Abstract
Purpose
To confirm whether flotillin 2 (FLOT2) is a direct target of miR-34a and miR-34a/FLOT2 pathway plays a key role in melanoma proliferation and metastasis.
Methods
First, miR-34a and FLOT2 expressions were both detected in human tissues and cell lines by qRT-PCR. Then, after transfection of mimics/inhibitor of miR-34a into melanoma cell lines, MTT, colony formation, scratch migration assays and transwell invasion assays were performed to evaluate the impact of miR-34a on cell proliferation and metastasis. Western blot, qRT-RCR and dual luciferase reporter gene assays were carried out to confirm whether FLOT2 is a direct target gene of miR-34a. In functional recovery experiments, proliferation and metastasis ability of WM35 and WM451 was tested after being co-transfected with miR-34a inhibitor/si-FLOT2 or miR-34a mimics/FLOT2 cDNA to confirm that FLOT2 is downregulated by miR-34a.
Results
The miR-34a significantly lower-expressed in metastasis melanoma tissues compared to in situ melanoma, nevi and normal skin whereas FLOT2 has an opposite trend. The level of miR-34a and FLOT2 in different melanoma cell lines was also texted and found that metastatic melanoma cell lines has lower miR-34a expression and higher FLOT2 expression compare to in situ melanoma cell line. MiR-34a overexpression profoundly inhibits WM451 cell proliferation and metastasis, whereas miR-34a reduction had a promoting effect to proliferation and metastasis of WM35. Results of Western blot, qRT-RCR and dual luciferase reporter gene assays revealed that FLOT2 is a direct target gene of miR-34a. Furthermore, overexpression/blockage of FLOT2 could attenuate effect of miR-34a overexpression/inhibition which indicated miR-34a suppresses melanoma biological behavior partially through FLOT2 inhibition.
Conclusions
Our study confirmed that miR-34a is involved in the tumor inhibition of melanoma by directly targeting FLOT2 gene. This finding provides potential novel strategies for therapeutic interventions of melanoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant melanoma is the most dangerous type of skin cancer, which begins in skin and spreads quickly. The ten-year survival rate drops dramatically with tumor metastasis (Bhatia and Thompson 2012) and the morbidity increases with age (Balch et al. 2013). Local skin lesions can be cured by surgery, but for the majority of patients, chemotherapy, immunotherapy or small molecule inhibitors of BRAF do not have satisfied curative effects. As a result, the prognosis for advanced melanoma remains very poor (Siegel et al. 2012).
MicroRNAs are small noncoding RNAs of approximately 22 nucleotides in length, which posttranscriptionally regulate the expression of thousands of genes (Lim et al. 2005). In cancer, microRNAs act as oncogenes (He et al. 2005) or oncosuppressor genes (Iorio and Croce 2012). They play important roles in melanoma proliferation and metastasis. For example, mir-155 can downregulate SKI protein and inhibits melanoma cell growth in vitro (Levati et al. 2011) and miR-211 regulates melanoma cell invasiveness (Boyle et al. 2011). Nemlich et al. found overexpression of miR-17 and miR-432 silences ADAR1 in melanoma (Nemlich et al. 2013) and promotes metastatic melanoma tumor growth. Furthermore, we previously found that let-7b, miR-199a and miR-33 are significantly associated with metastatic melanoma (Xu et al. 2012).
Mir-34a can be activated by TP53 and plays an important function in the TP53 tumor suppressor network (He et al. 2007a; Tarasov et al. 2007). Overexpression of miR-34a can inhibit the invasiveness and growth of melanoma with stimulating apoptosis (Yamazaki et al. 2012). However, the mechanism by which mir-34a inhibits melanoma metastasis has not been fully elucidated.
Flotillin 2 (FLOT2) was first identified in goldfish (Malaga-Trillo et al. 2002). It encodes an caveolae-associated membrane protein and is widely expressed in mammalian tissue (Volonte et al. 1999). FLOT2 was found to participate in various physiological and pathological processes such as axon regeneration, neuronal differentiation, tumor progression and endocytosis (Zhao et al. 2011). Upregulation of FLOT2 in SB2 melanoma cells can increase their tumorigenic and metastatic ability (Hazarika et al. 2004). Furthermore, Doherty found that high expression of FLOT2 is associated with melanoma lymph node metastasis and Breslow depth (Doherty et al. 2006). Thus, in summary, FLOT2 was regarded as a tumor-promoting factor in melanoma. And in our study before, we found that inhibition of FLOT2 can suppress the cell proliferation, migration and invasion in gastric carcinoma cells (Cao et al. 2014a).
There are three possible binding sites of miR-34a in the 3’ UTR of human FLOT2, at positions 206–213 bp, 1,171–1,177 bp and 1,179–1,185 bp, as identified using TargetScan bioinformatic software (Cambridge, MA). This indicates miR-34a may be an upstream regulator of FLOT2 and influences the biological behavior of melanoma through it. Focusing on posttranscriptional mechanisms, we provided evidence herein that miR-34a can inhibit melanoma cell proliferation and metastasis through inhibition of FLOT2.
Materials and methods
Tissue and cell lines
Melanoma in situ (six cases), metastatic melanoma (six cases), nevi tissue (six cases) and adjacent normal skin (located >5 cm from tumors/nevus, 18 cases) tissues were collected in our department through surgery. This work was approved by the Ethics Committee of third Xiangya Hospital of Central South University, and all tumor samples were collected after written informed consent was obtained.
Human WM35 melanoma cells (ATCC, Manassas, VA) were established from a primary superficial spreading melanoma (SSM) in radial growth phase (RGP)/vertical (VGP) growth phase, considered as “early-stage” of melanoma. WM451 melanoma cells (Jennio Biotical Company, Guangzhou, China) were established from distant tumor metastasis that was considered as “advanced-stage” melanoma (Lu et al. 1993). A375 (from Cell Bank of Xiangya Hospital) is a human metastasis cell line, and HM is the primary culture of human nevi tissue cell. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY) supplemented with 10 % fetal bovine serum (FBS, Gibco) and 1 % penicillin and streptomycin (Gibco) and incubated at 37 °C in a humidified incubator with 5 % CO2.
