Abstract
Objective
Renal cell carcinoma (RCC) is the most common malignancy of the urinary system, and it is a serious threat to human health. HOXA transcript at the distal tip (HOTTIP), located at the 5′ end of the HOXA locus, is a long non-coding RNA that has been newly discovered in recent years. It has been reported to promote the development of several types of tumors. Moreover, accumulating evidence has indicated that autophagy plays an important role in tumor cell survival or death. However, whether HOTTIP affects RCC development by regulating autophagy remains unknown.
Methods
In this study, we first measured HOTTIP expression in 42 paired RCC and adjacent non-tumor tissues, as well as in 4 RCC cell lines and 1 normal renal tubular epithelial cell line. Then, we selected 2 RCC cell lines to silence HOTTIP expression and 1 RCC cell line to overexpress HOTTIP, and we measured their proliferation, migration and invasion, as well as autophagy, after pretreatment with an autophagy inhibitor or inducer. In addition, we assessed the growth, metastasis and autophagy of tumors in nude mice and explored the mechanism involved.
Results
The results showed that HOTTIP expression was significantly upregulated in the RCC tissues and cell lines, and it was closely associated with TNM stage, histological grade, lymph node metastasis and patient prognosis. The in vitro and in vivo assays indicated that HOTTIP silencing inhibited RCC cell proliferation, migration and invasion and induced autophagy, and 3-MA (an autophagy inhibitor) reversed these effects. In contrast, HOTTIP overexpression and rapamycin (an autophagy inducer) yielded the opposite results. Further research revealed that HOTTIP modification could affect RCC cell autophagy via the PI3K/Akt/Atg13 signaling pathway.
Conclusions
Our study will help in finding a potential marker for RCC diagnosis and supply a target molecule for RCC treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Renal cell carcinoma (RCC, also known as kidney cancer), which originates from renal tubular epithelial cells, is a malignant tumor of the urinary system (Deng et al. 2013). RCC accounts for approximately 2–3% of all malignancies in adults; approximately 5–10 RCC patients are diagnosed per one million people (Chen et al. 2017; Xu et al. 2017), and the mortality rate reaches up to 40% (Turcotte et al. 2008). Unfortunately, morbidity and mortality continue to increase annually (Yu et al. 2012; Zhou et al. 2014), and a survey showed that in urinary systematic tumors, RCC morbidity and mortality (44.4% and 45.5%, respectively) were second-highest after bladder cancer in the US (Siegel et al. 2015). RCC is not sensitive to radiotherapy, chemotherapy or immunotherapy, and their effects are limited and poor. Surgery may be the only effective method, and recurrence or metastasis following surgery continues to occur in approximately 20–40% of patients (Williams et al. 2013; Nishikawa et al. 2015). Although some RCC patients receive two or more types of combination therapies, the 5-year survival rate is still 55%, and it is less than 10% in patients with metastasis (Li et al. 2017). Therefore, it is urgent to explore the molecular mechanism involved in RCC development and find an effective target to improve the survival rate and quality of life (QoL) for RCC patients.
Autophagy, a selective and caspase-independent programmed cell death (PCD) pathway, was found to be closely related to tumor progression (Shimizu et al. 2014). It is regarded as a ‘double-edged sword’, namely, it inhibits or promotes tumor cell growth under different stimuli (Thorburn et al. 2014; White 2015). Therefore, there has always been a debate on the role of autophagy in the regulation of tumor development.
Long non-coding RNAs (lncRNAs) are a class of RNAs that are more than 200 nucleotides in length and cannot be translated into proteins. Initially, they were thought to be non-functional; however, upon further study, they have been considered relevant by more and more researchers (Ponting et al. 2009; Hong et al. 2017). LncRNAs have been found to take part in the development of several diseases, including cardiovascular, autoimmune, infectious and neurological diseases, as well as tumors, such as pancreatic cancer, bladder cancer, lung cancer, liver cancer and gastric cancer (Uchida and Dimmeler 2015; Wu et al. 2015; Wang et al. 2014; Li et al. 2014). In addition, multiple studies have found that certain lncRNAs, such as HOTAIR, H19, MALAT1, and MEG3, play an important role in RCC development (Seles et al. 2016). These lncRNAs are differentially expressed in RCC and adjacent normal tissues, but only a few have been well characterized, and the underlying mechanisms have not yet been investigated.
Recently, a newly identified lncRNA, HOXA transcript at the distal tip (HOTTIP), was found to be located at the chromosomal locus 7p15.2. It was discovered that HOTTIP was significantly upregulated in several human cancers and is thus a functionally characterized lncRNA (Lian et al. 2016). Therefore, HOTTIP is likely to possess a high diagnostic value for cancer screening and great clinical potential for cancer therapy. However, its function and association with autophagy in RCC regulation have not yet been studied. Based on the above findings, we assessed HOTTIP expression in RCC and explored its roles in regulating autophagy and the molecular pathway involved in mediating RCC development.
Materials and methods
RCC samples
A total of 42 human RCC and adjacent normal tissue specimens were obtained from surgeries performed from April 2014 to January 2015 at Wuhan University Renmin Hospital (Wuhan, China), and all patients signed consent forms. The diagnosis of RCC was confirmed by two pathologists. The acquired tissue samples were immediately frozen in liquid nitrogen and stored at − 80 °C until RNA extraction. All protocols in this study were approved by the Hospital’s Protection of Human Ethics Committee.
RCC cell lines and transfection
The human RCC cell lines 786-O, A498, ACHN, and OSRC-2 were obtained from the Cell Bank of Wuhan University (Wuhan, China), and they were routinely cultured in RPMI 1640 media (Gibco, USA) with 10% fetal bovine serum (FBS, ScienCell, USA) at 37 °C and 5% CO2. To obtain RCC cell lines stably expressing HOTTIP, cDNA oligonucleotides were firstly designed and synthesized to suppress HOTTIP expression. shRNAs were designed with the help of Invitrogen Software. After annealing, the double-stranded oligonucleotides were inserted into the lentiviral vector (GeneChem, Shanghai, China). To produce the lentivirus suppressing HOTTIP expression, HEK-293T cells were transfected with the vector described above using Lipofectamine 2000 according to the manufacturer’s guidelines (GeneChem). The lentiviruses were harvested at 48 h after transfection. A scrambled shRNA (shRNA-NC) was used as the negative control for shRNA-HOTTIP. ACHN cells and OSRC-2 cells were then infected with shRNA-HOTTIP or shRNA-NC at a multiplicity of infection of 50. The supernatant was replaced with complete media after 24 h. The infected cells were cultured in fresh media in the presence of 5 µg/ml puromycin (Sigma, USA). Following monoclonal screening, HOTTIP expression in the cell lines was confirmed by qRT-PCR. Moreover, for the construction of the lentiviral vector over-expressing HOTTIP, full-length cDNA was amplified and cloned into the pcDNA3.1 vector. HEK-293T cells were then transfected with the vector described above. The lentiviruses were harvested at 48 h after transfection and designated as pcDNA-HOTTIP. The empty vector was used as a negative control (pcDNA-NC). The transfected A498 cells and stable clones were generated as described above. In addition, we added 3-methyladenine (3-MA, a common autophagy inhibitor, 0.5 µM) and rapamycin (RAPA, a common autophagy inducer, 200 nM) to the transfected RCC cell lines for 24 h of pretreatment (3-MA group and RAPA group hereafter) according to the experimental design. The HOTTIP shRNA sequences were as follows: 5′-GTCCTGACCAATGTAAGTGTC-3′ (shRNA-HOTTIP-1), 5′-GGAAAGTTTGCACTGAATTGA-3′ (shRNA-HOTTIP-2) and 5′-GGAAGAAATTCAATGCCATGC-3′ (shRNA-HOTTIP-3).
