% CO2 for 24 h. Then, the cells in the lower surface of the Transwell chamber were immersed in methanol for 30 min and stained in 0.1% crystal violet solution for 15 min. After air drying, the cells were placed under a microscope. Five fields at 200× magnification were randomly taken from each chamber, and the number of transferred/invaded cells in each field was counted.
Cell cycle assay
Following 48 h of transfection, CRC cells transfected with agomiR-144 or agomiR-NC were digested with trypsin without EDTA and gently dispersed into cell suspensions. 70% pre-cooled ethanol (1 mL) was gently added to the cell suspensions to fix the cells for 12 h at 4 °C, after which the cells were incubated with 1 mg/mL RNase A for 30 min at 37 °C. The cells were subsequently stained with propidium iodide (PI) (Beyotime, Shanghai, China) and detected using flow cytometry. The cell cycle phase was evaluated using ModFit software.
Cell apoptosis assay
After 48 h of transfection, CRC cells transfected with agomiR-144 or agomiR-NC were digested with trypsin without EDTA, washed with PBS, and gently re-suspended in 100 μL of 1× binding buffer. Then, the suspended cells were gently mixed with 5 μL each of Annexin-V-FITC (Vazyme, Nanjing, China) and PI (Vazyme, Nanjing, China) for staining. After 10 min of incubation at room temperature in darkness, 400 μL of 1× binding buffer was added to quench the staining. Stained cells were detected using flow cytometry; cell apoptosis was evaluated using CFlow Plus v1.0.
Cell apoptosis observation
Following 48 h of transfection, CRC cells transfected with agomiR-144 or agomiR-NC were collected and washed with PBS. The cells were stained with 10 μg/mL Hoechst33342 (Beyotime, Shanghai, China) for 15 min at 37 °C, washed with PBS, and stained with 10 μg/mL PI (Beyotime, Shanghai, China) for 20 min at 4 °C. After staining, PBS was used to remove excess dye. Cell apoptosis and necrosis were observed under a fluorescence microscope.
Statistical analysis
All data were processed using SPSS v18.0. Data are expressed as means ± standard errors of the mean (SEM). For data that were normally distributed and had homogenous variances, comparisons between two groups were carried out using Student’s t test, while comparisons between three or more groups were conducted using one-way ANOVA followed by Dunnett’s post hoc tests. For data that violated assumptions of normality or homogeneity of variances, comparisons between two groups were performed using the Mann–Whitney test followed by Tamhane's T2 test, while comparisons between three or more groups were conducted using the Kruskal–Wallis test followed by Dunnett's T3 test. P < 0.05 was considered statistically significant.
Results
miR-144 directly targets NUDCD1
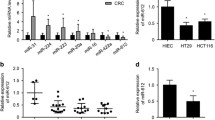
Potential regulators of NUDCD1 were searched in 11 miRNA target gene prediction databases, of which 7 predicted that NUDCD1 was a direct target gene of miR-144 (Fig. 1a). The GEO DataSet gene chip database analysis revealed that NUDCD1 was significantly upregulated in CRC tissues (Fig. 1b, c), as previously described (Han et al. 2018b). The 3′-UTR region of NUDCD1 was predicted as the target site of miR-144, so we examined relative miR-144 expression in tissue from 70 CRC patients. Indeed, miR-144 expression in CRC tissues was significantly lower than the adjacent normal tissues (P < 0.05; Fig. 1e). miR-144 and NUDCD1 expression profiles were consistent with the classical pattern of miRNA-regulated genes. Next, the binding site of miR-144 and NUDCD1 was mutated based on the sequences described in (Fig. 1d). The dual luciferase reporter assay demonstrated that the relative luciferase activity of pSicheck-NUDCD1-wt was significantly reduced by agomiR-144 co-transfection, while for pSicheck-NUDCD1-mut and pSicheck-NC, relative luciferase activity did not differ (Fig. 1f). Furthermore, miR-144 expression was significantly downregulated in SW620, HCT116, and HT29 cells compared to normal colon cells (NCM460) (Fig. 1g). AgomiR-144 transfection significantly upregulated miR-144 in HCT116 and HT29 cells (Fig. 1h), while NUDCD1 mRNA and protein expression were significantly downregulated (Fig. 1i, j). Altogether, the results indicate that miR-144 directly binds to NUDCD1 and can regulate its expression in CRC cells.
