Abstract
Purpose
Transcription factor 21 (TCF21) has been identified as a candidate tumor suppressor at 6q23–q24 that is epigenetically inactivated in many types of human cancers. This study aimed to determine the expression of TCF21 mRNA and protein in gastric cancer cell lines and tissue specimens and then investigate the prognostic impact of TCF21 expression in gastric cancer and analyze the relationship between TCF21 expression and methylation level.
Methods
We used real-time PCR and immunohistochemical staining to detect the expression of TCF21 and used methylation-specific-PCR to determine the methylation status of TCF21 in gastric cancer samples and gastric cancer cell lines.
Results
The results showed that TCF21 expression level in gastric cancer samples was significantly lower than in normal adjacent tissue samples. The Kaplan–Meier survival analysis demonstrated that TCF21 was a significant prognosticator of cancer-specific survival (p = 0.001). Furthermore, the methylation level of TCF21 in gastric cancer samples was much higher than the samples in normal adjacent tissue. Treatment with the DNA methyltransferase inhibitor 5-Aza-2′-deoxy-cytidine can upregulate the expression of TCF21 in gastric cancer cells.
Conclusions
These results suggest that the low expression of TCF21 was an independent prognostic factor for poor survival in patients with gastric cancer. Aberrant methylation was an important reason for the downregulation of TCF21 and may be associated with tumorigenesis in gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer remains a major public health issue as one of the most common digestive malignancies and the second leading cause of cancer death worldwide (Jemal et al. 2011; Crew and Neugut 2006). Most patients are diagnosed at an advanced stage, so the overall treatment response is poor and the 5-year survival rate is low. Detection of cancer cells at early stages could potentially increase survival rates in cancer patients. Understanding the molecular pathophysiology of gastric cancer is essential for determining how to effectively inhibit tumor progression.
The Transcription factor 21 (also referred to as capsulin/pod1/epicardin) is located on chromosome 6q23–q24 and encodes a cell type-specific class II basic helix-loop-helix transcription factor that binds DNA through the consensus E-box sequence (CANNTG) as a heterodimer. It mandates cell fate differentiation through mesenchymal–epithelial transition (MET) (Lu et al. 1998, 2002; Quaggin et al. 1999). Epithelial-to-mesenchymal transition (EMT) occurs normally during lung, kidney, and mammary gland development. Although essential for normal embryonic development, EMT can also be destructive if deregulated, and the latter has been proven essential for the dissemination and metastasis of malignant tumors (Baum et al. 2008; Hugo et al. 2007; Guarino et al. 2007). Normally, TCF21 is expressed at its highest levels during embryonic development and the expression levels rapidly decrease in postnatal tissues, with the exception of a subset of interstitial cells in several organs including kidney, lung and intestine. Accordingly, TCF21 knockout mice were found to be born alive, but to die shortly after birth due to underdeveloped lungs and kidneys (Guarino et al. 2007), while perinatal lethality is a classic feature of tumor suppressor activity (Meuwissen and Berns 2005).
The Transcription factor 21 as a candidate tumor suppressor that is epigenetically inactivated in many kinds of cancers, including lung and head and neck cancer, and further researches finding that aberrant methylation in lung cancer (Smith et al. 2006; Anglim et al. 2008; Shivapurkar et al. 2008; Tessema and Belinsky 2008; Tessema et al. 2008; Weiss et al. 2011), but has not been reported in gastric cancer. Aberrant methylation and decreased expression of TCF21 is tumor specific and very frequent in nonsmall-cell lung cancer (NSCLC), even early-stage disease, thus making TCF21 a potential candidate methylation biomarker for early-stage NSCLC screening (Richards et al. 2011).
In this study, we used immunohistochemistry to study the TCF21 in gastric cancer tissue samples and noncancerous tissue adjacent to tumor. The relationship between expression of TCF21 and survival time was evaluated. In addition, we confirmed the methylation of TCF21 gene in gastric cancer cell lines and tissues. The relationship between TCF21 methylation and expression level was also detected. In addition, the clinicopathologic significance of TCF21 was evaluated using archival tissue specimens and statistical analysis. We found that TCF21 is an independently prognostic factor. Our data will facilitate an understanding of gastric cancer carcinogenesis and mining biomarkers for the diagnosis and treatment of this disease.