CCK-8 assay
RCC cells in each group were plated in 96-well plates at approximately 7 × 103 cells/well and cultured in 100 µl complete media with 10% FBS. At the indicated time points (24 h, 48 h, 72 h, and 96 h), 10 µl CCK8 reagent was added to each well and incubated for 2 h according to the manufacturer’s instructions (Dojindo, Japan). Then, the absorbance at 450 nm was measured by a microplate reader (BioTek Elx9808, USA).
Ethynyldeoxyuridine (EdU) analysis
RCC cells in each group were plated in 96-well plates at 5 × 103 cells/well and cultured for 48 h. Then, 50 µM EdU medium was added to the 96-well plates and incubated for 2 h at 37 °C with 5% CO2. After treatment with 4% paraformaldehyde and 0.5% Triton X-100, the cells were stained with an anti-EdU working solution. Hoechst 33342 was used to label the nuclei according to the instructions of a 5-ethynyl-2-deoxyuridine labeling/detection kit (Ribobio, Guangzhou, China). The EdU-positive cells in 5 randomly selected fields were viewed by fluorescence microscopy (Olympus, Japan).
Plate clone formation assay
RCC cells in each group were digested, counted and plated into 6-well plates with complete media at a density of 1 × 103 cells per well; the cells were cultured at 37 °C and 5% CO2 for 8–10 days. Then, the media was discarded, and the cells were washed with phosphate buffered saline (PBS) for 2–3 times. After being fixed with 4% paraformaldehyde for 30 min, the cells were stained with a 0.1% crystal violet solution for another 30 min, and the cells were subsequently washed with PBS and air-dried. Finally, the number of colonies was counted manually.
Cell cycle analysis
The transfected RCC cells were digested with trypsin, collected in centrifuge tubes, and fixed with 70% ice-cold ethanol at 4 °C overnight after washing with PBS for 2–3 times. The cells were then centrifuged, re-suspended, washed, and stained with PI according to the manufacturer’s protocol (BD Biosciences, New Jersey, USA). Finally, the cell cycle was analyzed by flow cytometry (BD Biosciences, USA).
Scratch assay
RCC cells from each group were plated in 6-well plates for 48–72 h. When the cell density reached approximately 80–90%, the monolayer was wounded by scratching with a 200-µl sterile pipette tip lengthwise along the plate surface, and the media was removed. The cells were then washed 2–3 times with PBS and cultured in serum-free media. Images of cell migration were captured at 0 h and 24 h by an inverted microscope, and the area of migration (µm2) was measured with ImageJ software.
Transwell migration assay
RCC cells in each group were digested and counted, and 3 × 104 cells were plated in serum-free media on membranes with an 8-µm pore size in the upper Boyden chambers (Corning Costar, NY, USA), which were inserted into 24-well plates. Complete media with 10% FBS was added to the lower chambers. After incubation for 24 h, the cells that did not pass through the pores were removed, and the cells on the bottom of the membrane were fixed with 4% paraformaldehyde and stained with a crystal violet solution (Sigma, USA). The penetrated cells were counted under an inverted microscope (Olympus, Japan) in 3 randomly selected fields for each chamber.
Transwell invasion assay
RCC cells (1 × 105 per chamber) from each group were plated in serum-free media in upper Boyden chambers coated with 100 µl 10% Matrigel, and complete media containing 10% FBS was added to the lower chambers. After incubation for 24 h, the cells on the top side of each membrane were scraped off, and the penetrated cells on the basolateral side of the membranes were fixed and stained as the migration assay. Finally, the invasive cells were counted in 3 fields chosen at random under an inverted microscope (Olympus, Japan).