miR-144 directly targets NudC domain containing 1 (NUDCD1). a In the bioinformatics analysis, 7 of the 11 databases from miRWalk 2.0 predicted that NUDCD1 was a direct target of miR-144; b microarray data of NUDCD1 from the GDS4382 gene chip dataset; c Expression profile of NUDCD1 from the GDS4382 gene chip dataset; d miR-144 wild-type and mutant NUDCD1 binding sites as predicted by bioinformatics; e Relative expression of miR-144 in colorectal cancer (CRC) and adjacent peri-cancerous tissues (NC), *P < 0.05, compared with NC; f luciferase activity induced by miR-144, *P < 0.05, compared with pSicheck-NUDCD1-wt; g relative expression of miR-144 in the normal colon mucosal epithelial cell line, NCM460, and five CRC cell lines: SW620, HCT116, HT29, SW480, and DLD1, *P < 0.05; h relative expression of miR-144 in HCT116 and HT29 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC; i NUDCD1 mRNA expression in CRC cells after transfection, *P < 0.05, compared with agomiR-NC; j NUDCD1 protein in CRC cells after transfection, *P < 0.05, compared with agomiR-NC. Results display the mean ± standard error of the mean (SEM) from triplicate experiments
miR‑144 inhibits, in vitro and in vivo, the IGF1R-ERK1/2 signaling pathway by targeting NUDCD1
We previously reported that NUDCD1 inhibition in CRC cells can reduce IGF1R-ERK1/2 signaling (Han et al. 2018b). To confirm that the biological effects of miR-144/NUDCD1 were induced via the IGF1R-ERK1/2 signaling pathway, CRC cells were co-transfected with the NUDCD1 overexpression plasmid and agomiR-144 or agomiR-NC. miR-144 overexpression alone significantly reduced the relative protein expression of p-IGF1R and p-ERK1/2, while co-transfection with the NUDCD1 plasmid restored IGF1R-ERK1/2 signaling pathway activity (Fig. 2a). Next, a CRC xenograft model using the HCT116 cell line was established in nude mice. The tumor-bearing mice were transfected with agomiR-144 or agomiR-NC via intra-tumoral injection to evaluate whether miR-144 regulates the IGF1R-ERK1/2 signaling pathway via NUDCD1. As expected, both the growth volumes (Fig. 2b) and weights (Fig. 2b) of the tumor xenografts were significantly suppressed in mice that had received agomiR-144 versus those given agomiR-NC. miR-144, NUDCD1, p-IGF1R, and p-ERK1/2 expression in the xenografts was subsequently quantified. Relative to the agomiR-NC group, miR-144 expression was significantly upregulated in transplanted tumors transfected with agomiR-144 (Fig. 2d), whereas the protein expression of NUDCD1, p-IGF1R, and p-ERK1/2 was downregulated (Fig. 2e). These results indicate that miR-144 in the transplanted tumors decreases IGF1R-ERK1/2 signaling by downregulating NUDCD1, consistent with the in vitro results.
miR‑144 inhibits, in vitro and in vivo, the IGF1R-ERK1/2 signaling pathway by targeting NUDCD1. a Relative protein expression levels of p-IGF1R and p-ERK1/2 in CRC cells following miR-144 overexpression and subsequent co-transfection with NUDCD1, NC (agomiR-NC + pcDNA3.1-NC), agomiR-144 (agomiR-144 + pcDNA3.1-NC), *P < 0.05, compared with NC, #P < 0.05, compared with agomiR-144; volumes (b) and weights (c) of subcutaneously xenografted CRC tissue in nude mice, *P < 0.05, compared with agomiR-NC; d relative expression of miR-144 in xenografted CRC tissues, *P < 0.05, compared with agomiR-NC; e relative protein expression of p-IGF1R, p-ERK1/2, and NUDCD1 in xenografted CRC tissues, *P < 0.05, compared with agomiR-NC
The relative expression of miR‑144 in CRC tissues
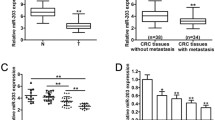