Materials and methods
Tissue samples and follow-up
A total of 200 patients who had surgery for gastric cancer between January 2007 and December 2010 at the Fourth affiliated hospital of China Medical University was selected for this study. All patients-derived specimens were collected and archived under protocols approved by the Institutional Review Boards of the Fourth affiliated Hospital China Medical University. The diagnosis was confirmed by at least two pathologists, and staging was based on pathological findings according to the 7th American Joint Committee on Cancer guidelines. The median duration of follow-up was 46 (range 2–73) months. The 200 patients who underwent gastrectomy were subjected to close clinical observation, including chest and abdominal CT imaging, CEA level, and blood testing at 2- to 3-month intervals and a yearly gastroscopy. Overall survival (OS) rates were defined as the interval from the initial surgery to clinically or radiologically proven recurrence or metastasis and death, respectively. The end date of the follow-up study for conducting the analysis was January 2013. Frozen tissues from 20 paired human gastric cancer tissues and their corresponding nonmalignant gastric tissues from patients with gastric carcinoma who underwent a gastrectomy were obtained from the Fourth affiliated hospital of China Medical University between May and December 2012.
Ethics statement
Ethical approval for this research was obtained from the Research Ethics Committee of China Medical University, China. All patients providing tumor tissue as well as normal gastric tissue samples signed a consent form prior to surgical removal of the gastric carcinoma to allow for this research to be undertaken.
TMA construction and immunohistochemistry
Hematoxylin and eosin (H&E)-stained slides were screened for optimal tumor tissue and noncancerous tissue adjacent to tumor (at least 2 cm from the tumor), and tissue microarrays (TMA) slides were constructed with a tissue manual arraying instrument. Puncture points were collected from each formalin-fixed, paraffin-embedded (FFPE) gastric cancer tissue sample and from each normal gastric mucosa sample using a 1.0-mm diameter punch instrument. Because of the heterogeneity of tumor cells, one point cannot represent the whole tumor. To minimize this error, we obtain four puncture points from core part of every case of gastric cancer tissue. Samples from the same patient were spotted next to each other to ensure similar reaction conditions for the normal and tumor tissue of that patient. Immunohistochemical analysis was performed on FFPE samples as described previously using an Envision kit (Dako Cytomation, Glostrup, Denmark) (Kononen et al. 1998). Pressure cooker-mediated antigen retrieval was performed in citrate buffer (pH 6.0) for 7 min. Sections were incubated with 1:50 dilution of polyclonal rabbit antihuman TCF21 antibody (ab32981, Abcam), overnight at 4 °C, and then incubated with goat antirabbit Envision System Plus-HRP (Dako Cytomation) for 30 min at room temperature. After rinsing three times in PBS for 10 min each, the sections were incubated with DAB for 1 min, counterstained with Mayer hematoxylin, dehydrated, and mounted.
Evaluation of immunohistochemical staining
Immunoreactivity was evaluated independently by two researchers who were blinded to patient outcome. Positive expression of TCF21 was defined as the brown staining in the cytoplasm and nucleus. The staining results for TCF21 were scored semiquantitatively. Intensity was estimated in comparison with the control and scored as follows: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). Staining extent was scored as 0 (0 %), 1 (1–25 %), 2 (26–50 %), 3 (51–75 %), and 4 (76–100 %), depending on the percentage of positive-stained cells. A final score was calculated by adding the scores for intensity and percentage. For statistical analysis, 0–3 was counted as low expression of TCF21, while 4–7 was counted as high expression of TCF21.
Gastric cells and culture
Six human gastric cancer cell lines, SGC-7901, BGC-823, MGC-803, AGS, MKN-45, and HGC-27, were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China), and one immortalized normal gastric cell line, GES-1 (as control), was obtained from the Oncology Institute of China Medical University. SGC-7901, BGC-823, MGC-803, MKN-45, and HGC-27 were propagated in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA), AGS was propagated in F-12 K medium (Invitrogen), and GES-1 was propagated in Dulbecco’s modified Eagle’s medium (Invitrogen). All the media were supplemented with 10 % fetal bovine serum. Cell lines were cultured at 37 °C in a humidified incubator of 5 % CO2.