Western blot analysis
RCC cell protein was extracted using RIPA lysis buffer with phosphatase inhibitors to prevent the degradation of the samples. The protein concentrations were measured using a bicinchoninic acid protein assay kit (Biyuntian Bio-Technology, Shanghai, China) and electrophoresed on 8%, 12% or 15% sodium dodecyl sulfate–polyacrylamide gels; subsequently, the proteins were transferred onto polyvinylidene difluoride membranes (Millipore, USA). After blocking with 5% skimmed milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room temperature for 1 h and washing with TBST for 2–3 times, the membranes were incubated with the indicated monoclonal primary antibodies (1:1000; Cell Signaling Technology, USA) overnight at 4 °C. The membranes were then washed 2–3 times with TBST and incubated with the corresponding goat-anti-rabbit secondary antibody (1:15,000; LI-COR, USA) at room temperature for 1 h. After washing 3 times with TBST, the membranes were scanned with a two-color infrared imaging system (Odyssey, LI-COR, USA). GAPDH was used as an internal reference.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from RCC tissues and cells was extracted using Trizol reagent (Invitrogen, USA). The reverse transcription reactions were performed using a PrimeScriptTM RT reagent kit (Takara, Japan) according to the manufacturer’s instructions, and quantitative PCR reactions were prepared at a final volume of 20 µl using a SYBR Green Mix (Takara, Japan); the reactions were measured on an AB7500 detection system (Thermo Fisher, USA). The 2−ΔΔCT method was used to determine the relative gene expression level. GAPDH was used as an internal control to normalize the data. Each reaction was performed in triplicate. The primers used for these reactions were as follows:
HOTTIP
Forward, 5′-CCTAAAGCCACGCTTCTTTG-3′,
Reverse, 5′-TGCAGGCTGGAGATCCTACT-3′;
LC3B
Forward, 5′-TGCGTCTGTCCACTTGGCTA-3′,
Reverse, 5′-AGGAATCTTACTGATCACACGG-3′;
Beclin1
Forward, 5′-GGCGGCTCCTATTCCATC-3′,
Reverse, 5′-CCCAAGCAAGACCCCACT-3′;
p62
Forward, 5′-AGAATCAGCTTCTGGTCCATCG-3′,
Reverse, 5′-CCTCCGTGCTCCACATCG-3′;
Atg3
Forward, 5′-TCACCTAGTCCACCACTGTCCA-3′,
Reverse, 5′-CCATCCGCCATCACCATC-3′;
Atg4
Forward, 5′-GGGGATGTATGCTACGCTGTG-3′,
Reverse, 5′-GGATGCGTTGGTATTCTTTGG-3′;
Atg5
Forward, 5′-AATCAGGTTTGGTGGAGGCA-3′,
Reverse, 5′-CAGTGGAGGAAAGCAGAGGTG-3′;
Atg7
Forward, 5′-TAGGACGTTGATGGGTTGGG-3′,
Reverse, 5′-TGGAGAAGTTCACTGGATGC-3′;
Atg10
Forward, 5′-TGCAGCAGCGTCCGAAGT-3′,
Reverse, 5′-TCCTGTTGCGTAATAGTGTCCC-3′;
Atg12
Forward, 5′-TGACCTGCTGGCTGAATACC-3′,
Reverse, 5′-GTGAAACCAAAACGCCTAACC-3′;
Atg13
Forward, 5′-CTCTTTCCCTGGTCCCCTATC-3′,
Reverse, 5′-TTGCTCAGCCACAAATCCC-3′;
Atg14
Forward, 5′-AACCCACAGGCACCAAACA-3′,
Reverse, 5′-AGTCCCGTCCACTCTACCCA-3′;
GAPDH
Forward, 5′-AATCCCATCACCATCTTCCAG-3′,
Reverse, 5′-GAGCCCCAGCCTTCTCCAT-3′.
Transmission electron microscopy (TEM)
The transfected RCC cell lines (ACHN and A498) were harvested and fixed with 2.5% glutaraldehyde and 1% osmium tetroxide and then dehydrated in a graded series of ethanol. The cells were embedded in epoxy resin and sectioned at 70-nm thickness using an ultramicrotome. The sections were stained with 0.2% lead citrate and 2% uranyl acetate. The images were captured with a Tecnai Spirit electron microscope (FEI, USA).
Xenografts and tumor metastasis model
Four-week-old female BALB/c nude mice weighing 20 ± 2 g were obtained from the Animal Experiment Center of Wuhan University (Wuhan, China). All animals were divided randomly into two groups (n = 6/group) and housed in a specific pathogen-free environment with an appropriate temperature and humidity. 200 µl of PBS containing approximately 1 × 107 ACHN cells and OSRC-2 cells transfected with sh-NC or sh-HOTTIP and 2 × 106 A498 cells transfected with pc-NC or pc-HOTTIP were injected into the left flank of each mouse. The tumor volumes were measured every 3 days following inoculation and calculated by the following formula: (length × width2)/2. After 1 month, all mice were killed with an overdose of carbon dioxide, and the weights of the tumors were measured immediately. In addition, a tumor metastasis model was established by tail vein injection of transfected ACHN, OSRC-2 and A498 cells into additional BALB/c nude mice, namely, each mouse in two groups (n = 3/group) was injected with 1 × 106 cells mixed with PBS; after 1 month, all of these mice were also killed with carbon dioxide. Finally, the tumors and lung tissues of mice were collected for further research. All animal studies were approved by the Animal Care and Use Committee of Wuhan University Renmin Hospital, and all experimentation on animals was performed in agreement with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Hematoxylin–eosin (HE) staining
The acquired lung tissues were fixed in 4% paraformaldehyde. All samples were dehydrated, embedded in paraffin, and cut into 4-µm-thick slices. The sections were then stained with hematoxylin and eosin and examined by light microscopy (Olympus, Japan).
Immunohistochemistry (IHC) assay
The paraffin-embedded tumor sections were deparaffinized in xylene for 15 min, rehydrated with graded alcohol solutions and heated at 105 °C for 10 min in a citric acid buffer (0.01 M) for antigen retrieval. The slides were subsequently rinsed with PBS for 10 min and then transferred into a 3% hydrogen peroxide solution for 5 min to block endogenous peroxidase activity. After another washing with PBS for 5 min, the sections were blocked with blocking buffer for 1 h at room temperature and incubated with the relevant antibody for 1 h at room temperature. Finally, the immune complexes were detected using an immunochemistry kit (ZSGB-Bio, Beijing, China).
Statistical analysis
The data in this study are expressed as the mean ± standard deviation (SD), and the statistical analyses were performed using SPSS 19.0 software (IBM, USA). The differences between groups were analyzed using Student’s t-test, one-way ANOVA and the Chi-square test. P < 0.05 was considered to indicate a statistically significant difference. All experiments were repeated 3 times independently.
Results
HOTTIP is highly expressed in RCC, and its high expression predicts a poor prognosis
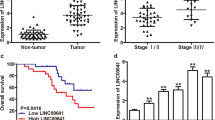
To assess HOTTIP expression in RCC tissues, we collected 42 paired human RCC and adjacent normal samples. The qRT-PCR results indicated that HOTTIP expression in RCC tissues was significantly higher than that in the adjacent normal tissues (Fig. 1a). Subsequently, we further analyzed the relationship between HOTTIP levels and the clinicopathological characteristics of RCC patients, and evaluated the overall survival of patients with different HOTTIP levels. As shown in Table 1, there was a significant correlation between HOTTIP expression and TNM stage, histological grade and lymph node metastasis (P < 0.05), except for sex, age and tumor size. Moreover, we found that the prognosis of RCC patients with high HOTTIP expression was much worse than that for RCC patients with low expression (Fig. 1b). In addition, HOTTIP expression levels in 4 RCC cell lines (786-O, A498, ACHN, and OSRC-2) and 1 normal renal tubular epithelial cell line (HK-2) were also measured. The qRT-PCR results showed that HOTTIP expression in 3 of the RCC cell lines, but not A498 cells, was much higher than that in HK-2 cells (Fig. 1c). These results indicated that HOTTIP expression was significantly upregulated in RCC tissues and cell lines and that high HOTTIP expression predicted a poor prognosis.