We examined the relative expression of miR-144 in cancer tissues from 70 CRC patients as well as patient clinical and pathological features. The results are summarized in Table 1. We found that miR-144 expression was associated with CRC Duke grade, primary range (T grade), and lymph node invasion (N grade) (P < 0.05; Fig. 3a–d). The online bioinformatics analysis revealed that for four cancer types, survival was significantly associated with miR-144. More specifically, miR-144 was upregulated in living colon adenocarcinoma (COAD) patients (Fig. S1a). miR-144 expression was also associated with pathologic M and N statuses as well as sex (Fig. S1b). A risk score formula for COAD based on pre-calculated survival and custom signatures indicated that low levels of miR-144 were associated with higher risk (Fig. 3e). Survival outcome analysis revealed that rectum adenocarcinoma patients with low miR-144 expression had poorer outcomes (Fig. 3f), especially those at stage three (Fig. 3g) or that have low mutation burdens (Fig. 3h). Next, based on the above results and the suppressor role of miR-144 in cancers, a pan-cancer survival analysis was conducted by Kaplan–Meier Plotter. Notably, in breast cancer (Fig. S2b), pancreatic ductal adenocarcinoma (Fig. S2g), as well as carcinomas of the bladder (Fig. S2a), clear cell renal cell (Fig. S2c), papillary renal cell (Fig. S2d), hepatocellular (Fig. S2e), squamous cell (Fig. S2f), and thyroid (Fig. S2h), Kaplan–Meier survival analysis indicated that overall survival rates were significantly higher for patients with high miR-144 expression. As a target of miR-144, NUDCD1 can promote CRC metastasis by inducing epithelial mesenchymal transition (EMT) and inhibiting apoptosis (Han et al. 2018b). Accordingly, we used FunRich to perform an enrichment analysis for miR-144 and found that miR-144 was enriched in cell growth, apoptosis, cell proliferation, and cell cycle regulation (Fig. 3g). Altogether, the data indicate that miR-144 is associated with certain pathological features of CRC, and it may play a role as a tumor suppressor gene in CRC by targeting NUDCD1.
The relative expression of miR‑144 in CRC tissues. a Relative miR-144 expression in CRC and adjacent peri-cancerous tissues from patients at Duke stages I–II and III–IV, *P < 0.05; b relative miR-144 expression in CRC and adjacent peri-cancerous tissues from patients at stages T1–T2 and T3–T4, *P < 0.05; c relative miR-144 expression in CRC and adjacent peri-cancerous tissues from patients with lymph metastasis (N1–N2) and without lymph metastasis (N0), *P < 0.05; d relative miR-144 expression in CRC and adjacent peri-cancerous tissues from patients with (M1) and without metastasis (M0), *P < 0.05; e risk score formula from OncomiR based on pre-calculated survival and custom signatures, including miR-144; f survival outcome analysis of miR-144 in rectum adenocarcinoma (READ) patients using the Kaplan–Meier Plotter. Survival analysis of miR-144 in READ patients at stage three (g) or that have low mutation burdens (h). i FunRich biological process enrichment analysis for miR-144
miR-144 influences CRC cell proliferation, cell cycle, migration, and invasion
HCT116 and HT29 cells were first transfected with agomiR-144 and agomiR-NC. To evaluate the effect of miR-144 on cell proliferation, the MTS assay was used. Growth curves were drawn to evaluate proliferation activity. The results indicated that miR-144 overexpression significantly inhibited the growth of HCT116 and HT29 cells (Fig. 4a, d). miR-144 has been previously observed to promote tumor cell cycle arrest (Sun et al. 2020); therefore, we assessed its effect on the CRC cell cycle. After transfection with agomiR-144 or agomiR-NC, HCT116 and HT29 cells were stained with PI, and cell cycle changes were analyzed using flow cytometry. In HCT116 cells, miR overexpression significantly increased and decreased, respectively, the proportions of cells in G0/G1 versus the S and G2 phases (Fig. 4b). For HT29 cells, the proportion of G0/G1 phase cells was significantly increased, while only the proportion of cells in the S phase was significantly decreased (Fig. 4e). These results suggest that miR-144 overexpression can arrest CRC cells in the G0/G1 phase and delay the cell cycle. To evaluate the effect of miR-144 on cell migration and invasion, a Transwell chamber transfer assay was performed with or without matrigel. miR-144 overexpression significantly reduced the number of HCT116 and HT29 cells that penetrated the filter via transfer and invasion (Fig. 4c, f). Altogether, the results imply that miR-144 overexpression can reduce the proliferation, delay the cell cycle, and suppress the invasion and migration of CRC cells.