RNA extraction and real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cultured cells or tissues using a Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and reversely transcribed into cDNA using an Expand Reverse Transcriptase Kit (Takara, Japan). The expression of TCF21 mRNA was detected using real-time PCR. The total reaction volume was 20 μl including 1 μg RNA. The reaction mixture was incubated at 42 °C for 60 min, heated at 95 °C for 10 min and then cooled on ice. The reaction was diluted 1:1 with water and aliquoted for further analysis. Real-time PCR was carried out with SYBR Green dyeII in 7500 sequence detection system (Applied Biosystems). Oligonucleotide primers were designed for human TCF21 and GAPDH gene, using the Beacon Designer 7, based on their mRNA sequences. One microliter diluted cDNA aliquot was used as template for PCR in a total volume of 20 μl including TaqMan Universal PCR Master Mix and the corresponding probes and primers. The mixture was pre-incubated at 95 °C for 30 min, followed by 40 cycles of two step incubations at 95 °C for 5 s and 60 °C for 34 s. All samples were measured in triplicates. The Ct value of TCF21 mRNA was normalized to the reference gene Ct (GAPDH), and the relative quantification was performed according to Pfaff mathematical model.
Protein isolation and Western blot
Total protein was extracted from cultured cells and tissue specimens in a lysate buffer containing 50 mM Tris–HCl, 350 mM NaCl, 0.1 % NP-40, 5 mM EDTA, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 10 lg/ml aprotinin, 10 lg/ml leupeptin, and 1 mM Na3VO4. Protein samples were then quantified and loaded onto 10 % sodium dodecyl sulfate polyacrylamide gels for electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore Corp, Bedford, MA, USA). After that, the membranes were incubated with 5 % skim milk in phosphate buffered saline (PBS) for 2 h at room temperature and then incubated with a polyclonal rabbit antihuman TCF21 antibody at a dilution of 1:1,000 or a GAPDH antibody overnight at 4 °C. The next day, the membranes were washed with PBS thrice and incubated with a horseradish peroxidase-conjugated antibody against rabbit IgG (Sigma-Aldrich, St. Louis, MO, USA) for 2 h at room temperature. After washing, the immunoreactive protein bands were visualized using an electrochemiluminescence (ECL) detection kit (Thermol Biotech Inc, Rockford, IL, USA). Each experiment was repeated three times. Protein bands were scanned and quantified using densitometric software (Bio-Rad, CA, USA).
DNA extraction and methylation-specific PCR (MSP)
Genomic DNA was extracted from cultured cell lines and 40 specimens by SDS/proteinase K treatment, followed by phenol–chloroform extraction and ethanol precipitation. The bisulfite treatment was performed using the kit EZ DNA Methylation-Gold Kit (Zymo Research, CA, USA) according to the manufacturer’s protocol. The methylated primer was 5′-TTTGGTTAACGATAAATACGAGAACG-3′ (sense), 5′-CCTAAAAACTCTAAACCCGCGAT-3′ (antisense), which produced a 198-bp band; the unmethylated primer was 5′-TTTGGTTAATGATAAAT ATGAGAATGG-3′ (sense), 5′-TCCCTAAAAACTCTAAACCCACAAT-3′ (antisense), which produced a 200-bp band. The reaction mixture contained 1 μl DNA in a volume of 50 μl containing 1 μl of each primer, 2×GC buffer I, 2.5 mM dNTP Mix, and 2.5 U LA Taq (Takara, Japan). Complete MSP conditions were as follows: 94 °C for 5 min, 40 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 45 s, and 72 °C for 10 min. Human placental genomic DNA (gDNA; Sigma-Aldrich) methylated in vitro with SssI methylase (New England Biolabs, Ipswich, USA) was used, after sodium bisulfite conversion, as fully methylated (100 %) MSP-positive control. The same unmethylated placental gDNA was used, after sodium bisulfite conversion, as a negative control. Water was run with every MSP. The analyst repeated the procedures on three different days. PCR products were subjected to 2 % agarose gel electrophoresis at 120 V for 40 min.
5-Aza-cytidine treatment
Gastric cancer cells were seeded at 5 × 105 cells per well in 6-well culture plates and cultured with RPMI 1640 medium (Invitrogen) containing 10 % fetal bovine serum in a humidified atmosphere of 5 % CO2 at 37 °C and allowed to attach for 24 h. The cells were treated with or without 5 or 10 μM of 5-Aza-2′-deoxy-cytidine (5-Aza-dC) (Sigma) for 3 days. After that, total RNA and protein were isolated from these treated cells for qRT-PCR and Western blot analysis, respectively.