HOTTIP is significantly upregulated in RCC, and its high expression predicts a poor prognosis. a Relative HOTTIP expression in 42 paired RCC and adjacent non-tumor tissues was measured by qRT-PCR. b The overall survival percentages of RCC patients with high and low HOTTIP expression are displayed in Kaplan–Meier plots. c Relative HOTTIP expression in 4 RCC cell lines and 1 normal tubular epithelial cell line was measured by qRT-PCR. d–i Representative images of transfected ACHN cells (d), OSRC-2 cells (f) and A498 cells (h) were captured with a fluorescence microscope (200 × magnification), and HOTTIP silencing efficiency in ACHN cells (e) and OSRC-2 cells (g) and its overexpression efficiency in A498 cells (i) were verified by qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 tumor vs normal, high expression group vs low expression group, sh-HOTTIP group vs sh-NC group and pc-HOTTIP group vs pc-NC group
HOTTIP expression affects RCC cell proliferation by regulating autophagy in vitro
To assess the effects of HOTTIP expression on the biological behaviors of RCC cells, we silenced and overexpressed HOTTIP in RCC cells and determined their proliferation and cell cycle status. Firstly, HOTTIP silencing efficiency in ACHN cells (Fig. 1d, e) and OSRC-2 cells (Fig. 1f, g) and HOTTIP overexpression efficiency in A498 cells (Fig. 1h, i) were verified to be significant. The CCK-8 assay results showed that the viability of ACHN cells (Fig. 2a) and OSRC-2 cells (Fig. 2b) was significantly lower in the sh-HOTTIP group than in the sh-NC group, while the viability of A498 cells (Fig. 2c) in the pc-NC and pc-HOTTIP groups was opposite to that in ACHN cells and OSRC-2 cells. According to the plate cloning assay, there were fewer numbers of colonies of ACHN cells (Fig. 2d, e) and OSRC-2 cells (Fig. 2f, g) in the sh-HOTTIP group than in the sh-NC group, but the trend was opposite for A498 cells (Fig. 2h, i) in the pc-NC and pc-HOTTIP groups. According to the EdU results, there were significantly fewer fluorescent dots indicating nuclei in ACHN cells (Fig. 2j) and OSRC-2 cells (Fig. 2k) in the sh-HOTTIP group than in the sh-NC group, and the trend for A498 cells (Fig. 2l) in the pc-NC and pc-HOTTIP groups was opposite to that for ACHN cells and OSRC-2 cells. The cell cycle analysis results indicated that compared with that in the sh-NC group, the number of ACHN cells (Fig. 2m, n) and OSRC-2 cells (Fig. 2o, p) in the G1 phase in the sh-HOTTIP group was significantly increased, but the number of cells in the S phase was significantly decreased; the number of A498 cells (Fig. 2q, r) in the G1 phase was decreased and in the S and G2 phases was increased in the pc-HOTTIP group compared to those in the pc-NC group. To further verify our results, we added 3-MA (a common autophagy inhibitor) or RAPA (a common autophagy inducer); we found that 3-MA could reverse the effects of HOTTIP silencing on RCC cell proliferation, and RAPA could reverse the effects of HOTTIP overexpression on RCC cell proliferation (Fig. 2). These results indicated that HOTTIP silencing suppressed RCC cell proliferation by inducing autophagy, while HOTTIP overexpression promoted RCC cell proliferation by inhibiting autophagy.
HOTTIP levels affect RCC cell proliferation by modulating autophagy in vitro. a–c ACHN (a), OSRC-2 (b) and A498 (c) cell viability for each group was measured by CCK-8 assays. d–i Representative images and quantitative analyses of ACHN (d, e), OSRC-2 (f, g) and A498 (h, i) cell colonies in each group are displayed according to plate cloning assays. j–l Representative fluorescent images of proliferating ACHN (j), OSRC-2 (k) and A498 (l) cell nuclei are shown by EdU (200 × magnification). m–r Representative images and quantitative analyses of ACHN (m, n), OSRC-2 (o, p) and A498 (q, r) cell cycle status were assessed by flow cytometry. *P < 0.05, **P < 0.01 sh-HOTTIP group vs sh-NC group and pc-HOTTIP group vs pc-NC group; #P < 0.05, ##P < 0.01 3-MA group vs sh-HOTTIP group and RAPA group vs pc-HOTTIP group
HOTTIP expression affects RCC cell migration and invasion by regulating autophagy in vitro
To further assess the effects of HOTTIP expression on the biological behaviors of RCC cells, we detected the migration and invasion of RCC cells in different groups. The scratch assay results showed that compared with that in the sh-NC group, the migration rate of ACHN cells (Fig. 3a, b) and OSRC-2 cells (Fig. 3c, d) was significantly inhibited in the sh-HOTTIP group, and the trend was opposite for A498 cells (Fig. 3e, f) in the pc-NC and pc-HOTTIP groups. Moreover, the transwell migration assay results indicated that compared with that in the sh-NC group, the number of migrated ACHN cells (Fig. 3g, h) and OSRC-2 cells (Fig. 3i, j) was significantly decreased in the sh-HOTTIP group, and the results for A498 cells (Fig. 3k, l) in the pc-NC and pc-HOTTIP groups were opposite to those for ACHN cells and OSRC-2 cells. In addition, the transwell invasion assay results indicated that there were fewer invasive ACHN cells (Fig. 3m, n) and OSRC-2 cells (Fig. 3o, p) in the sh-HOTTIP group than in the sh-NC group, while the invasion results for A498 cells (Fig. 3q, r) were opposite to those for ACHN cells and OSRC-2 cells. Likewise, we found that 3-MA could reverse the effects of HOTTIP silencing on RCC cell migration and invasion, and RAPA could reverse the effects of HOTTIP overexpression on RCC cell migration and invasion (Fig. 3). These results indicated that HOTTIP silencing suppressed RCC cell migration and invasion by inducing autophagy, while HOTTIP overexpression promoted RCC cell migration and invasion by inhibiting autophagy.