miR‑144 influences CRC cell proliferation, cell cycle, migration, and invasion. a Proliferative capacity of HCT116 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC; b cell cycle phase of HCT116 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC; c the migration and invasion of HCT116 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC; d proliferative capacity of HT29 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC; e cell cycle phase of HT29 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC; f the migration and invasion of HT29 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC
miR-144 induces CRC cell apoptosis
miR-144 has been reported to induce apoptosis in tumor cells (Chen et al. 2018; Li et al. 2017). To determine if it can also induce apoptosis in CRC cells, HCT116 and HT29 cells were stained with Hochest 33342/PI and observed under a fluorescence microscope. The number of PI-stained cells (PI-stained nuclei indicate severe apoptosis or necrosis) was significantly increased following agomiR-144 transfection compared to agomiR-NC (Fig. 5a). Similarly, in cells stained with Annexin V-FITC/PI and analyzed using flow cytometry, miR-144 overexpression significantly increased the proportion of early and late apoptotic HCT116 and HT29 cells (Fig. 5b). These findings indicate that miR-144 overexpression may induce the apoptosis of CRC cells.
miR-144 induces CRC cell apoptosis. a HCT116 and HT29 cells were stained with Hoechst 33342 and propidium iodide. Cells with severe apoptosis or necrosis were detected using fluorescence microscopy; b The ratio of apoptotic cells as detected by flow cytometry in HCT116 and HT29 cells, *P < 0.05, compared with agomiR-NC
The molecular mechanism underlying miR-144 regulation of the cell cycle, apoptosis, and EMT
To explore the molecular mechanisms underlying the phenotypic changes in CRC cells following miR-144 overexpression, we evaluated classical protein markers related to metastasis, invasion, the cell cycle, and apoptosis (Cao et al. 2019; Davis et al. 2016; Gu et al. 2018; Padmanaban et al. 2019). These markers were not only closely related to NUDCD1 but also to the IGF1R-ERK1/2 signaling pathway (Han et al. 2018b). NUDCD1 has unique interaction partners, with its first isoform binding to DHX15, a putative RNA helicase involved in mRNA splicing (Asselin-Mullen et al. 2017). The PPI network of these genes was established (Fig. 6a), and their protein expression was quantified using western blotting. In HCT116 and HT29 cells, following miR-144 overexpression, E-cadherin and N-cadherin protein were upregulated and downregulated, respectively, indicating inhibition of the EMT process. Moreover, cyclin D1 protein was downregulated, suggesting that the cell cycle was attenuated. Finally, Bcl2 protein was downregulated, while cleaved-caspase3 protein was upregulated, indicating an increase in cell apoptosis (Fig. 6b, c). In CRC cells, the changes in these markers following miR-144 overexpression were similar to those of NUDCD1 knockdown in our previous studies, further implying that miR-144 specifically cleaves NUDCD1 to cause phenotypic changes in cell metastasis, invasion, the cell cycle, and apoptosis.
The molecular mechanism underlying miR-144 regulation of the cell cycle, apoptosis, and epithelial-mesenchymal transition. a Protein–protein interaction network of NUDCD1 and classical protein markers established by the Search Tool for the Retrieval of Interacting Genes/Proteins database; b Western blots of classical protein markers in HCT116 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC; c Western blots of classical protein markers in HT29 cells transfected with agomiR-144 or agomiR-NC, *P < 0.05, compared with agomiR-NC
Discussion
NUDCD1, a cancer antigen, is abnormally upregulated in multiple tumors and acts as an oncogene. In this study, we identified miR-144 as an upstream modulator of NUDCD1 and revealed that it suppresses the IGF1R-ERK1/2 signaling pathway via its target, NUDCD1. This inhibition mediates CRC cell proliferation and metastasis and may serve as a potential biomarker for CRC diagnosis and prognosis.
Classically, miRNA regulates its target genes by binding to their 3′-UTRs (Bartel 2009; Brodersen and Voinnet 2009). Here, we demonstrated via the dual-luciferase reporter experiments that miR-144 binds to NUDCD1’s 3′-UTR region. In mammals, members of the miR-144/451 gene cluster regulate hematopoietic function (Rasmussen et al. 2010). Moreover, they are involved in many physiological activities and diseases (Gao et al. 2016; Jiang et al. 2015; Wang et al. 2012). miR-144 is generally downregulated in numerous tumor tissues and exhibits tumor suppressor gene characteristics (Kooshkaki et al. 2020). With respect to CRC, miR-144 acts as a tumor suppressor by regulating the ROCK1, mTOR, and Notch-1 signaling pathways (Iwaya et al. 2012; Liu et al. 2019; Sureban et al. 2011). In CRC cells, miR-144 directly binds to genes such as NRAS, BCL6, PBX3, and CXCL11 (Cheng et al. 2020; Han et al. 2018a; Ji et al. 2019; Sun et al. 2020). Here, we found that increases in miR-144 expression induce NUDCD1 downregulation; this inverse relationship suggests that miR-144 may affect CRC cell function by directly binding to NUDCD1.