Cell transfection
MiR-34a mimics (Hsa-miR-34a-mimics:5′-UGGCAGUGUCUUAGCUGGUUGU-3′), miR-34a inhibitor (Hsa-miR-34a-inhibitor:5′-ACAACCAGCUAAGACACUGCCA-3′), siRNA against FLOT2 (5′-GCCUGUCCCUUCUGGUAAATT-3′) and their negative control were purchased from GenePharma Biotechnology Company (Shanghai, China). The GV230-FLOT2-cDNA plasmid (POSE 230002185) and the negative control were bought from GeneChem Biotechnology Company (Shanghai, China). Cells were transfected using Lipofectamine RNAiMAX (Invitrogen, Grand Island, NY) when cell confluence had reached 40–50 % in 6 cm culture dishes. Forty-eight hours after transfection, qRT-PCR was performed to check transfection efficiency.
Quantitative real-time PCR
MiR-34a and FLOT2 sequences were acquired from GenBank. The primer sequence for FLOT2 was as follows: forward, 5′-GGCAGTAGGAAACTGAGGAAGCT-3′, and reverse, 5′-GGACTGGTCTTCCCAGCCCTAAA-3′. The primer sequence of miR-34a was as follows: forward, 5′-AGCCGCTGGCAGTGTCTTA-3′, and reverse, 5′-CAGAGCAGGGTCCGAGGTA-3′. Total RNA was extracted from cells and tissue using TRIzol (Dingguo Biotechnology Company, Beijing, China). The purity and integrity of RNA were detected using micro-ultraviolet spectrophotometry and RNA electrophoresis after purification. After DNase treatment (RQ1 RNase-free DNase, Promega, Madison, WI), RNA was reverse transcribed into cDNA using a TOYOBO Reverse Transcription System (Japan) on a ABI-9600 PCR Amplifier (Grand Island, NY) and detected with a ABI PRISM 7700 (Grand Island, NY). Samples were compared using the relative CT method, the relative expression of miR-34a was normalized against U6 and FLOT2 was normalized against ACTB (β-actin). The fold change was determined relative to a control after normalizing to a housekeeping gene using 2−ΔΔCT.
Western blot
Cells were harvested 48 h after transfection, and proteins were extracted and then quantified with a BCA protein assay kit and separated by 10 % sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Gels were transferred to nitrocellulose membranes and probed with 0.5 mg/ml anti-FLOT2/anti- PAR1 antibody (Bioss Biotechnology Company, Beijing, China). Following incubation with a rabbit radish peroxidase-conjugated secondary antibody, immunoreactive bands were visualized by electrochemiluminescence. The relative intensity of interest protein level is count by the formula: the gray value of interest protein band/the gray value of internal reference protein (β-actin) band.
MTT assay and colony formation assay
For MTT proliferation assays, cells were seeded into 96-well plates (3,000/well) after transfection. MTT (5 mg/ml) was added to the cells followed by 150 μl DMSO (4 h after MTT application) at 24, 48, 72, 96 and 120 h. Then the OD value was tested at 490 nm to produce a growth curve indicative of the ability of the cells to proliferate.
For colony formation assays, cells were seeded in 10 cm dishes (1,000/plates) after transfection and maintained in complete culture medium for 21 days. Then, cells were fixed in 4 % paraformaldehyde for 15 min and stained with Gimsa dye. The number of clones was record and calculated by the formula: colony formation rate = total colony number/1,000 × 100 %.
Scratch migration assays and transwell invasion assays
Cells were seeded in 6-well plates (5 × 105/well) after transfection and cultured for 12 h. Then a straight scratch was made in the cell monolayer using a sterile 1 μl tip. Cells were continuously cultured and imaged at 0 and 24 h. The relative migration area was calculated by the formula: relative migration area at 24 h = (scratch area at 0 h—remaining scratch area at 24 h)/scratch area at 0 h × 100 %.
To measure invasive capacity, transwell invasion assays were performed using transfected cells plated on 8 µm pore size matrigel-coated membranes, at 1 × 105/well in serum-free medium, which were in turn placed in the top chamber of 24-well transwell plates. DMEM containing 10 % FBS was added to the bottom chamber as a chemo-attractant. After 24 h, cells on the upper surface were removed, while cells attached on the bottom membranes were fixed in 4 % paraformaldehyde for 20 min and stained with hematoxylin then fixed and stained. Cells were imaged and the number of cells was calculated under a microscope (choose five high power fields to count the number). Invaded cell number per field was calculated by the formula: invaded cell number per field = the total count of five high power fields/5.
Dual luciferase reporter assays
Luciferase reporter assays were performed using the psiCHECK2-FLOT2-3′-UTR/MUT1/MUT2 vector. WM451 cells were grown to approximately 80 % confluence in 6-well plates and co-transfected with 1 μg psiCHECK2-FLOT2-3′-UTR/MUT1/MUT1 vector (sequence as follows, FLOT2-UTR: forward, AGGCCCACTCTCTTCAGCA(XhoI); reverse, GAGTTCTTTATTTTACCAGAAGGGAC(NotI); FLOT2-MUT1: forward, ACTTTCATCACCACTTTCATCAGTTTGTG, reverse, ACTTTCATCACCACTTTCATCAGTTTGTG; FLOT2-MUT2: forward, ATGTTGCATCACCCCATCACCTGTCCCTTCTGGTA, reverse, AAGGGACAGGTGATGGGGTGATGCAACATCCAAGCC) plus Hsa-miR-34a-mimics/Hsa-miR-34a-mimics-NC (1 μg). Cells were incubated with transfection reagent/DNA complex for 3 h. In a separate set of experiments, cells were incubated for 48 h after transfection. Firefly and Renilla luciferase activities were evaluated using the Dual Luciferase Reporter Assay system (Promega), and Renilla luciferase activity was normalized to firefly luciferase activity.
Statistical analysis
All experiments were repeated independently at least in triplicate, and the results were expressed as the mean ± SD. The results were assessed by a one-way ANOVA or Student’s t test. A value of P < 0.05 was accepted to indicate statistical significance.