HOTTIP levels affect RCC cell migration and invasion by modulating autophagy in vitro. a–f Representative images and quantitative analyses of ACHN (a, b), OSRC-2 (c, d) and A498 (e, f) cell migration rates in each group are shown according to scratch assays (×200 magnification). g–l Representative images and quantitative analyses of penetrated ACHN cells (g, h), OSRC-2 cells (i, j) and A498 cells (k, l) in each group are shown according to transwell migration assays (×200 magnification). m–r Representative images and quantitative analyses of invasive ACHN cells (m, n), OSRC-2 cells (o, p) and A498 cells (q, r) in each group are displayed according to transwell invasion assays (×200 magnification). *P < 0.05, **P < 0.01 sh-HOTTIP group vs sh-NC group and pc-HOTTIP group vs pc-NC group; #P < 0.05, ##P < 0.01 3-MA group vs sh-HOTTIP group and RAPA group vs pc-HOTTIP group
HOTTIP expression affects RCC growth and metastasis in vivo
To assess the effects of HOTTIP expression on RCC growth and metastasis in vivo, we established a xenograft model and a tumor metastasis model, respectively. For ACHN cells (Fig. 4a–d), OSRC-2 cells (Fig. 4e–h) and A498 cells (Fig. 4i–l), xenografts were found in all mice. Moreover, the weights and volumes of the tumors were significantly lower in the sh-HOTTIP group than in the sh-NC group (Fig. 4a–h), while they were significantly higher in the pc-HOTTIP group than in the pc-NC group (Fig. 4i–l). In addition, the IHC results indicated that Ki-67 (a proliferation marker) and MMP-2 (a metastasis marker) expression was significantly lower in the sh-HOTTIP group than in the sh-NC group for ACHN cells (Fig. 4m, n) and OSRC-2 cells (Fig. 4q, r), while their expression levels in the pc-NC and pc-HOTTIP groups for A498 cells (Fig. 4u, v) were opposite to those for ACHN cells and OSRC-2 cells. Finally, the HE staining results showed that compared with that in the sh-NC group, RCC cell (ACHN and OSRC-2) metastasis was also suppressed in the sh-HOTTIP group (Fig. 4o, s), while compared with that in the pc-NC group, RCC cell (A498) metastasis was enhanced in the pc-HOTTIP group (Fig. 4w). These results indicated that RCC growth and metastasis were inhibited by HOTTIP silencing but promoted by HOTTIP overexpression.
HOTTIP levels affect RCC growth, metastasis and autophagy in vivo. a–d Representative images of mice with xenografts (a) and the isolated tumors (b) as well as the weight (c) and volume (d) analyses of the tumors from ACHN cells in the sh-NC and sh-HOTTIP groups are presented. e–h Representative images of mice with xenografts (e) and the isolated tumors (f) as well as the weight (g) and volume (h) analyses of the tumors from OSRC-2 cells in the sh-NC and sh-HOTTIP groups are presented. i–l Representative images of mice with xenografts (i) and the isolated tumors (j) as well as the weight (k) and volume (l) analyses of the tumors from A498 cells in the pc-NC and pc-HOTTIP groups are presented. Representative IHC images of Ki-67 and MMP-2 expression in the tumors from ACHN cells (m, n), OSRC-2 cells (q, r) and A498 cells (u, v) from different groups are displayed (×200 magnification, bar = 50 µm). Representative HE staining images of ACHN (o), OSRC-2 (s) and A498 (w) cell metastases into the lungs of mice from different groups are shown (×100 magnification, bar = 100 µm). Representative IHC images of LC3B expression in the tumors from ACHN cells (p), OSRC-2 cells (t) and A498 cells (x) in the different groups are shown (×200 magnification, bar = 50 µm). *P < 0.05, **P < 0.01 sh-HOTTIP group vs sh-NC group and pc-HOTTIP group vs pc-NC group
HOTTIP expression regulates RCC autophagy in vivo and in vitro
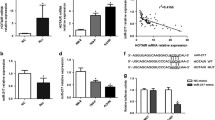
To explore the relationship between HOTTIP expression and autophagy in RCC, we assessed the expression of autophagy-related markers in different groups. Firstly, the IHC results indicated that LC3B expression was much higher in the tumors in the sh-HOTTIP group than in the tumors in the sh-NC group (Fig. 4p, t), while LC3B expression in the tumors in the pc-HOTTIP group was much lower than that in the tumors in the pc-NC group (Fig. 4x). Subsequently, the western blot results showed that compared with that in the sh-NC group, LC3B (Fig. 5a, b), Beclin1 (Fig. 5a, c) and LAMP2 (Fig. 5a, e) expression was significantly increased, while p62 (Fig. 5a, d) expression was significantly decreased in the sh-HOTTIP group. Moreover, LC3B, Beclin1 and LAMP2 protein expression was significantly lower and p62 protein expression was significantly higher in the 3-MA pretreatment group than in the sh-HOTTIP group (Fig. 5a–e). However, the western blot results for RCC cells with HOTTIP overexpression and RAPA pretreatment (Fig. 5f–j) showed an opposite trend to the above. Furthermore, the TEM results indicated that the number of autophagosomes was much higher in the sh-HOTTIP group than in the sh-NC group (Fig. 5k), while the number of autophagosomes was much lower in the pc-HOTTIP group than in the pc-NC group (Fig. 5l). In addition, qRT-PCR results indicated significantly higher LC3B, Beclin1 and Atg13 gene expression and significantly lower p62 gene expression in the sh-HOTTIP group than in the sh-NC group (Fig. 5m), and the trend was opposite for their expression in RCC cells in the pc-NC and pc-HOTTIP groups (Fig. 5n). These results further indicated that HOTTIP silencing induced RCC autophagy, while HOTTIP overexpression inhibited RCC autophagy in vivo and in vitro.
HOTTIP levels regulate RCC cell autophagy. a–e Representative western blot images and quantitative analyses of autophagy-related proteins, including LC3B-II (a, b), Beclin1 (a, c), p62 (a, d) and LAMP2 (a, e), in transfected ACHN cells in the different groups. f–j Representative western blot images and quantitative analyses of autophagy-related proteins, including LC3B-II (f, g), Beclin1 (f, h), p62 (f, i) and LAMP2 (f, j), in transfected A498 cells in the different groups. Representative ultrastructural images of autophagosomes in transfected ACHN cells (k) and A498 cells (l) in the different groups are shown by TEM. The red rectangles indicate the magnified area (1700 × magnification, bar = 2 µm), and the black arrows indicate autophagosomes (5000 × magnification, bar = 1 µm). Relative mRNA expression levels of autophagy-related genes in transfected ACHN cells (m) and A498 cells (n) in the different groups were measured by qRT-PCR. *P < 0.05, **P < 0.01 sh-HOTTIP group vs sh-NC group and pc-HOTTIP group vs pc-NC group; #P < 0.05, ##P < 0.01 3-MA group vs sh-HOTTIP group and RAPA group vs pc-HOTTIP group
HOTTIP expression regulates RCC cell autophagy via the PI3K/Akt/Atg13 signaling pathway
To explore the mechanism involved in the regulation of RCC cell autophagy by HOTTIP expression, we assessed the expression of several critical proteins in transfected RCC cells. The western blot results showed that compared with that in the sh-NC group, the protein expression of p-PI3K (Fig. 6a, b) and p-Akt (Fig. 6a, c) was significantly decreased, while the protein expression of Atg13 (Fig. 6a, d) was significantly increased in the sh-HOTTIP group. Moreover, no significant differences in the protein expression of Atg3 (Fig. 6a, e), Atg5 (Fig. 6a, f) and Atg7 (Fig. 6a, g) were found between the above two groups. However, the western blot results showed opposite trends for the protein expression of p-PI3K (Fig. 6h, i), p-Akt (Fig. 6h, j) and Atg13 (Fig. 6h, k) in RCC cells with HOTTIP overexpression compared to that in RCC cells with HOTTIP silencing, but the protein expression of Atg3 (Fig. 6h, l), Atg5 (Fig. 6h, m) and Atg7 (Fig. 6h, n) was the same as that in RCC cells with HOTTIP silencing. These results indicated that HOTTIP expression could regulate RCC cell autophagy via the PI3K/Akt/Atg13 signaling pathway.
