Abstract
Purpose
To determine the interaction between insulin receptor substrate-1 (IRS-1) and miR-23a on the migration and invasion of non-small cell lung cancer (NSCLC) cells, and to examine IRS-1 expression in NSCLC tissues and its correlation with clinicopathologic characteristics.
Methods
The migration and invasion of A549 cells were measured using transwell assay. miR-23a levels were examined by quantitative reverse transcription-PCR and IRS-1 expression by Western blotting. The interaction between miR-23a and IRS-1 was examined by luciferase reporter assay. IRS-1 expression in 105 NSCLC specimens was determined by immunohistochemistry and its correlation with patient clinicopathologic characteristics was evaluated.
Results
Transwell assay revealed that miR-23a significantly promoted the migration and invasion of A549 cells with a 44.0 and 44.6 % increase in the number of migrated and invading cells, respectively. Luciferase assay showed that miR-23a markedly reduced luciferase activities of A549 cells co-transfected with plasmids overexpressing the 3′ UTR of IRS-1 mRNA (P < 0.05). Co-transfection of A549 cells with miR-23a and plasmids overexpressing IRS-1 significantly reduced the increase in the number of migrated and invading cells mediated by miR-23a. Immunohistochemistry showed low IRS-1 expression in 26.7 % and high IRS-1 expression in 73.3 % of the NSCLC specimens. Kaplan–Meier analysis revealed that the overall survival and disease-free survival of NSCLC were markedly longer in patients with high IRS-1 expression than those with low IRS-1 expression (P = 0.002). Multivariate Cox regression analysis showed that IRS-1 was an independent prognostic factor for the overall survival of NSCLC patients (RR 0.413 CI 0.238–0.718, P = 0.002).
Conclusions
There is an interaction between miR-23a and IRS-1 in the modulation of the migration and invasion of NSCLC cells. IRS-1 is variably expressed in NSCLC patients and correlates with NSCLC patient survival.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is one of the most frequent and lethal types of tumors and accounts for 17 % of total new cancer cases and approximately one-fourth of total cancer deaths (Jemal et al. 2011). Non-small cell lung cancers (NSCLCs) are composed of adenocarcinoma, squamous cell carcinoma, and large cell carcinoma and are responsible for approximately 85 % of newly found lung cancer cases (Moran 2011). NSCLC patients are often diagnosed at advanced stages that are not amenable to surgical intervention. Despite recent advances in chemotherapies and molecular-targeted therapies for lung cancer, the prognosis of NSCLC patients is still dismal (Schiller et al. 2002; Ohe et al. 2007; Maemondo et al. 2010).
MicroRNAs (miRNAs) are a class of short (21–24 nucleotides) single-stranded non-coding endogenous RNAs which post-transcriptionally modulate gene expression by either inhibiting translation or inducing mRNA degradation (Chekulaeva and Filipowicz 2009). MiRNAs are involved in diverse processes including cellular development, proliferation, differentiation, apoptosis, and metabolism (Garzon et al. 2009). An inverse correlation has been demonstrated between miRNA expression and oncogene expression in lung cancer (Johnson et al. 2005). Abundant evidence shows that miRNAs may act as oncogenes or tumor suppressors and play a critical role in tumorigenesis and metastasis (Ortholan et al. 2009; Yanaihara et al. 2006; Hayashita et al. 2005). MiRNAs such as miR-155 and let7a are of prognostic value for lung cancer (Yanaihara et al. 2006). The miR-17-92 cluster is recognized as oncogenes that can promote lung cancer carcinogenesis (Hayashita et al. 2005).
Insulin receptor substrate-1 (IRS-1) is a major member of the insulin receptor substrate family (IRS) and acts as an important adaptor in insulin and insulin-like growth factors signaling (White 2003). It functions as a mediator molecule in signal transduction and is regulated by certain cytokines, hormones, and growth factor receptors (Gual et al. 2005). The effect of IRS-1 on tumor progression and metastasis has been widely described in recent years. Decreased IRS-1 expression has been correlated with the progression of breast cancer (Schnarr et al. 2000). The loss of IRS-1 enhances metastasis of head and neck cancer (Luo et al. 2012). IRS-1 also suppresses transforming growth factor-β (TGF-β)-induced epithelial mesenchymal transition (EMT) in lung cancer (Shi et al. 2009; Reiss et al. 2000; Ma et al. 2006; Gibson et al. 2007).