Results
MiR-34a and FLOT2 expression in melanoma tissue and cell lines
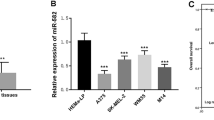
To validate the expression levels of miR-34a in melanoma, we performed quantitative real-time PCR (qRT-PCR) of miR-34a and FLOT2 mRNA generated from nevi, in situ melanoma and metastasis melanoma tissue, and corresponding normal skin tissue (located >5 cm away from the tumor/nevi) (Table 1). The results of miR-34a expression are similar to the results of Satzger (Satzger et al. 2010), its expression was significantly lowered in melanoma tissues, both in in situ (p < 0.01) and metastatic tumor (p < 0.001), whereas there was no obvious change in nevi (p > 0.05). And metastatic melanoma tissues have lower miR-34a level compared to in situ melanoma (p < 0.05) (Fig. 1a). The results of FLOT2 expression show an opposite trend to miR-34a. FLOT2 expresses higher in nevi (p < 0.05), in situ melanoma (p < 0.001) and metastatic melanoma (p < 0.001) comparing to normal tissue, and metastatic melanoma has higher FLOT2 than in situ tumor (p < 0.001) (Fig. 1b). We make a correlation analysis between the FLOT2 and miR-34a expression and found it has an inverse relationship (r = −0.6140, p = 0.0148) (Fig. 1c).
MiR-34a and FLOT2 expression in melanoma tissue and cell lines. a qRT-PCR for miR-34a was performed using six in situ melanoma tissue samples, six metastatic melanoma tissue samples, six nevi samples and matched adjacent non-tumor normal tissues; b qRT-PCR for FLOT2 mRNA was performed on the same 18 samples above; c the correlativity between miR-34a and FLOT2 using the same 18 samples above. d qRT-PCR for miR-34a was performed on HM (set as 1), WM35, A375 and WM451 cell lines. e qRT-PCR for FLOT2 mRNA was performed on the HM (set as 1), WM35, A375 and WM451 cell lines. (ns: p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001)
We also tested miR-34a/FLOT2 expression in metastasis melanoma cell lines A375/WM451, in situ melanoma WM35 cell lines and primary culture of human nevi tissue cell line HM. We found miR-34a exhibited lower expression in all the melanoma cell lines (A375 and WM451 with p < 0.001 and WM35 with p < 0.05) comparing to normal nevi cell HM. What’s more, metastasis melanoma cell lines (A375 and WM451 with p < 0.001) express lower miR-34a than in situ melanoma cell (WM35) (Fig. 1d). The results similar to previous reports (Greenberg et al. 2011, Yamazaki et al. 2012) indicate that miR-34a has an important function in melanoma and may be associated with malignancy grade. Also FLOT2 has an opposite trend to miR-34a, it expresses higher in melanoma cell (WM35 with p < 0.01 and A375, WM451 with p < 0.001) compare to normal nevi cell HM, and higher in advanced melanoma cell (A375 with p < 0.01 and WM451 with p < 0.001) compare to early-stage melanoma cell WM35 (Fig. 1e).
Change of FLOT2 after overexpression/inhibition of miR-34a in melanoma
We choose two cell lines for the further experiments: WM451, a metastatic melanoma cell line which has lower miR-34a/higher FLOT2, and WM35, an in situ melanoma cell line which has high miR-34a/low FLOT2 expression. The WM35 was transfected with miR-34a inhibitor, and the negative control (NC) and WM451 was transfected with miR-34a mimics and the negative control. After transfection, qRT-PCR for miR-34a was performed to make sure the success of transfection. The cells which were transfected successfully have miR-34a overexpression in miR-34a-mimics-WM451 (p < 0.05, Fig. 2a) and miR-34a inhibition in miR-34a-inhibitor-WM35 (p < 0.05, Fig. 2d).
Change of FLOT2 after overexpression/inhibition of miR-34a in melanoma. a miR-34a qRT-PCR results of the 3 Groups: WM451 (a blank group without any treatment), NC-WM451 (a control group transfected with Hsa-miR34a-mimics-NC) and miR-34a-mimics-WM451 (a treatment group transfected with Hsa-miR34a-mimics). We set the expression level of WM451 group as 1, and then calculated the 2−ΔΔCT value for each group to show the relative expression level. b FLOT2 mRNA qRT-PCR results of WM451, NC-WM451 and miR-34a-mimics-WM451 groups after miR-34a transfection. c Western blot band image of the WM451, NC-WM451 and miR-34a-mimics-WM451 group, the relative intensity of FLOT2/β-actin was showed under the band image. d miR-34a qRT-PCR results of the 3 Groups: WM35 (a blank group without any treatment), NC-WM35 (a control group transfected with Hsa-miR34a-inhibitor-NC) and miR-34a-inhibitor-WM35 (a treatment group transfected with Hsa-miR34a-inhibitor). We set the expression level of WM35 group as 1, and then calculated the 2−ΔΔCT value for each group to show the relative expression level. e FLOT2 mRNA qRT-PCR results of WM35, NC-WM35 and miR-34a-inhibitor-WM35 groups after miR-34a transfection. f Western blot band image of the WM35, NC-WM35 and miR-34a-inhibitor-WM35 group, the relative intensity of FLOT2/β-actin was showed under the band image. (ns: p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001)
FLOT2 can promote the growth and metastasis of multiple tumor types. And in previous study, we found it can promote tumor cell proliferation, migration and invasion in gastric carcinoma cells (Cao et al. 2014a). Using microRNA target gene prediction software, we identified FLOT2 as a target gene of miR-34a. So we used Western blot and qRT-PCR to confirm the level of FLOT2 in the above mentioned six groups. The results of qRT-PCR demonstrated FLOT2 was downregulated when miR-34a was overexpressed in miR-34a-mimics-WM451 group comparing to the control group NC-WM451 (Set blank treatment group WM451 as 1, p < 0.05, Fig. 2b), and increased in miR-34a-mimics-WM35 when miR-34a was inhibited (comparing to the control group NC-WM35, set blank treatment group WM35 as 1, p < 0.001, Fig. 2e). We take β-actin as a reference gene to test the protein level of FLOT2 in the 6 groups of transfected cells, and the results are similar to the qRT-PCR results (Fig. 2c, f). Taken together indicated that FLOT2 was inhibited by miR-34a and may be a target gene of miR-34a.