Previous studies have shown that in lung, ovarian, and colorectal cancers, NUDCD1 downregulation can inhibit IGF1R-ERK1/2 activity (Han et al. 2018b; He et al. 2020a; Rao et al. 2014a). Activation of this pathway promotes malignant biological behaviors such as metastasis, invasion, excessive proliferation, and apoptosis (Roskoski 2012, 2019). In CRC cells, the changes in cell phenotype and their related proteins following the inactivation of ERK1/2 signaling are consistent with those induced by miR-144 overexpression or NUDCD1 inhibition (Han et al. 2018b; Li et al. 2017). Therefore, we speculate the existence of a miR-144/NUDCD1/IGF1R-ERK1/2 axis, whereby miR-144 overexpression downregulates p-IGF1R and p-ERK1/2 and conversely, p-IGF1R and p-ERK1/2 downregulation can be restored by NUDCD1 overexpression. Given the negative regulation of NUDCD1 by miR-144, miR-144 likely affects the IGF1R-ERK1/2 signaling pathway via NUDCD1; the miR-144/NUDCD1/IGF1R-ERK1/2 axis is present in CRC cells. This axis was confirmed in vivo, as the results from the animal experiments were similar to those in vitro: intra-tumoral miR-144 overexpression attenuated IGF1R-ERK1/2 activity in transplanted tumor tissues by downregulating NUDCD1 and also slowed the growth of CRC cell xenografts.
NUDCD1 is not only upregulated in CRC but is also strongly associated with cancer metastasis (Han et al. 2018b; He et al. 2020a). Gene chip data analysis confirmed that NUDCD1 was highly expressed in CRC tissues. To verify our hypothesis regarding miR-144 and NUDCD1 in humans, we analyzed the correlation between miR-144 expression and pathological characteristics in CRC patients. miR-144 expression was reduced in CRC tissues compared to surrounding peri-cancerous tissues, was downregulated in early-stage tumor tissues, and was at its lowest level in advanced-stage invasive tumor tissues. Moreover, miR-144 levels were correlated with poor clinical features including advanced stage and lymph node metastasis. Compared to the normal colon epithelial cell line NCM460, miR-144 expression was reduced in the CRC cells SW620, HCT116, and HT29. This progressively decreasing expression profile mirrors the degree of malignancy in tumor cells and suggests a suppressive role of miR-144 in CRC development. More importantly, the online survival analysis revealed that deceased COAD patients had reduced miR-144 expression, and a risk score formula for COAD indicated that low levels of miR-144 were associated with higher risk. Interestingly, in some other cancer types such as breast cancer, pancreatic ductal adenocarcinoma, or bladder carcinoma, the overall survival rates were also significantly higher for patients with high miR-144 expression. This indicates the potential of miR-144 as a biomarker for survival estimates was not only in CRC, but also in some other cancer types. Here, we determined the role of miR-144 in CRC cells and characterized the mechanism in vitro.
We previously confirmed that NUDCD1 as an oncogene promotes CRC cell proliferation, migration, invasion, and survival (Han et al. 2018b). The FunRich results demonstrated that miR-144 is enriched in cell growth, apoptosis, proliferation, and cell cycle regulation, which was confirmed by our findings. In trophoblast, breast cancer, and esophageal squamous carcinoma cells as well as malignant gliomas, miR-144 is associated with cancer cell proliferation. miR-144 is also closely related to tumor cell metastasis and invasion (He et al. 2020b; Sheng et al. 2019). It can regulate the cell cycle of T and acute lymphoblastic leukemia cells (Jin et al. 2017; Sun et al. 2017) and is involved in the apoptotic process in liver and salivary gland cancers (Huo et al. 2016; Zhou et al. 2016). The in vitro data here indicate that miR-144 can inhibit CRC cell proliferation, migration, and invasion. More specifically, following miR-144 transfection, more CRC cells were in the G0/G1 phase, and their apoptotic rate was increased. These results agree with those of the FunRich biological process analysis as well as those from previous studies in CRC and other cancers. Furthermore, we evaluated classical proteins associated with CRC metastasis (Hu et al. 2018; Zhao et al. 2020), invasion (Dino et al. 2019; Zhao et al. 2020), cell cycle (Davis et al. 2016; Kim et al. 2020), and apoptosis (Gu et al. 2018; Zhou et al. 2019). Notably, our PPI network revealed that these classical proteins were also partly enriched in the ERK/MAPK signaling pathway, which is regulated by miR-144/NUDCD1. More specifically, miR-144 overexpression increased the expression of E-cadherin and cleaved-caspase3 and decreased the expression of N-cadherin, cyclin D1, and Bcl2. These results indicate that miR-144 acts as a tumor suppressor by repressing IGF1R-ERK1/2 activity, a key driving factor in CRC development.