HOTTIP levels regulate RCC cell autophagy via the PI3K/Akt/Atg13 signaling pathway. a–g Representative western blot images and quantitative analyses of several critical proteins, including p-PI3K (a, b), p-Akt (a, c), Atg13 (a, d), Atg3 (a, e), Atg5 (a, f) and Atg7 (a, g), in transfected ACHN cells from the different groups. h–n Representative western blot images and quantitative analyses of several critical proteins, including p-PI3K (h, i), p-Akt (h, j), Atg13 (h, k), Atg3 (h, l), Atg5 (h, m) and Atg7 (h, n), in transfected A498 cells from the different groups. **P < 0.01, ***P < 0.001 sh-HOTTIP group vs sh-NC group and pc-HOTTIP group vs pc-NC group
Discussion
RCC, a malignant tumor with a poor prognosis, is a leading cause of cancer-related death (Turcotte et al. 2008). More than 100,000 people die from RCC worldwide each year (Wu et al. 2014; Ellinger et al. 2016). Similar to most tumors, the tumorigenesis and progression of RCC is a multifactorial, multistep and multistage biological process, but its specific pathogenesis is still unclear (Murai and Oya 2004). As the early diagnosis and treatment of RCC may offer a great help in improving prognosis (Pastore et al. 2015), it is necessary to find a related molecular-targeted therapeutic site for this disease.
It is well known that mechanism research has reached the gene level. Genomic studies have shown that most human genes can be transcribed, but only 2% of the transcripts encode proteins; most transcripts are non-coding RNAs (ncRNAs), including lncRNAs (Qin et al. 2014). Thus, the discovery of lncRNAs is an important addition to genetics. In most cases, lncRNAs possess an mRNA-like structure; some have a poly (A) tail, while others do not. lncRNAs occupy a larger portion of the human transcriptome than protein-coding genes, and most are transcribed by RNA polymerase II. Compared with coding genes, lncRNA expression is diverse in different tissues. Dynamic expression and different splicing methods are involved in cell differentiation (Prensner and Chinnaiyan 2011; Khorkova et al. 2015). Numerous studies have shown that lncRNAs can modulate the development of cancers, including RCC. These lncRNAs either promote cancer as an oncogene or inhibit cancer as an anti-oncogene and have progressively become the focus of contemporary molecular medicine research (Yang et al. 2014).
HOTTIP is a lncRNA newly identified in recent years. It was reported for the first time in the peripheral fibroblasts of humans. It displays a looped gene structure that is adjacent to the promoter and terminator of 5′ on the HOXA site, and it can activate development-related gene expression on the HOXA locus (Zhang et al. 2016; Fu et al. 2017). Furthermore, subsequent studies found that it was associated with the development of several types of cancer. For example, Quagliata et al. (2014) demonstrated that high HOTTIP expression was closely associated with the metastasis and poor prognosis of hepatocellular carcinoma patients. Sun et al. (2017) found that HOTTIP was overexpressed in small cell lung cancer (SCLC) tissues, and its knockdown suppressed cell proliferation, affected the cell cycle and inhibited tumor growth in mice. Li et al. (2015) verified that HOTTIP expression was increased in pancreatic ductal adenocarcinoma (PDAC) tissues and cell lines, and its silencing resulted in proliferation arrest via cell cycle progression alterations and impaired cell invasion via epithelial–mesenchymal transition (EMT) inhibition. In addition, Chang et al. (2016) showed that HOTTIP levels were markedly increased in gastric cancer tissues and cell lines and were correlated with TNM stage and lymph node metastasis, and HOTTIP knockdown inhibited gastric cancer cell proliferation, migration and invasion. These studies indicate the facilitation of HOTTIP in the progression of several types of cancer. Moreover, HOTTIP expression is required for 5′ HOXA gene (including HOXA11) activation (Pradeepa et al. 2017), and HOXA11 was reported to be closely associated with the kidney development and the progression of RCC (Schwab et al. 2006; Wang et al. 2017). These results offer a solid theoretical foundation for studying the role of HOTTIP in RCC. To investigate the expression and function of HOTTIP in RCC, we firstly measured its expression in RCC tissues and cell lines. We found that HOTTIP expression was significantly upregulated in RCC, and its levels were closely correlated with the histological grade, TNM stage, lymph node metastasis and RCC patient prognosis. Subsequently, we silenced and overexpressed HOTTIP in 3 RCC cell lines (ACHN, OSRC-2 and A498), and we found that HOTTIP silencing inhibited RCC cell (ACHN and OSRC-2) proliferation, migration and invasion, as well as G1 phase arrest in the cell cycle, while HOTTIP overexpression in A498 cells yielded the opposite results. These results showed that HOTTIP is significantly upregulated in RCC and it could predict RCC patient prognosis. Moreover, its silencing suppressed RCC progression, and its overexpression promoted RCC progression in vitro.