Effect of miR-34a on melanoma cell proliferation
MiR-34a expression was significantly reduced in melanoma tissue and advanced-stage melanoma cells, thus we sought to confirm if it acted as a tumor suppressor in WM35 and WM451 cells.
We used MTT assays to detect the cell proliferation of the six groups described before, from which we constructed proliferation curves in 120 h. Cells transfected with miR-34a precursor (miR-34a-mimics-WM451) grew much slower than the control group (NC-WM451) and blank group (WM451). To the opposite, cells transfected with miR-34a inhibitor (miR-34a-inhibitor-WM35) grew more rapidly than the control group (NC-WM35) and blank group (WM35) (Fig. 3a, b. The proliferative ability of miR-34a-mimics-WM451 was clearly restrained while the proliferation of miR-34a-inhibitor-WM35 was promoted.
Effect of miR-34a on melanoma cell proliferation. WM451 or WM35 cells were transfected with miR-34a precursor/inhibitor or the corresponding control. a, b At 24, 48, 36, 72, 96 and 120 h after transfection, MTT assays were performed to examine WM451 and WM35 proliferation, the OD value was record and was made a time-OD value curve to show the trend of growth; c, d colony formation assay in WM451 and WM35 cells transfected with miR-34a precursor/inhibitor, corresponding control or blank control. The colony formation rate was calculated as formula describe before. (ns: p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001)
We use colony formation assays to further confirm the inhibitory effect of miR-34a on melanoma growth. The colony formation rate of miR-34a-mimics-WM451 was much lower than WM451 and NC-WM451, whereas miR-34a-inhibitor-WM35 demonstrated increased colony formation capacity compared with WM35 and NC-WM35 (Fig. 3c, d).
Thus, MTT and colony formation assays showed that miR-34a overexpression can limit WM451 cell proliferation and weakness of miR-34a can promote WM35 cell proliferation, indicating that miR-34a can inhibit melanoma growth.
Effect of miR-34a on melanoma cell migration and invasion
After confirming miR-34a inhibition to melanoma cell proliferation, we wanted to investigate its effect on cell migration and invasion. We use a scratch migration assay to test the migratory ability of the six groups. The upper image showed the scratch area of each group at 0 or 24 h after scratch was made. And the migration area of WM451 and NC-WM451 cells at 24 h after scratch are much larger than miR-34a-mimics-WM451 cells. Moreover, the migration area of WM35 and NC-WM35 cells were smaller than miR-34a-inhibitor-WM35 cells (Fig. 4a, b), indicating that the migratory ability of melanoma cells was inhibited by miR-34a and promoted by miR-34a inhibition.
Effect of miR-34a on melanoma cell migration and invasion. a, b The scratch migration assay of WM451 and WM35 cells transfected with miR-34a precursor/inhibitor. Representative microscopic images (×40) are shown on the upper row. The edge of scratch was showed by the black line on images. The relative migration area at 24 h was calculated as formula mentioned before and showed under the image. c, d Transwell invasion assay of WM451 and WM35 cells transfected with miR-34a precursor/inhibitor. Representative microscopic images (×200) are shown on the upper row. The invaded cell number per field was calculated as formula mentioned before and showed under the image. (ns: p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001)
Also, transwell invasion assays were performed to explore the impact of miR-34a on the invasive ability of WM451 and WM35 cells. The upper images are cells invaded on the bottom of membrane. And the invaded cell number per field was showed below. The results shows that cells overexpressing miR-34a (miR-34a-mimics-WM451) exhibited much lower invasion compared with WM451 and NC-WM451 cells, but the number of invading WM35 cells with inhibited miR-34a (miR-34a-inhibitor-WM35) was much higher than WM35 and NC-WM35 cells (Fig. 4c, d). These results are similar to the scratch assay findings.
In summary, miR-34a could inhibit WM451 migration and invasion, while inhibition of miR-34a could promote WM35 migration and invasion, indicating its ability to suppress the metastasis of melanoma cells.
FLOT2 is directly inhibited by miR-34a acting on the 3′-UTR
As we see that FLOT2 has an inverse relationship to miR-34a in melanoma tissue, and inhibition of miR-34a increased FLOT2 in WM35 while overexpression of miR-34a inhibit FLOT2 in WM451. We have the hypothesis that FLOT2 is a downstream target gene of miR-34a, which means miR-34a can directly inhibit FLOT2 expression. Using TargetScan software, we found that there might be three possible binding site of miR-34a to FLOT2-3′UTR. We then use dual luciferase reporter assays to confirm that miR-34a can directly affect FLOT2-3′UTR. We amplified the FLOT2 3′-UTR (Supplement Fig. 1(a)), which has three possible biding sites (Supplement Fig. 1(b)), then inserted it into a psiCHECK2 luciferase reporter vector (Fig. 5a) as wild type (WT). Also, we constructed two mutation sequences as MUT1 and MUT2 (Supplement Fig. 1(c)) which also be inserted into the psiCHECK2 luciferase reporter vector. The vectors were, respectively, co-transfected into WM451 cell with miR-34a-mimics or the negative control. Then luciferase activity was tested (Fig. 5b, set the FLOT2-WT-miR-34a-NC group as 1, p < 0.05), the relative luciferase activity show only FLOT2-WT-miR-34a-mimics has a reduced luciferase activity, and other groups have no significant difference, which means miR-34a could bind to FLOT2-3′UTR and can directly suppress its expression. Mutations in FLOT2-3′UTR will lead to failed combination and make miR-34a have no suppressed effect. In conclusion, these results show FLOT2 is a direct target gene of miR-34a, which acting on the 3′-UTR of FLOT2 at the posttranscriptional level.