Statistical analysis
The χ2 test was used to analyze the relationship between TCF21 expression and clinicopathologic characteristics. The survival rates were calculated by the Kaplan–Meier method, and the differences between the survival curves were examined by the log-rank test. Univariate Cox proportional hazards regressions were applied to estimate the individual hazard ratio (HR). The significant variables in the univariate analyses (p < 0.05) were then put into the multivariate analysis. The HR with 95 % confidence interval (CI) was measured to estimate the hazard risk of individual factors. Fisher’s exact test was used to determine the relationship between methylation levels and mRNA expression levels in gastric cancer tissues and corresponding nontumorous tissues. p < 0.05 was considered to be statistically significant. Analyses were performed using the SPSS statistical software program version 19.0 (SPSS Inc., Chicago, IL).
Results
Analysis of expression of TCF21 protein by immunohistochemistry
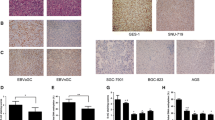
Stained sections of TMA of 200 tissue cores were graded for their immunohistochemical staining intensity against TCF21 protein. The 200 readable samples included 200 gastric carcinoma, and 72 samples of peri-carcinoma tissue. The expression of TCF21 by immunoreactivity was mainly localized in the epithelial cells of gastric tissues (Fig. 1a–c). The percentage of high expression (4–7 staining score) of TCF21 in gastric carcinoma samples is 30 % (60/200), which is significantly less than those in peri-carcinoma samples (86.5 %, 173/200) (Table 1, p < 0.001).
Immunohistochemical detection of TCF21 protein. a High expression in normal gastric mucosa sample. b Low expression in gastric cancer tissue sample. c High expression in gastric cancer tissue sample. Original magnification: ×200. d Kaplan–Meier survival analysis of overall survival duration in 200 gastric cancer patients according to TCF21 protein expression. Low TCF21 expression are significantly correlated with poor patient outcome (p < 0.05) in gastric cancer
Association between TCF21 and clinicopathological factors
In this study, 200 gastric cancer patients with sufficient primary tumors materials and follow-up time were available, whose tissues were collected over the last 6 years. Table 2 gave the descriptive statistics for parameters measured for these patients. The 5-year overall survival (OS) rate was 46.5 %. We found that the expression of TCF21 was significantly associated with macroscopic type (p = 0.004), tumor differentiation (p = 0.01), Lauren grade (p = 0.01), T stage (p = 0.01), lymph node metastasis (p = 0.01) and Lymphovascular invasion (p = 0.013, Table 2). However, no significant correlation was observed between TCF21 expression and other parameters including age, gender, location, and size.
Expression of TCF21 in relation to prognosis
Using Cox’s proportional hazards regression model, the univariate relationships between tumor characteristics and patients’ outcome were obtained (Supplementary Table S3). Of the 200 patients analyzed, statistically significant differences in OS were seen, with a poor outcome for patients with low expression of TCF21. Other predictive factors that were found to be correlated with OS were size (p = 0.002), location (p = 0.006), macroscopic type (p = 0.001), tumor differentiation (p < 0.001), Lauren grade (p = 0.01), T stage (p < 0.001), lymph node metastasis (p < 0.001), and Lymphovascular invasion (p < 0.001) (Supplementary Table S1). Using log-rank test, there were significant differences in OS between positive and negative patients of TCF21 (p < 0.05) in all patients were shown in Fig. 1d. Cox multivariate proportional hazard regression model was then used to determine which factors were jointly predicative of OS. Variables which were thought to be significant in univariate analysis were included in the analysis. The significance, adjusted for other co-variates, was given in Table 3. Multivariate analysis of the effect of TCF21 with other prognostic factors showed that T stage (p = 0.008), location (p = 0.017) and N stage (p < 0.001), TCF21 (HR 0.565, 95 % CI 0.341–0.936, p = 0.027) was a independently prognostic marker for OS (Table 3).