In conclusion, we investigated the biological role of miR-144 in CRC in vitro and in vivo, evaluated the regulatory relationship between miR-144 and NUDCD1, and confirmed the presence of a miR-144/NUDCD1/IGF1R-ERK1/2 axis. Moreover, based on the axis, we explored the tumor suppressor role of miR-144 and found that it can inhibit CRC cell proliferation, metastasis, and invasion (Fig. 7). Our results provide new directions for the prevention, treatment, and drug development of CRC.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Asselin-Mullen P, Chauvin A, Dubois ML, Drissi R, Levesque D, Boisvert FM (2017) Protein interaction network of alternatively spliced NudCD1 isoforms. Sci Rep 7:12987. https://doi.org/10.1038/s41598-017-13441-w
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. https://doi.org/10.1016/j.cell.2009.01.002
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Brenner H, Kloor M, Pox CP (2014) Colorectal Cancer. Lancet 383:1490–1502. https://doi.org/10.1016/S0140-6736(13)61649-9
Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10:141–148. https://doi.org/10.1038/nrm2619
Cao ZQ, Wang Z, Leng P (2019) Aberrant N-cadherin expression in cancer. Biomed Pharmacother 118:109320. https://doi.org/10.1016/j.biopha.2019.109320
Cao HL, Gu MQ, Sun Z, Chen ZJ (2020) miR-144-3p contributes to the development of thyroid tumors through the PTEN/PI3K/AKT pathway. Cancer Manag Res 12:9845–9855. https://doi.org/10.2147/CMAR.S265196
Chen W et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66:115–132. https://doi.org/10.3322/caac.21338
Chen G et al (2018) Lico A causes er stress and apoptosis via up-regulating miR-144-3p in human lung cancer cell line H292. Front Pharmacol 9:837. https://doi.org/10.3389/fphar.2018.00837
Cheng B, Zhang Y, Wu ZW, Cui ZC, Li WL (2020) MiR-144 inhibits colorectal cancer cell migration and invasion by regulating PBX3. Eur Rev Med Pharmacol Sci 24:9361–9369. https://doi.org/10.26355/eurrev_202009_23019
Davis MI et al (2016) Small molecule inhibition of the ubiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J Biol Chem 291:24628–24640. https://doi.org/10.1074/jbc.M116.738567
Dino P, D’Anna C, Sangiorgi C, Di Sano C, Di Vincenzo S, Ferraro M, Pace E (2019) Cigarette smoke extract modulates E-cadherin, claudin-1 and miR-21 and promotes cancer invasiveness in human colorectal adenocarcinoma cells. Toxicol Lett 317:102–109. https://doi.org/10.1016/j.toxlet.2019.09.020
Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44:839–847. https://doi.org/10.1016/j.jbi.2011.05.002
Fonseka P, Pathan M, Chitti SV, Kang T, Mathivanan S (2020) FunRich enables enrichment analysis of OMICs datasets. J Mol Biol. https://doi.org/10.1016/j.jmb.2020.166747
Gao Z, Liu R, Liao J, Yang M, Pan E, Yin L, Pu Y (2016) Possible tumor suppressive role of the miR-144/451 cluster in esophageal carcinoma as determined by principal component regression analysis. Mol Med Rep 14:3805–3813. https://doi.org/10.3892/mmr.2016.5691
Goodall GJ, Wickramasinghe VO (2021) RNA in cancer. Nat Rev Cancer 21:22–36. https://doi.org/10.1038/s41568-020-00306-0
Gu YY et al (2018) Matrine induces apoptosis in multiple colorectal cancer cell lines in vitro and inhibits tumour growth with minimum side effects in vivo via Bcl-2 and caspase-3. Phytomedicine 51:214–225. https://doi.org/10.1016/j.phymed.2018.10.004
Guo W et al (2015) MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology 62:1132–1144. https://doi.org/10.1002/hep.27929
Han B et al (2018a) MicroRNA-144 mediates chronic inflammation and tumorigenesis in colorectal cancer progression via regulating C-X-C motif chemokine ligand 11. Exp Ther Med 16:1935–1943. https://doi.org/10.3892/etm.2018a.6389
Han B et al (2018b) NUDCD1 promotes metastasis through inducing EMT and inhibiting apoptosis in colorectal cancer. Am J Cancer Res 8:810–823