FLOT2 can promote melanoma progression and miR-34a inhibit melanoma progression by targeting FLOT2
To further confirm that in melanoma, miR-34a inhibit tumor progression trough FLOT2 inhibition, we carried out functional recovery experiments. We co-transfected miR-34a-mimics/GV230-FLOT2-cDNA and their negative control into the WM451, which formed 6 groups: blank treatment group (WM451), two negative control groups (miR-34a-mimics-NC-WM451 and FLOT2-NC-WM451), the group only transfected with miR-34a-mimics (miR-34a-mimics-WM451), the group only transfected GV230-FLOT2-cDNA (FLOT2-WM451) and the co-transfection group (miR-34a-mimics-FLOT2-WM451). We also made a co-transfection of miR-34a-inhibitor/siFLOT2 into WM35, which formed similar 6 groups as in WM451 (WM35, miR-34a-inhibitor-NC-WM35, siFLOT2-NC-WM35, miR-34a-inhibitor-WM35, siFLOT2-WM35 and miR-34a-inhibitor-siFLOT2-WM35).
After transfection, qRT-PCR (Fig. 6a, b) and Western blot (Fig. 6c, d) of FLOT2 were performed on each group. FLOT2 was downregulated when miR-34a overexpression in WM451 and co-transfection of FLOT2 cDNA could raise FLOT2 level. FLOT2 was upregulated when miR-34a was inhibited in WM35. However, co-transfection of siFLOT2 can block the FLOT2 increasing caused by miR-34a inhibition.
Expression of FLOT2 in functional recovery experiments. a qRT-PCR of FLOT2 mRNA in WM451, miR-34a-mimics-NC-WM451, FLOT2-NC-WM451, miR-34a-mimics-WM451, FLOT2-WM451 and miR-34a-mimics-FLOT2-WM451. b qRT-PCR of FLOT2 mRNA in WM35, miR-34a-inhibitor-NC-WM35, siFLOT2-NC-WM35, miR-34a-inhibitor-WM35, siFLOT2-WM35 and miR-34a-inhibitor-siFLOT2-WM35. c Western blot of WM451, miR-34a-mimics-NC-WM451, FLOT2-NC-WM451, miR-34a-mimics-WM451, FLOT2-WM451 and miR-34a-mimics-FLOT2-WM451. d Western blot of WM35, miR-34a-inhibitor-NC-WM35, siFLOT2-NC-WM35, miR-34a-inhibitor-WM35, siFLOT2-WM35 and miR-34a-inhibitor-siFLOT2-WM35. (Set the FLOT2-WT-miR-34a-NC group as 1, *p < 0.05)
We next used MTT and colony formation assays to detect the proliferative ability of the groups. As shown in the MTT growth curve (Fig. 7a, b) and the colony formation rate (Fig. 7c, d), both in WM451 and WM35, the blank group and the two negative control groups had a similar proliferation curve or the colony formation rate. Comparing to the blank and control group, miR-34a-mimics inhibit WM451′s proliferation while miR-34a-inhibitor promotes WM35’s proliferation, and FLOT2 cDNA promotes WM451’s proliferation while siFLOT2 inhibit WM35’s proliferation. Compare the co-transfection group to the miR-34a-mimics/inhibitor group: In WM451, overexpression of FLOT2 could attenuate the anti-tumor effect of miR-34a overexpression, in WM35, inhibition of FLOT2 could attenuate the tumor-promoting effect of miR-34a inhibition. These data show miR-34a inhibit melanoma’s growth targeting FLOT2 inhibiting.
Mir-34a inhibit melanoma proliferation targeting FLOT2 inhibition. a MTT proliferation value curve for 120 h of WM451, miR-34a-mimics-NC-WM451, FLOT2-NC-WM451, miR-34a-mimics-WM451, FLOT2-WM451 and miR-34a-mimics-FLOT2-WM451. b MTT proliferation value curve for 120 h of WM35, miR-34a-inhibitor-NC-WM35, siFLOT2-NC-WM35, miR-34a-inhibitor-WM35, siFLOT2-WM35 and miR-34a-inhibitor-siFLOT2-WM35. c Results of colony formation assays of WM451, miR-34a-mimics-NC-WM451, FLOT2-NC-WM451, miR-34a-mimics-WM451, FLOT2-WM451 and miR-34a-mimics-FLOT2-WM451. d Results of colony formation assay of WM35, miR-34a-inhibitor-NC-WM35, siFLOT2-NC-WM35, miR-34a-inhibitor-WM35, siFLOT2-WM35 and miR-34a-inhibitor-siFLOT2-WM35. (ns: p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001)
Then scratch migration assays (Fig. 8a, b) and transwell invasion assays (Fig. 8c, d) also performed on the groups to detect the metastatic ability. Both in WM451 and WM35, the blank group and the two negative control groups had a similar migration area or invaded cell number. Comparing to the blank and control group, miR-34a-mimics inhibit WM451’s migration and invasion while miR-34a-inhibitor promotes WM35’s migration and invasion, and FLOT2 cDNA promotes WM451’s metastasis while siFLOT2 inhibit WM35’s metastasis. Compare the co-transfection group to the miR-34a-mimics/inhibitor group: In WM451, overexpression of FLOT2 could attenuate the anti-metastasis effect of miR-34a overexpression, in WM35, inhibition of FLOT2 could attenuate the metastasis-promoting effect of miR-34a inhibition. These data show miR-34a inhibit melanoma’s metastasis ability by targeting FLOT2 inhibition.
Mir-34a inhibits melanoma metastasis targeting FLOT2 inhibition. a The relative migration area at 24 h of WM451, miR-34a-mimics-NC-WM451, FLOT2-NC-WM451, miR-34a-mimics-WM451, FLOT2-WM451 and miR-34a-mimics-FLOT2-WM451. b The relative migration area at 24 h of WM35, miR-34a-inhibitor-NC-WM35, siFLOT2-NC-WM35, miR-34a-inhibitor-WM35, siFLOT2-WM35 and miR-34a-inhibitor-siFLOT2-WM35. c Results of transwell invasion assay of WM451, miR-34a-mimics-NC-WM451, FLOT2-NC-WM451, miR-34a-mimics-WM451, FLOT2-WM451 and miR-34a-mimics-FLOT2-WM451. d Results of transwell invasion assay of WM35, miR-34a-inhibitor-NC-WM35, siFLOT2-NC-WM35, miR-34a-inhibitor-WM35, siFLOT2-WM35 and miR-34a-inhibitor-siFLOT2-WM35. (ns: p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001)
All the data of functional recovery experiments indicate miR-34a inhibit melanoma’s biography behavior of proliferation and metastasis by targeting FLOT2 inhibition.