Downregulation of TCF21 expression in gastric cancer cells and tissues
In this study, we assessed TCF21 expression in gastric cell lines and found that the expression of TCF21 mRNA was substantially downregulated in MGC-803 (0.095-fold), AGS (0.128-fold), HGC-27 (0.196-fold), SGC-7901 (0.27-fold), BGC-823 (0.69-fold), and MKN-45 (0.84-fold) cells compared with GES-1 (onefold as the control) (p < 0.05; Fig. 2a). Furthermore, Western blot analysis confirmed our data on TCF21 mRNA expression in gastric cell lines (Fig. 2c). We also analyzed TCF21 expression in 20 gastric cancer and corresponding normal tissue specimens, and our data showed that 16 out of 20 gastric cancer tissues reduced TCF21 expression compared to the corresponding adjacent normal tissues (Fig. 2b).
Downregulation of TCF21 in gastric cancer lines and tissues. Expression of TCF21 mRNA (a) and protein (c) in human gastric cancer cells, taking GAPDH as control. Expression of TCF21 mRNA (b) in 6 cases of gastric cancer tissues and corresponding nontumorous tissues. Total RNA and protein are extracted from gastric cells and tissue specimens and then subjected to qRT-PCR and Western blot analysis of TCF21 mRNA and protein expression, respectively. Data are presented as mean ± SD for three independent experiments. *p < 0.05, **p < 0.001, statistically significant difference
Methylation status of TCF21 in gastric cancer cells and tissues
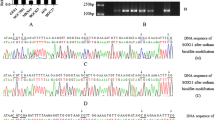
We assessed DNA methylation of TCF21 gene promoter CpG islands using a MSP. Our data showed that TCF21 was hypermethylated in all gastric cancer cells, but not methylated in GES-1 cell (Fig. 3a). In gastric cancer tissue specimens, methylation of the TCF21 gene promoter was observed in 65 % (13/20) of gastric cancer tissues, while 15 % (3/20) of nonmalignant gastric tissues also showed TCF21 gene promoter methylation (Fig. 3b). MSP products were sequenced, which confirmed that sodium bisulfite modification was sufficient for DNA (Fig. 3c, d). The difference between primary gastric cancer and nonmalignant gastric tissue specimens is significant (p < 0.01; Supplementary Table S2).
DNA methylation of TCF21 in gastric cancer cell lines and tissues. a, b MSP analysis of TCF21 gene promoter methylation in gastric cancer cell lines and tissues, respectively. c, d The promoter sequence of TCF21 after treatment with sodium bisulfate. Unmethylation of cytosine was transformed to uracil, while methylated cytosine unchanged. M methylation lane, U unmethylation lane, UP unmethylation positive control, MP methylation positive control, N nonmalignant gastric tissue, T tumor specimens. The case number (Jemal et al. 2011; Crew and Neugut 2006; Lu et al. 1998, 2002; Quaggin et al. 1999; Baum et al. 2008) in Figs. 2b and 3b shows same patient
Upregulation of TCF21 expression after treatment with 5-Aza-dC
To confirm this, epigenetic change was responsible for downregulation of TCF21 expression, we treated MGC-803 and AGS gastric cancer cells (low expression of TCF21) with 5-Aza-dC to inhibit DNA methylation. We found that 5-Aza-dC was able to significantly upregulate TCF21 expression (p < 0.05) (Fig. 4).
Relationship between TCF21 mRNA and DNA methylation
We assessed the association between the relative TCF21 mRNA levels and relative DNA methylation level in gastric cancer samples compared to nonmalignant gastric samples using Fisher’s exact test, but no significant correlation was observed between TCF21 mRNA levels and DNA methylation (p > 0.05) (Supplementary Table S3).
Discussion
The assessment of biological prognostic factors is of clinical importance, especially for a disease with poor outcome, such as gastric cancer. In the past, several studies have been aimed at assessing the role of TCF21 as a potential biomarker in lung cancer, head and neck squamous cell carcinoma and metastatic melanoma (Smith et al. 2006; Weiss et al. 2013; Arab et al. 2011). In NSCLC and prostate cancers, a higher tumor stage was associated with a higher TCF21 promoter methylation rate (Anglim et al. 2008; Costa et al. 2011). Our studies demonstrate that TCF21 is frequently down expressed in gastric cancer samples compared to nonmalignant gastric tissue samples. According to Kaplan–Meier analysis, TCF21 protein expression in gastric cancer was correlated with patient’s overall survival. Patients with lower expression of TCF21 protein had shorter survival time. Cox multivariate analysis showed that low expression of TCF21 was an independent poor prognostic factor of gastric cancer patients. These results are consistent with the report which showed that low expression of TCF21 was an independent prognostic factor for poor survival in patients with clear cell renal cell carcinoma (Ye et al. 2012). It suggests that this value may serve as a prognostic biomarker for gastric cancer patients after surgery.