Autophagy was initially defined as ‘self-eating’, but later studies noted that it was a highly regulated cell process. Specifically, non-lysosomal membrane-like materials with a two-layer structure package the long half-life proteins and degenerative and necrotic organelles due to the external or self-factors to form autophagic vacuoles and autophagosomes; and these structures are subsequently digested and degraded by lysosomes in the eukaryotic cell cytoplasm (Hale et al. 2013; Parzych and Klionsky 2014). During this process, a series of autophagy-related proteins are involved, such as two forms of LC3, LC3-I and LC3-II. LC3-I is cytosolic, while LC3-II is membrane-bound, and the increased levels of LC3-II can be viewed as a biomarker of autophagy. Beclin-1 can be observed during the formation of autophagosomes. Atg3 is an E2-like enzyme that is essential for autophagy vesicle elongation, and its interaction with Atg12 is critical for LC3 lipidation. Atg5 and Atg7 are two key components in the ubiquitin-like system of autophagy. p62 acts as a molecular chaperone during the degradation of autophagosomes. LAMP2 is essential for the degradation of autophagosomal contents upon the fusion of lysosomes with autophagosomes in the late stage of autophagy (Shibutani et al. 2015; Li et al. 2016). Atg13, a protein involved in the initiation of autophagy, is correlated with autophagic flux. Previous studies support the idea that Atg13 acts as a central hub for the complex in autophagy (Chen et al. 2016). Follo et al. (2018) have also identified Atg13 as a static marker of autophagy that closely correlates with the autophagic flux in tumors and found that compared to the protein derived from it, Atg13 maintained its positivity in formalin-fixed tumors, which indicated that it is a relatively stable marker. Moreover, it was found that Atg13 knockdown could inhibit autophagosome formation (Gao et al. 2015). As such, Atg13 is a vital regulatory component of autophagy, and it may be a more clinically significant autophagy marker (Puente et al. 2016). Autophagy not only plays a key role in cellular dynamic balance by degrading and destroying senescent proteins and organelles, but also collaborates with a variety of tumor suppressor genes, so it is considered to play a dual role in tumor progression (Avalos et al. 2014). However, the role of autophagy in these processes remains controversial. Although autophagy plays a protective role for cancers in unfavorable conditions, in several cancers, including human breast, ovarian and prostate cancers, autophagy plays a tumor suppressor role to some extent (Ikeda et al. 2013).
Although an increasing number of studies have focused on the regulation of the biological actions of tumors, such as proliferation, invasion, migration, apoptosis, cell cycle arrest and drug resistance, by lncRNAs (Li et al. 2017; Hong et al. 2017), few reports have examined the relationship between lncRNAs and autophagy in tumors. For example, Ying et al. (2013) demonstrated that downregulated lncRNA-MEG3 could activate autophagy in bladder cancer cells via the p53 pathway. Zhao et al. (2014) reported that lncRNA-HULC induced autophagy in gastric cancer cells and played an important role in the tumorigenesis and growth of gastric cancer. Li et al. (2016) indicated that lncRNA-MALAT1 promoted pancreatic cancer proliferation and metastasis via regulating autophagy. Although Banreti et al. (2014) has reported that HOX genes are potent repressors of autophagy, none of the cancer studies to date have addressed the association of HOTTIP with autophagy, not to mention in RCC. In this study, the nude mouse xenograft assay and the tumor metastasis assay showed that HOTTIP silencing induced tumor autophagy and inhibited tumor growth and metastasis, while its overexpression inhibited tumor autophagy and promoted tumor growth and metastasis in vivo. In addition, the protein and gene expression levels of several autophagy-related molecules as well as autophagosome formation were assessed. The results indicated that the downregulation of HOTTIP induced RCC cell autophagy, while the upregulation of HOTTIP inhibited RCC cell autophagy. Therefore, we added 3-MA (a common autophagy inhibitor) (Roux et al. 2014) or RAPA (a common autophagy inducer) (Kroemer et al. 2010) to regulate autophagy to further verify our results, and we again found that RCC cell proliferation, migration and invasion were significantly reversed. Finally, to explore the specific mechanism through which HOTTIP regulates RCC cell autophagy, we measured the expression of several critical proteins and found that HOTTIP could regulate RCC cell autophagy through the PI3K/Akt/Atg13 signaling pathway. All these results suggested that HOTTIP was highly expressed in RCC, was closely related to RCC patient prognosis, and could affect RCC development by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway.
In conclusion, our findings are consistent with the HOTTIP data from other tumors. Furthermore, we studied the relationship between HOTTIP and autophagy in RCC and explored the relevant mechanism. Our findings may provide a potential marker for the accurate diagnosis of RCC and supply a new molecular target for effective RCC therapy. However, the stability and specificity of HOTTIP, as well as the deeper mechanism involved in RCC progression, still need further exploration and discussion.
References
Avalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF (2014) Tumor suppression and promotion by autophagy. Biomed Res Int 2014:603980
Banreti A, Hudry B, Sass M, Saurin AJ, Graba Y (2014) Hox proteins mediate developmental and environmental control of autophagy. Dev Cell 28:56–69
Chang S, Liu J, Guo S, He S, Qiu G, Lu J, Wang J, Fan L, Zhao W, Che X (2016) HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol Rep 35:3577–3585
Chen S, Wang C, Yeo S, Liang CC, Okamoto T, Sun S, Wen J, Guan JL (2016) Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev 30:856–869
Chen S, Ma P, Zhao Y, Li B, Jiang S, Xiong H, Wang Z, Wang H, Jin X, Liu C (2017) Biological function and mechanism of MALAT-1 in renal cell carcinoma proliferation and apoptosis: Role of the MALAT-1-Livin protein interaction. J Physiol Sci 67:577–585
Deng Q, Wang Z, Wang L, Zhang L, Xiang X, Wang Z, Chong T (2013) Lower mRNA and protein expression levels of LC3 and Beclin1, markers of autophagy, were correlated with progression of renal clear cell carcinoma. Jpn J Clin Oncol 43:1261–1268
Ellinger J, Gevensleben H, Muller SC, Dietrich D (2016) The emerging role of non-coding circulating RNA as a biomarker in renal cell carcinoma. Expert Rev Mol Diagn 16:1059–1065
Follo C, Cheng Y, Richards WG, Bueno R, Broaddus VC (2018) Inhibition of autophagy initiation potentiates chemosensitivity in mesothelioma. Mol Carcinog 57:319–332
Fu Z et al (2017) LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett 410:68–81