Discussion
Melanoma, the malignant transformation of melanocytes, has the highest fatality and malignancy of all the skin cancers. Melanoma also can occur in some rare sites such as the uvea (Spagnolo et al. 2012), mouth (Bhullar et al. 2012), central nervous system (Shang et al. 2013) and vagina (Kahr et al. 2013). Cancer statistics in 2013 reports an estimated 9,710 deaths owing to melanoma in 2014 compared with 3,270 from non-epithelial skin cancer (Siegel et al. 2014). From 1992 to 2010, the incidence of melanoma, which is on the increase, ranked 5th in males and 7th in females of all malignancies (Siegel et al. 2014). The mortality rate of melanoma has a close relationship with tumor metastasis. According to Cancer Facts and Figures 2013, in early-stage melanoma without lymph and organ metastasis the 5-year survival rate is 98 %; this drops to 62 % for lymph metastasis and to 15 % for metastasis to distant organs (Cancer Facts and Figures 2013). Hence, understanding the mechanism of melanoma metastasis is critical to improve survival rates.
MicroRNAs are small noncoding RNAs of approximately 22 nucleotides that posttranscriptionally regulate the expression of thousands of genes (Cao et al. 2014b). Many studies have found specific microRNAs not only affect the development and invasion of tumors, but also play roles as tumor suppressor genes (Lu et al. 2014) or oncogenes (Pritchard et al. 2012). The study of microRNAs has also become a hotspot in melanoma research. A growing number of microRNAs have been found to be involved in melanoma progression, and associated signaling pathways and downstream genes have been identified. In our previous study, endostatin (Xiao et al. 2005), miR-199a, miR-33 and let-7b (Xu et al. 2012) were found to inhibit the growth and metastasis of melanoma.
The miR-34 family has an inhibitory effect on melanoma, and in humans, the family includes miR-34a, miR-34b and miR-34c (He et al. 2007b). MiR-34a is located at 1p36.23 and contains 110 nucleotides and can be triggered by TP53 (Tarasov et al. 2007) to participate in a variety of pathological processes such as apoptosis of pancreas islet B cells (Nesca et al. 2013), obesity and aging (Choi et al. 2013), glucose metabolism (Kim et al. 2013), tumor development [including breast cancer (Eichelser et al. 2013), pancreatic cancer (Chakraborty et al. 2013), cervical cancer (Gocze et al. 2013), melanoma, lung cancer (Garofalo et al. 2013)] and heart failure caused by acute myocardial infarction (Matsumoto et al. 2013). In a study in 2008, miR-34a was found to be deactivated owing to abnormal methylation in a variety of cancers including melanoma (Lodygin et al. 2008). Furthermore, it was also reported that compared with melanin nevus, expression of miR-34a in melanoma is much lower (Satzger et al. 2010). Greenberg (Greenberg et al. 2011) and Yamasaki (Yamazaki et al. 2012) also found that in melanoma, lower expression of miR-34a indicated increased tumor aggressiveness and higher proliferative capacity. Ectopically overexpressed miR-34a can inhibit the growth of melanoma and promote melanoma cell apoptosis (Chen et al. 2010).
Our experiments verify that sequence of miR-34a’s expression level is nevi > in situ melanoma > metastasis melanoma, which of confirmed by tissue and cell line qRT-PCR examination. As WM35 has the highest expression of miR-34a in tumor cell line and WM451 has the lowest miR-34a expression. We choose WM35 and WM451 for the follow-up experiments which, respectively, to stand for in situ melanoma and metastasis melanoma. We further used gene overexpression and silencing to upregulate miR-34a in WM451 and downregulate miR-34a in WM35, respectively. The experiment results of cell proliferation and metastasis tests suggest that miR-34a expression can significantly inhibit the proliferation and metastasis of WM451, while silencing of miR-34a can promote the proliferation and metastasis of WM35.
The mechanism by which miR-34a inhibits melanoma growth and metastasis has been explored but not fully illustrated. A previous report suggested that C-MET is a downstream target gene of miR-34a, through which miR-34a inhibits the proliferation and migration of uveal melanoma (Yan et al. 2009). Furthermore, other researchers found that miR-34a/c may inhibit melanoma through inhibition of ULBP2 expression (Heinemann et al. 2012).
In the former experiments, we found that FLOT2 can suppress the cell proliferation, migration and invasion in gastric carcinoma cells (Cao et al. 2014a). FLOT2 belongs to the flotillin family, which was first identified in goldfish (Malaga-Trillo et al. 2002), and is a conserved protein expressed in vertebrates and invertebrates (Rivera-Milla et al. 2006). FLOT2 is widely expressed in tissue (Volonte et al. 1999) and encodes a cell membrane valve protein that participates in a variety of physiological and pathological process such as axonal regeneration and neural differentiation, endocytosis, insulin signals, cell proliferation and tumor progression. FLOT2 is highly expressed in metastatic melanoma cell lines and melanoma tumor tissues. In an earlier study, SB2 melanoma cells transfected with FLOT2 exhibited significant upregulation of tumorigenicity and metastasis, as reflected in a nude mice transplantation tumor model (Hazarika et al. 2004). In 2006, Doherty et al. found high FLOT2 expression was associated with lymph node metastasis of melanoma and Breslow depth (Doherty et al. 2006). As show previously, qRT-PCR of tissues shows an increase FLOT2 from normal nevi to metastatic melanoma. Same situation occurs in melanoma cell lines. Therefore, FLOT2 acts as a promoter of melanoma metastasis and invasion.
We found that there are possible miR-34a binding sites in FLOT2-3’UTR in vertebrate, located in the FLOT2 3′ UTR at 206–213 bps, 213–1177 bps and 1179–1179 bps, using TargetScan. What’s more an correlativity of miR-34a to FLOT2 was found in the melanoma tissue and cell lines, which make us propose that in melanoma, miR-34a inhibits FLOT2, thereby inhibiting the invasion and metastasis of melanoma cells. So we test FLOT2 mRNA and protein after miR-34a-mimics/inhibitor was transfected into WM451/WM35. The results of qRT-PCR and Western blot show that after overexpression of miR-34a in WM451, FLOT2 expression decreased; conversely, when miR-34a was inhibited in WM35 cells, FLOT2 mRNA and protein expression increased. To further confirm that miR-34a have a direct bind at FLOT2-3’-UTR, the dual luciferase reporter assays also performed and confirmed that miR-34a actually combined to FLOT2-3’-UTR and inhibit FLOT2 expression. Mutation at FLOT2-3’-UTR could stop this bind and make inhibition affect lose.