Methylation of DNA is an important reason for the down expression of genes. Aberrant DNA methylation has been reported to play a major role in carcinogenesis (Gopalakrishnan et al. 2008). The data from our current study showed the loss of TCF21 expression in gastric cancer cell lines and tissue samples, which was mainly due to methylation of TCF21 gene promoter. Treatment with the DNA methyltransferase inhibitor 5-Aza-dC can upregulate the expression of TCF21 in gastric cancer cells. While we found TCF21 mRNA levels did not completely match with methylation levels between gastric cancer samples and nonmalignant gastric tissue samples in our results. This may be due to these reasons: (1) the limited number of cases, (2) due to the limited conditions, the insufficiency of sodium bisulfite modification was inevitable, (3) the methylation status of TCF21 promoter was partially changed in these tissues, and (4) we also speculated that there are some other reasons for the low expression of TCF21 in gastric cancer samples, such as the regulation by miRNA (Su et al. 2011; Zhang et al. 2012) or other genes (Bhandari et al. 2011 May). Interestingly, the sole predicted regulator of TCF21 is miR-92a (Griffiths-Jones et al. 2006), which is overexpressed in a variety of cancers (Matsubara et al. 2007; Volinia et al. 2006).
Loss of the TCF21 transcription factor results in a failure of mesenchymal epithelialization, a process known as mesenchymal-to-epithelial transition. EMT is a normal process by which a differentiated epithelial cell acquires characteristics that allow for dedifferentiation into a mobile mesenchymal cell (Lu et al. 2000; Prindull and Zipori 2004). Transition between epithelial cell and mesenchymal cell is known to occur during tumorigenesis (Hugo et al. 2007; Guarino et al. 2007). EMT has been described in many cancers and correlates with clinical outcome (Hugo et al. 2007). In general, less differentiated tumors are more aggressive (Welch and Rinker-Schaeffer 1999; Yoshida et al. 2000). Malignant lesions are often defined by their differentiation status, where benign tumors typically retain their epithelial phenotype and malignant cells acquire a more fibroblastic mesenchymal phenotype (Kiemer et al. 2001). Along the invasive front of a carcinoma, epithelial cells often gain mesenchymal cell characteristics and gene expression profiles (Miyazawa et al. 2004). As mentioned in the results, Smith et al. (2006) previously reported significant downregulation of mesenchymal markers (SNAI1 and VIM) expression as well as upregulation of epithelial markers (WNT4 and CDH1) in lung cancer cells when transfected with TCF21. Shivapurkar et al. (2008) also observed significant expression of WNT4 in the lung cancer cells of TCF21 expression positive. It suggest that WNT4 might be one of the proximate downstream targets of TCF21. This possibility should be further explored. The putative causal link between TCF21 expression and EMT requires further clarification and is a main subject of our ongoing research.
In summary, our study showed that TCF21 expressed at lower level in gastric cancer samples than in nonmalignant gastric tissues samples. TCF21 may represent a predictive biomarker of prognosis in patients with gastric cancer. Aberrant methylation is an important reason for the downregulation of TCF21 and may be associated with development and progression in gastric cancer. However, the functional role and mechanisms of TCF21 in gastric cancer are unclear and require further investigation.