We have recently reported that miR-23a promotes TGF-β-induced EMT in NSCLC in a Smad-dependent manner (Cao et al. 2012). Shi et al. (2009) have shown that TGF-β1 induces EMT by modulating IRS-1 signaling and that IRS-1 may function as a critical EMT suppressor in NSCLC cells. However, there is no report on the interaction between miR-23a and IRS-1. In this study, we hypothesized that IRS-1 could be a target of miR-23a and be involved in the migration and invasion of NSCLC cells. Here, we examined the role of miR-23a in the migration and invasion of NSCLC cells in vitro and whether IRS-1 was a direct target of miR-23a. Moreover, we examined the expression of IRS-1 in NSCLC tissues and its correlation with the clinicopathologic characteristics of NSCLC. Our results demonstrated that IRS-1 was expressed variably in NSCLC patients and high IRS-1 expression was associated with a better prognosis for NSCLC patients.
Patients and methods
Tissue specimens
One hundred and five surgically resected NSCLC specimens, provided by the Tumor Hospital of Harbin Medical University, Harbin, Heilongjiang, China, were fixed with formalin and embedded in paraffin. These specimens were obtained between January 2006 and January 2007, and the patients were followed up to October 2012 or until death. Primary cancers were evaluated according to the American Joint Committee on Cancer staging system (7th ed.) (Tsim et al. 2010). No patient received chemotherapy or radiotherapy prior to surgery. Acquisition of all tissue specimens was approved by the ethical committee at Tumor Hospital of Harbin Medical University and was carried out in accordance with the established national and institutional ethical guidelines regarding the use of human tissues for research.
Cells
A549 cells were obtained from Heilongjiang Cancer Institute (Harbin, China) and maintained in RPMI-1640 culture medium supplemented with 10 % fetal bovine serum (FBS) at 37 °C in 5 % CO2. To stimulate miR-23a expression, we treated A549 cells with 5 ng/ml TGF-β1 (R&D System, Minneapolis, MN, USA) for the indicated periods of time as detailed elsewhere in the text.
Quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was extracted from A549 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). miR-23a expression levels were quantified by qRT-PCR using TaqMan MicroRNA Assay System (Applied Biosystems, Foster City, CA, USA). RNU66 was used as an internal control. MiRNA expression was quantified using the 2−∆∆Ct method (Bustin 2000).
Western blotting
Cellular lysate of A549 cells was prepared with lysis buffer containing 50 mM Tris–HCl (pH 7.6), 150 mM NaCl, 0.1 % sodium dodecyl sulfate, 1 % Nonidet P-40, and 0.5 % sodium deoxycholate. Western blotting was performed as previously described (Cao et al. 2012) and the following antibodies were used: anti-IRS-1 and anti-β-actin antibodies (Cell Signaling Technology, Beverley, MA, USA). Blots were visualized using the ECL chemiluminescence detection system (GE Healthcare Bio-Science, Piscataway, NJ, USA).
Transfection
Small interfering RNA for IRS-1 (Si-IRS-1) (5′-GUCAGUCUGUCGUCCAGUATT-3′) and scrambled siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Dharmacon (Lafayette, CO). miR-23a inhibitor (anti-miR-23a) (5′-GGAAAUCCCUGGCAAUGUGAU-3′) and its negative control (5′-CAGUACUUUUGUGUAGUACAA-3′) were synthesized by Ambion (Ambion, Austin, TX). miR-23a mimic (5′-AUCACAUUGCCAGGGAUUUCC-3′), and control mimic (5′-UUCUCCGAACGUGUCACGUUU-3′), IRS-1-expressing plasmid (p-EZ-M02-IRS-1) and p-EZ-M02 vector were obtained from GenePharma (Shanghai, China). A549 cells were transfected using the Lipofectamine 2000 reagent as instructed by the manufacturer (Invitrogen, Carlsbad, CA). IRS-1 3′ UTR with mutations in the region complementary to the miR-23 seed region were generated by mutagenesis. The primers used for mutagenesis were as follows: 5′-TCTCTTTGGAATGAATCCGGTGGGCGTTTGTAAA-3′ (forward) and 5′-TTTACAAACGCCCACCGGATTCATTCCAAAGAGA-3′ (reverse).
Transwell cell migration and invasion assays
Migration and invasion of A549 cells were measured using a 24-well Transwell plate (8 μm pore size, Corning Costar, USA). For migration assays, 5 × 104 A549 cells were seeded into the non-coated membrane in the top chamber and the lower chamber was filled with 0.6 ml complete medium containing 10 % FBS. For invasion assays, the chamber inserts were coated with 200 mg/ml BD Matrigel (BD Biosciences, San Jose, CA, USA) and dried overnight. A549 cells (5 × 104) were incubated at 37 °C for 48 h and stationary cells were removed from the upper surface of the membranes. The cells that had migrated to the lower surface were fixed and stained with 0.1 % crystal violet. The stained cells were counted from five random fields under microscope.