We then applied functional recovery experiments on WM451 and WM35 by co-transfection miR-34a-mimics/FLOT2-cDNA to WM451 and miR-34a-inhibitor/siFLOT2 to WM35, and then detect the change of melanoma cell proliferation ability and metastasis ability for confirming that miR-34a has an actually functional inhibition through FLOT2 to melanoma. These results of MTT assays, colony formation assays, scratch assays and transwell assays suggest that miR-34a inhibits melanoma progression through FLOT2.
As PAR-1 (Saleiban et al. 2014; Villares et al. 2011; Zigler et al. 2011) is famous metastasis promoter in melanoma. PAR-1(thrombin receptor protease activated receptor-1) could be inhibited when FLOT2 was inhibited (Hazarika et al. 2004) but has no report about its relation to miR-34a. To further investigate the fact that whether it plays a role in miR-34a/FLOT2 pathway in melanoma inhibition. We performed Western blot to test the PAR-1 expression in miR-34a-mimics-WM451 and in siFLOT-WM451 to see whether miR-34a could inhibit PAR-1 through FLOT2. We found that PAR-1 was significantly inhibited by siFLOT2 (Suppl Fig. 2(a)), whereas miR-34a overexpression can also inhibit it, but less strong than siFLOT2 (Suppl Fig. 2(b)). That may because miR-34a affect to PAR-1 through FLOT2 inhibition. These results suggest that PAR-1 may play a role in miR-34a/FLOT2 pathway.
In summary, we confirmed that miR-34a plays a role as a tumor suppressor in melanoma. As a target gene of miR-34a, FLOT2-3’UTR could act with miR-34a and be inhibited by miR-34a at the mRNA and protein levels then to affect the biology behavior of tumor cell. These findings also imply that miR-34a may regulate cell proliferation and invasion through lipid rafts on the cell membranes. PAR-1 may play a role in this inhibition. Further experiments are required to expand upon these findings.
Conclusion
MiR-34a express at low levels in invasive melanoma, while its higher expression can inhibit the proliferation and metastasis of melanoma though suppressing FLOT2 expression. Mir-34a directly binds to FLOT2-3’-UTR and inhibit its expression.
References
Balch CM et al (2013) Age as a prognostic factor in patients with localized melanoma and regional metastases. Ann Surg Oncol 20:3961–3968. doi:10.1245/s10434-013-3100-9
Bhatia S, Thompson JA (2012) Systemic therapy for metastatic melanoma in 2012: dawn of a new era. J Natl Compr Cancer Netw JNCCN 10:403–412
Bhullar RP, Bhullar A, Vanaki SS, Puranik RS, Sudhakara M, Kamat MS (2012) Primary melanoma of oral mucosa: A case report and review of literature. Dent Res J 9:353–356
Boyle GM et al (2011) Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res 24:525–537. doi:10.1111/j.1755-148X.2011.00849.x
Cancer Facts and Figures (2013). American Cancer Society. http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-036845.pdf
Cao K et al (2014a) SiRNA-mediated flotillin-2 (Flot2) downregulation inhibits cell proliferation, migration, and invasion in gastric carcinoma cells. Oncol Res 21:271–279. doi:10.3727/096504014X13946737557031
Cao M, Li Y, Lu H, Meng Q, Wang L, Cai L, Dong X (2014b) miR-23a-mediated migration/invasion is rescued by its target, IRS-1, in non-small cell lung cancer cells. J Cancer Res Clin Oncol. doi:10.1007/s00432-014-1725-0
Chakraborty C, George Priya Doss C, Bandyopadhyay S (2013) miRNAs in insulin resistance and diabetes-associated pancreatic cancer: the ‘minute and miracle’ molecule moving as a monitor in the ‘genomic galaxy’ Curr Drug Targets 14:1110–1117
Chen Y, Zhu X, Zhang X, Liu B, Huang L (2010) Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Therapy J Am Soc Gene Therapy 18:1650–1656. doi:10.1038/mt.2010.136
Choi SE et al (2013) Elevated microRNA-34a in obesity reduces NAD + levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 12:1062–1072. doi:10.1111/acel.12135
Doherty SD, Prieto VG, George S, Hazarika P, Duvic M (2006) High flotillin-2 expression is associated with lymph node metastasis and Breslow depth in melanoma. Melanoma Res 16:461–463. doi:10.1097/01.cmr.0000222592.75858.20
Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H (2013) Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem 59:1489–1496. doi:10.1373/clinchem.2013.205161
Garofalo M et al (2013) MiR-34a/c-Dependent PDGFR-alpha/beta downregulation inhibits tumorigenesis and enhances TRAIL-induced apoptosis in lung cancer. PLoS ONE 8:e67581. doi:10.1371/journal.pone.0067581
Gocze K et al (2013) Unique microRNA expression profiles in cervical cancer. Anticancer Res 33:2561–2567
Greenberg E et al (2011) Regulation of cancer aggressive features in melanoma cells by microRNAs. PLoS ONE 6:e18936. doi:10.1371/journal.pone.0018936
Hazarika P et al (2004) Up-regulation of Flotillin-2 is associated with melanoma progression and modulates expression of the thrombin receptor protease activated receptor 1. Cancer Res 64:7361–7369. doi:10.1158/0008-5472.can-04-0823
He L et al (2005) A microRNA polycistron as a potential human oncogene. Nature 435:828–833. doi:10.1038/nature03552
He L et al (2007a) A microRNA component of the p53 tumour suppressor network. Nature 447:1130–1134. doi:10.1038/nature05939
He X, He L, Hannon GJ (2007b) The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res 67:11099–11101. doi:10.1158/0008-5472.CAN-07-2672
Heinemann A et al (2012) Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res 72:460–471. doi:10.1158/0008-5472.CAN-11-1977
Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4:143–159. doi:10.1002/emmm.201100209
Kahr HS, Mejlgaard E, Lund B (2013) Primary malignant melanoma of the vagina. Ugeskr Laeger 175:133–134