He B, Xia S, Zhang Z (2020a) NudCD1 promotes the proliferation and metastasis of non-small cell lung cancer cells through the activation of IGF1R-ERK1/2. Pathobiology 87:244–253. https://doi.org/10.1159/000505159
He L, Liao L, Du L (2020b) miR1443p inhibits tumor cell growth and invasion in oral squamous cell carcinoma through the downregulation of the oncogenic gene, EZH2. Int J Mol Med 46:828–838. https://doi.org/10.3892/ijmm.2020.4638
Hu Y et al (2018) Epigenetic suppression of E-cadherin expression by Snail2 during the metastasis of colorectal cancer. Clin Epigenet 10:154. https://doi.org/10.1186/s13148-018-0592-y
Huo F, Zhang C, He H, Wang Y (2016) MicroRNA-144-3p inhibits proliferation and induces apoptosis of human salivary adenoid carcinoma cells via targeting of mTOR. Biotechnol Lett 38:409–416. https://doi.org/10.1007/s10529-015-2007-x
Iwaya T et al (2012) Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis 33:2391–2397. https://doi.org/10.1093/carcin/bgs288
Ji X et al (2019) miR-144 suppresses cell proliferation and migration in colorectal cancer by targeting NRAS. J Cell Biochem. https://doi.org/10.1002/jcb.29543
Jiang X et al (2015) miR-144/451 promote cell proliferation via targeting PTEN/AKT pathway in insulinomas. Endocrinology 156:2429–2439. https://doi.org/10.1210/en.2014-1966
Jin J, Wang Y, Xu Y, Zhou X, Liu Y, Li X, Wang J (2017) MicroRNA-144 regulates cancer cell proliferation and cell-cycle transition in acute lymphoblastic leukemia through the interaction of FMN2. J Gene Med. https://doi.org/10.1002/jgm.2898
Kim MJ et al (2020) Novel SIRT inhibitor, MHY2256, induces cell cycle arrest, apoptosis, and autophagic cell death in HCT116 human colorectal cancer cells. Biomol Ther (seoul) 28:561–568. https://doi.org/10.4062/biomolther.2020.153
Kooshkaki O et al (2020) MiR-144: a new possible therapeutic target and diagnostic/prognostic tool in cancers. Int J Mol Sci. https://doi.org/10.3390/ijms21072578
Li J, Sun P, Yue Z, Zhang D, You K, Wang J (2017) miR-144–3p induces cell cycle arrest and apoptosis in pancreatic cancer cells by targeting proline-rich protein 11 expression via the mitogen-activated protein kinase signaling pathway. DNA Cell Biol 36:619–626. https://doi.org/10.1089/dna.2017.3656
Liu F, Chen N, Xiao R, Wang W, Pan Z (2016) miR-144–3p serves as a tumor suppressor for renal cell carcinoma and inhibits its invasion and metastasis by targeting MAP3K8. Biochem Biophys Res Commun 480:87–93. https://doi.org/10.1016/j.bbrc.2016.10.004
Liu JL et al (2019) MiR-144 inhibits tumor growth and metastasis in osteosarcoma via dual-suppressing RhoA/ROCK1 signaling pathway. Mol Pharmacol 95:451–461. https://doi.org/10.1124/mol.118.114207
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Nagy A, Lanczky A, Menyhart O, Gyorffy B (2018) Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 8:9227. https://doi.org/10.1038/s41598-018-27521-y
Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D (2007) Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res 67:10782–10788. https://doi.org/10.1158/0008-5472.CAN-07-1484
Padmanaban V, Krol I, Suhail Y, Szczerba BM, Aceto N, Bader JS, Ewald AJ (2019) E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573:439–444. https://doi.org/10.1038/s41586-019-1526-3
Rao W et al (2014a) OVA66 increases cell growth, invasion and survival via regulation of IGF-1R-MAPK signaling in human cancer cells. Carcinogenesis 35:1573–1581. https://doi.org/10.1093/carcin/bgu070
Rao W et al (2014b) OVA66, a tumor associated protein, induces oncogenic transformation of NIH3T3 cells. PLoS ONE 9:e85705. https://doi.org/10.1371/journal.pone.0085705
Rasmussen KD et al (2010) The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 207:1351–1358. https://doi.org/10.1084/jem.20100458
Roskoski R Jr (2012) ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res 66:105–143. https://doi.org/10.1016/j.phrs.2012.04.005
Roskoski R Jr (2019) Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol Res 142:151–168. https://doi.org/10.1016/j.phrs.2019.01.039