Gao Q, Liu H, Yao Y, Geng L, Zhang X, Jiang L, Shi B, Yang F (2015) Carnosic acid induces autophagic cell death through inhibition of the Akt/mTOR pathway in human hepatoma cells. J Appl Toxicol 35:485–492
Hale AN, Ledbetter DJ, Gawriluk TR, Rucker EB (2013) Autophagy: Regulation and role in development. Autophagy 9:951–972
Hong Q, Li O, Zheng W, Xiao WZ, Zhang L, Wu D, Cai GY, He JC, Chen XM (2017) LncRNA HOTAIR regulates HIF-1α/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis 8:e2772
Ikeda T, Ishii KA, Saito Y, Miura M, Otagiri A, Kawakami Y, Shimano H, Hara H, Takekoshi K (2013) Inhibition of autophagy enhances sunitinib-induced cytotoxicity in rat pheochromocytoma PC12 cells. J Pharmacol Sci 121:67–73
Khorkova O, Hsiao J, Wahlestedt C (2015) Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev 87:15–24
Kroemer G, Marino G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40:280–293
Li X, Wu Z, Fu X, Han W (2014) LncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res 762:1–21
Li Z et al (2015) The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med 13:84
Li L et al (2016) Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther 15:2232–2243
Li JK et al (2017) Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer 16:111
Lian Y, Cai Z, Gong H, Xue S, Wu D, Wang K (2016) HOTTIP: A critical oncogenic long non-coding RNA in human cancers. Mol Biosyst 12:3247–3253
Murai M, Oya M (2004) Renal cell carcinoma: Etiology, incidence and epidemiology. Curr Opin Urol 14:229–233
Nishikawa M, Miyake H, Liu B, Fujisawa M (2015) Expression pattern of autophagy-related markers in non-metastatic clear cell renal cell carcinoma: Association with disease recurrence following radical nephrectomy. J Cancer Res Clin Oncol 141:1585–1591
Parzych KR, Klionsky DJ (2014) An overview of autophagy: Morphology, mechanism, and regulation. Antioxid Redox Signal 20:460–473
Pastore AL, Palleschi G, Silvestri L, Moschese D, Ricci S, Petrozza V, Carbone A, Di Carlo A (2015) Serum and urine biomarkers for human renal cell carcinoma. Dis Markers 2015:251403
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641
Pradeepa MM, McKenna F, Taylor GC, Bengani H, Grimes GR, Wood AJ, Bhatia S, Bickmore WA (2017) Psip1/p52 regulates posterior Hoxa genes through activation of lncRNA Hottip. PLoS Genet 13:e1006677
Prensner JR, Chinnaiyan AM (2011) The emergence of lncRNAs in cancer biology. Cancer Discov 1:391–407
Puente C, Hendrickson RC, Jiang X (2016) Nutrient-regulated phosphorylation of ATG13 inhibits starvation-induced autophagy. J Biol Chem 291:6026–6035
Qin C et al (2014) Expression pattern of long non-coding RNAs in renal cell carcinoma revealed by microarray. PLoS One 9:e99372
Quagliata L et al (2014) Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 59:911–923
Roux C, Lesueur C, Aligny C, Brasse-Lagnel C, Genty D, Marret S, Laquerriere A, Bekri S, Gonzalez BJ (2014) 3-MA inhibits autophagy and favors long-term integration of grafted Gad67-GFP GABAergic precursors in the developing neocortex by preventing apoptosis. Cell Transplant 23:1425–1450
Schwab K, Hartman HA, Liang HC, Aronow BJ, Patterson LT, Potter SS (2006) Comprehensive microarray analysis of Hoxa11/Hoxd11 mutant kidney development. Dev Biol 293:540–554
Seles M, Hutterer GC, Kiesslich T, Pummer K, Berindan-Neagoe I, Perakis S, Schwarzenbacher D, Stotz M, Gerger A, Pichler M (2016) Current insights into long non-coding RNAs in renal cell carcinoma. Int J Mol Sci 17:573
Shibutani ST, Saitoh T, Nowag H, Munz C, Yoshimori T (2015) Autophagy and autophagy-related proteins in the immune system. Nat Immunol 16:1014–1024
Shimizu S, Yoshida T, Tsujioka M, Arakawa S (2014) Autophagic cell death and cancer. Int J Mol Sci 15:3145–3153
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65:5–29
Sun Y, Zhou Y, Bai Y, Wang Q, Bao J, Luo Y, Guo Y, Guo L (2017) A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer 16:162
Thorburn A, Thamm DH, Gustafson DL (2014) Autophagy and cancer therapy. Mol Pharmacol 85:830–838
Turcotte S, Sutphin PD, Giaccia AJ (2008) Targeted therapy for the loss of von Hippel-Lindau in renal cell carcinoma: a novel molecule that induces autophagic cell death. Autophagy 4:944–946
Uchida S, Dimmeler S (2015) Long noncoding RNAs in cardiovascular diseases. Circ Res 116:737–750
Wang H et al (2014) LncRNA-regulated infection and inflammation pathways associated with pregnancy loss: genome wide differential expression of lncRNAs in early spontaneous abortion. Am J Reprod Immunol 72:359–375
Wang L, Cui Y, Sheng J, Yang Y, Kuang G, Fan Y, Jin J, Zhang Q (2017) Epigenetic inactivation of HOXA11, a novel functional tumor suppressor for renal cell carcinoma, is associated with RCC TNM classification. Oncotarget 8:21861–21870
White E (2015) The role for autophagy in cancer. J Clin Invest 125:42–46
Williams RT, Yu AL, Diccianni MB, Theodorakis EA, Batova A (2013) Renal cancer-selective Englerin a induces multiple mechanisms of cell death and autophagy. J Exp Clin Cancer Res 32:57
Wu Y, Liu J, Zheng Y, You L, Kuang D, Liu T (2014) Suppressed expression of long non-coding RNA HOTAIR inhibits proliferation and tumourigenicity of renal carcinoma cells. Tumour Biol 35:11887–11894
Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, Ye DQ (2015) Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev 14:798–805
Xu Y, Tong Y, Zhu J, Lei Z, Wan L, Zhu X, Ye F, Xie L (2017) An increase in long non-coding RNA PANDAR is associated with poor prognosis in clear cell renal cell carcinoma. BMC Cancer 17:373
Yang G, Lu X, Yuan L (2014) LncRNA: A link between RNA and cancer. Biochim Biophys Acta 1839:1097–1109
Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F (2013) Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst 9:407–411
Yu G et al (2012) LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One 7:e42377
Zhang SR, Yang JK, Xie JK, Zhao LC (2016) Long noncoding RNA HOTTIP contributes to the progression of prostate cancer by regulating HOXA13. Cell Mol Biol (Noisy-le-grand) 62:84–88
Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y (2014) Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep 31:358–364
Zhou S, Wang J, Zhang Z (2014) An emerging understanding of long noncoding RNAs in kidney cancer. J Cancer Res Clin Oncol 140:1989–1995
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81470923, 81470986, 81770078 and 81770688).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that no conflict of interest exists.
Ethical approval
This study was approved by the Ethical Committee of Renmin Hospital of Wuhan University. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed, and all procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in this study.
Rights and permissions
About this article
Cite this article
Su, Y., Lu, J., Chen, X. et al. Long non-coding RNA HOTTIP affects renal cell carcinoma progression by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin Oncol 145, 573–588 (2019). https://doi.org/10.1007/s00432-018-2808-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-018-2808-0