References
Anglim PP, Galler JS, Koss MN, Hagen JA, Turla S et al (2008) Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer 7:62. doi:10.1186/1476-4598-7-62
Arab K, Smith LT, Gast A, Weichenhan D, Huang JP et al (2011) Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis 32:1467–1473
Baum B, Settleman J, Quinlan MP (2008) Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol 19:294–308
Bhandari RK, Sadler-Riggleman I, Clement TM (2011) Skinner MK (2011) Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY. PLoS One 6(e19935):17. doi:10.1371/journal.pone.0019935
Costa VL, Henrique R, Danielsen SA, Eknaes M, Patrício P et al (2011) TCF21 and PCDH17 methylation: an innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics 6:1120–1130
Crew KD, Neugut AI (2006) Epidemiology of gastric cancer. World J Gastroenterol 12:354–362
Gopalakrishnan S, Van Emburgh BO, Robertson KD (2008) DNA methylation in development and human disease. Mutat Res 647:30–38
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Guarino M, Rubino B, Ballabio G (2007) The role of epithelial-mesenchymal transition in cancer pathology. Pathology 39:305–318
Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA et al (2007) Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol 213:374–383
Jemal A, Bray F, Center MM, Ferlay J, Ward E et al (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Kiemer AK, Takeuchi K, Quinlan MP (2001) Identification of genes involved in epithelial-mesenchymal transition and tumor progression. Oncogene 20:6679–6688
Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P et al (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4:844–847
Lu J, Richardson JA, Olson EN (1998) Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech Dev 73:23–32
Lu J, Chang P, Richardson JA, Gan L, Weiler H et al (2000) The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc Natl Acad Sci USA 97:9525–9530
Lu JR, Bassel-Duby R, Hawkins A, Chang P, Valdez R et al (2002) Control of facial muscle development by MyoR and capsulin. Science 298:2378–2381
Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y et al (2007) Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene 26:6099–6105
Meuwissen R, Berns A (2005) Mouse models for human lung cancer. Genes Dev 19:643–664
Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K (2004) Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res 64:2024–2029
Prindull G, Zipori D (2004) Environmental guidance of normal and tumor cell plasticity: epithelial mesenchymal transitions as a paradigm. Blood 103:2892–2899
Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J et al (1999) The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development 126:5771–5783
Richards KL, Zhang B, Sun M, Dong W, Churchill J et al (2011) Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer 117:606–617
Shivapurkar N, Stastny V, Xie Y, Prinsen C, Frenkel E et al (2008) Differential methylation of a short CpG-rich sequence within exon 1 of TCF21 gene: a promising cancer biomarker assay. Cancer Epidemiol Biomarkers Prev 17:995–1000
Smith LT, Lin M, Brena RM, Lang JC, Schuller DE et al (2006) Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci USA 103:982–987
Su Z, Xia J, Zhao Z (2011) Functional complementation between transcriptional methylation regulation and post-transcriptional microRNA regulation in the human genome. BMC Genom 12(Suppl 5):S15
Tessema M, Belinsky SA (2008) Mining the epigenome for methylated genes in lung cancer. Proc Am Thorac Soc 5:806–810
Tessema M, Willink R, Do K, Yu YY, Yu W et al (2008) Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res 68:1707–1714
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A et al (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103:2257–2261
Weiss D, Basel T, Sachse F, Braeuninger A, Rudack C (2011) Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol Carcinog 50:680–688
Weiss D, Stockmann C, Schrödter K, Rudack C (2013) Protein expression and promoter methylation of the candidate biomarker TCF21 in head and neck squamous cell carcinoma. Cell Oncol 36:213–224
Welch DR, Rinker-Schaeffer CW (1999) What defines a useful marker of metastasis in human cancer? J Natl Cancer Inst 91:1351–1353
Ye YW, Jiang ZM, Li WH, Li ZS, Han YH et al (2012) Down-regulation of TCF21 is associated with poor survival in clear cell renal cell carcinoma. Neoplasma 59:599–605
Yoshida BA, Sokoloff MM, Welch DR, Rinker-Schaeffer CW (2000) Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst 92:1717–1730
Zhang H, Guo Y, Shang C, Song Y, Wu B (2012) miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology 80:1298–1302
Acknowledgments
This work was supported by the Higher Specialized Research Fund from the Doctoral Program of the Ministry of Education of China (#20102104110001), the Liao ning Province science and technology plan project (#2013225021) and the Young Innovation and Development Foundation from the Fourth Affiliated Hospital of China Medical University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Z., Li, D.M., Xie, Q. et al. Protein expression and promoter methylation of the candidate biomarker TCF21 in gastric cancer. J Cancer Res Clin Oncol 141, 211–220 (2015). https://doi.org/10.1007/s00432-014-1809-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-014-1809-x