Shi C et al (2021) NUDCD1 knockdown inhibits the proliferation, migration, and invasion of pancreatic cancer via the EMT process. Aging (albany NY) 13:18298–18309. https://doi.org/10.18632/aging.203276
Sheng S, Xie L, Wu Y, Ding M, Zhang T, Wang X (2019) MiR-144 inhibits growth and metastasis in colon cancer by down-regulating SMAD4. Biosci Rep. https://doi.org/10.1042/BSR20181895
Song F, Chen Q, Rao W, Zhang R, Wang Y, Ge H, Wei Q (2019) OVA66 promotes tumour angiogenesis and progression through enhancing autocrine VEGF-VEGFR2 signalling. EBioMedicine 41:156–166. https://doi.org/10.1016/j.ebiom.2019.02.051
Sun J et al (2017) E2F8, a direct target of miR-144, promotes papillary thyroid cancer progression via regulating cell cycle. J Exp Clin Cancer Res 36:40. https://doi.org/10.1186/s13046-017-0504-6
Sun N, Zhang L, Zhang C, Yuan Y (2020) miR-144-3p inhibits cell proliferation of colorectal cancer cells by targeting BCL6 via inhibition of Wnt/beta-catenin signaling. Cell Mol Biol Lett 25:19. https://doi.org/10.1186/s11658-020-00210-3
Sureban SM et al (2011) Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. J Nanobiotechnol 9:40. https://doi.org/10.1186/1477-3155-9-40
Szklarczyk D, Jensen LJ (2015) Protein–protein interaction databases. Methods Mol Biol 1278:39–56. https://doi.org/10.1007/978-1-4939-2425-7_3
Wang Q et al (2008) RNA interference targeting CML66, a novel tumor antigen, inhibits proliferation, invasion and metastasis of HeLa cells. Cancer Lett 269:127–138. https://doi.org/10.1016/j.canlet.2008.04.035
Wang X et al (2012) Loss of the miR-144/451 cluster impairs ischaemic preconditioning-mediated cardioprotection by targeting Rac-1. Cardiovasc Res 94:379–390. https://doi.org/10.1093/cvr/cvs096
Wong NW, Chen Y, Chen S, Wang X (2018) OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 34:713–715. https://doi.org/10.1093/bioinformatics/btx627
Zhao S, Xue H, Hao CL, Jiang HM, Zheng HC (2020) BTG1 overexpression might promote invasion and metastasis of colorectal cancer via decreasing adhesion and inducing epithelial-mesenchymal transition. Front Oncol 10:598192. https://doi.org/10.3389/fonc.2020.598192
Zhou S, Ye W, Zhang Y, Yu D, Shao Q, Liang J, Zhang M (2016) miR-144 reverses chemoresistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Am J Transl Res 8:2992–3002
Zhou J et al (2019) Antitumor activity in colorectal cancer induced by hinokiflavone. J Gastroenterol Hepatol 34:1571–1580. https://doi.org/10.1111/jgh.14581
Zhou M, Wu Y, Li H, Zha X (2020) MicroRNA-144: a novel biological marker and potential therapeutic target in human solid cancers. J Cancer 11:6716–6726. https://doi.org/10.7150/jca.46293
Acknowledgements
The authors thank Fu Liu, Qiang Ma and Daiyuan Ma for providing valuable technical supports.
Funding
This study was supported by National Natural Science Foundation of China (819θ3660); Sichuan Science and Technology Plan Project (2019YJ0386); Sichuan Education Plan Project (18ZB0219); Nanchong City and School Cooperation Project (NSMC20170447, 18SXHZ0324, 20SXQT0101); Affiliated Hospital of North Sichuan Medical College Plan Projects (2019ZD005).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by BH and KX. DF collected the samples and contributed to the design of this study. YB and YL were involved in the animal experiments. The first draft of the manuscript was written by LZ and YZ, all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflict of interests.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, B., Xu, K., Feng, D. et al. miR-144 inhibits the IGF1R-ERK1/2 signaling pathway via NUDCD1 to suppress the proliferation and metastasis of colorectal cancer cells: a study based on bioinformatics and in vitro and in vivo verification. J Cancer Res Clin Oncol 148, 1903–1918 (2022). https://doi.org/10.1007/s00432-022-03951-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-03951-0