Kim HR, Roe JS, Lee JE, Cho EJ, Youn HD (2013) p53 regulates glucose metabolism by miR-34a. Biochem biophysical research Commun 437:225–231. doi:10.1016/j.bbrc.2013.06.043
Levati L et al (2011) MicroRNA-155 targets the SKI gene in human melanoma cell lines. Pigment Cell Melanoma Res 24:538–550. doi:10.1111/j.1755-148X.2011.00857.x
Lim LP et al (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773. doi:10.1038/nature03315
Lodygin D et al. (2008) Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell cycle (Georgetown, Tex) 7:2591–2600
Tarasov V et al (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle (Georgetown, Tex) 6:1586–1593
Lu C, Rak JW, Kobayashi H, Kerbel RS (1993) Increased resistance to oncostatin M-induced growth inhibition of human melanoma cell lines derived from advanced-stage lesions. Cancer Res 53:2708–2711
Lu R et al (2014) miR-145 functions as tumor suppressor and targets two oncogenes, ANGPT2 and NEDD9, in renal cell carcinoma. J Cancer Res Clin Oncol 140:387–397. doi:10.1007/s00432-013-1577-z
Malaga-Trillo E, Laessing U, Lang DM, Meyer A, Stuermer CA (2002) Evolution of duplicated reggie genes in zebrafish and goldfish. J Mol Evol 54:235–245. doi:10.1007/s00239-001-0005-1
Matsumoto S et al (2013) Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 113:322–326. doi:10.1161/CIRCRESAHA.113.301209
Nemlich Y et al (2013) MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Investig 123:2703–2718. doi:10.1172/JCI62980
Nesca V et al (2013) Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia 56:2203–2212. doi:10.1007/s00125-013-2993-y
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13:358–369. doi:10.1038/nrg3198
Rivera-Milla E, Stuermer CA, Malaga-Trillo E (2006) Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci CMLS 63:343–357. doi:10.1007/s00018-005-5434-3
Saleiban A, Faxalv L, Claesson K, Jonsson JI, Osman A (2014) miR-20b regulates expression of proteinase-activated receptor-1 (PAR-1) thrombin receptor in melanoma cells. Pigment Cell Melanoma Res 27:431–441. doi:10.1111/pcmr.12217
Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R (2010) MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer J Int Cancer 126:2553–2562. doi:10.1002/ijc.24960
Shang JX, Zhang D, Zhang JT, Zhao JZ, Zhang Y (2013) Melanocytic neoplasms of central nervous system analysis. Zhonghua yi xue za zhi 93:34–36
Siegel R et al (2012) Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 62:220–241. doi:10.3322/caac.21149
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64:9–29. doi:10.3322/caac.21208
Spagnolo F, Caltabiano G, Queirolo P (2012) Uveal melanoma. Cancer Treat Rev 38:549–553. doi:10.1016/j.ctrv.2012.01.002
Villares GJ, Zigler M, Bar-Eli M (2011) The emerging role of the thrombin receptor (PAR-1) in melanoma metastasis—a possible therapeutic target. Oncotarget 2:8–17
Volonte D, Galbiati F, Li S, Nishiyama K, Okamoto T, Lisanti MP (1999) Flotillins/cavatellins are differentially expressed in cells and tissues and form a hetero-oligomeric complex with caveolins in vivo. Characterization and epitope-mapping of a novel flotillin-1 monoclonal antibody probe. J Biol Chem 274:12702–12709
Xiao GH, Luo CQ, Tang GM, Zhou JD (2005) [Human endostatin gene transfected adult skin melanoma cells]. Zhong nan da xue xue bao Yi xue ban= J Central South Univ Med Sci 30:677–681
Xu D et al (2012) Let-7b and microRNA-199a inhibit the proliferation of B16F10 melanoma cells. Oncol Lett 4:941–946. doi:10.3892/ol.2012.878
Yamazaki H et al (2012) Overexpression of the miR-34 family suppresses invasive growth of malignant melanoma with the wild-type p53 gene. Exp Ther Med 3:793–796. doi:10.3892/etm.2012.497
Yan D et al (2009) MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci 50:1559–1565. doi:10.1167/iovs.08-2681
Zhao F, Zhang J, Liu YS, Li L, He YL (2011) Research advances on flotillins. Virol J 8:479. doi:10.1186/1743-422X-8-479
Zigler M, Kamiya T, Brantley EC, Villares GJ, Bar-Eli M (2011) PAR-1 and thrombin: the ties that bind the microenvironment to melanoma metastasis. Cancer Res 71:6561–6566. doi:10.1158/0008-5472.can-11-1432
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81372140, 81301688, 81272192, 81171882), PhD. Programs Foundation of Ministry of Education of China (No. 20130162110050 and 20130162120093), Natural Science Foundation of Hunan Province (No. 13JJ4028), Project of the Department of Science and Technology of Hunan Province (No. 2013FJ6003), Program for New Century Excellent Talents in University (NCET-11-0527), Postdoctoral Foundation of Central South University (No. 131425), and 125 Talent Project of the third Xiangya Hospital of Central South University.
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
432_2014_1874_MOESM1_ESM.tif
Supplement Figure 1. (a) Sequence of FLOT2-3’-UTR. (b) Possible bind sites of miR-34a to FLOT2-3’-UTR. (c) Primer Sequences of Mutations (TIFF 1303 kb)
432_2014_1874_MOESM2_ESM.tif
Supplement Figure 2. PAR-1 expression (a) Western blot results of PAR-1 in WM451 transfected with siFLOT2 and the negative control. (b) Western blot results of PAR-1 in WM451 transfected with miR-34a-mimics and the negative control. (ns: p > 0.05;*:p < 0.05,**:p < 0.01,***:p < 0.001) (TIFF 471 kb)
Rights and permissions
About this article
Cite this article
Liu, R., Xie, H., Luo, C. et al. Identification of FLOT2 as a novel target for microRNA-34a in melanoma. J Cancer Res Clin Oncol 141, 993–1006 (2015). https://doi.org/10.1007/s00432-014-1874-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-014-1874-1