Luciferase reporter assay
Bioinformatic target gene predictions were performed using web-based compilation of prediction algorithms FindTar3 (http://bio.sz.tsinghua.edu.cn/). The cDNA of IRS-1 mRNA 3′-UTR was cloned at the 3′-end of Firefly luciferase open reading frame (ORF) in dual reporter (Firefly and Renilla luciferases) vector pmirGLO (Promega, San Luis Obispo, CA, USA) using SacI and XhoI. A549 cells were grown in 96-well plates and co-transfected with 2.5 pmol miR-23a mimics or control mimics and 100 ng pmirGlo/3′-UTR using Lipofectamine 2000. The cells were then lysed and measured 48 h post-transfection. Firefly luciferase activities were measured using dual-luciferase reporter reagents (Promega) with a GloMax luminometer (Promega), and the results were normalized against Renilla luciferase according to the manufacturer’s protocol. All experiments were performed in triplicate at least thrice independently.
Immunohistochemistry assay
Formalin-fixed, paraffin-embedded, 3-μm tissue sections were cut, and three adjacent sections were chosen from each sample for routine immunohistochemical staining to detect the expression of IRS-1. Briefly, after deparaffinization and rehydration, the sections were boiled in citrate buffer for 10 min for antigen retrieval. Endogenous peroxidase was inactivated with 3 % H2O2. Non-specific binding was blocked by incubation with non-immune serum for 30 min followed by overnight incubation at 4 °C with anti-IRS-1 and epidermal growth factor receptor (EGFR) antibodies (Cell Signaling, Danvers, MA, USA). After washing with phosphate-buffered saline-Tween buffer (PBS-T) thrice, the sections were incubated with secondary antibody for 1 h at room temperature, followed by additional three washes with PBS-T. 3,3′-Diaminobenzidine (DAB) solution was used to visualize the samples. Totally 500 cells were counted in five random areas of each slide to determine the ratio of positive cells. All slides were analyzed by two independent observers blind to patient clinicopathologic data. Immunohistochemistry staining was examined for staining positive ratio. The stained cells were scored as following: (−) represented 0–5 % of the cells stained; (+) represented 6–25 % of the cells stained; (++) represented 26–50 % of the cells stained; (+++) represented 51–75 % of the cells stained; (++++) represented 76–100 % of the cells stained. The immunohistochemistry score of IRS-1 and EGFR expression was 0 (− ≤ score <2+) and 1 (2+ ≤ score ≤4+), which represented low and high expression, respectively.
Statistical analysis
Data was analyzed using the SPSS software version 18.0 (SPSS, Chicago, IL). Differences between groups were analyzed using Student’s t test and the χ 2 test. The relationship between IRS-1 and age was tested using U test. Overall survival (OS) was defined as the time from the date of surgery to the date of death or the last follow-up. The OS and disease-free survival (DFS) curves were drawn by Kaplan–Meier method, and the correlation between each of the variables and survival was evaluated by the log-rank test of univariate analysis. Then, the parameters were tested by multivariate Cox analysis, which was performed to estimate independent variables for predicting prognosis. P < 0.05 was considered statistically significant.
Results
miR-23a enhances the migration and invasion of A549 cells in vitro
Since miR-23a promotes TGF-β-induced EMT, we speculated that miR-23a might influence the migration and invasion of NSCLC cells. Transwell assay revealed that miR-23a significantly increased the number of the migrated A549 cells by 44.01 % (P = 0.005, Fig. 1b, d, f). The addition of anti-miR-23a in the presence of miR-23a mimics noticeably attenuated the miR-23a-mediated increase of the migrated cell number by 32.62 % compared with the cells transfected with miR-23a mimics only (P = 0.0044, Fig. 1d, e, f). Anti-miR-23a further suppressed the migration of A549 cells in the absence of miR-23a mimics by 68.89 % compared with controls (P = 0.001, Fig. 1b, c, f). Consistently, transwell assay showed a similar invasion pattern of A549 cells. miR-23a significantly increased the invasion of A549 cells by 44.64 % (P = 0.0005, Fig. 1h, j, l). The addition of anti-miR-23a in the presence of miR-23a mimics noticeably attenuated the miR-23a-mediated increase of invasion by 30.78 % compared with the cells transfected with miR-23a mimics only (P = 0.033, Fig. 1j, k, l). Anti-miR-23a further suppressed the invasion of A549 cells in the absence of miR-23a mimics by 75.11 % compared with controls (P < 0.0001, Fig. 1h, i, l). Our observation indicated that miR-23a could upregulate the migration and invasion of A549 cells.
miR-23a contributes to the migration and invasion of NSCLC cells. The motility ability of A549 cells was detected by transwell assay. F and L show the cell counts of migration and invasion. The numbers (1–5) under the horizontal axes mean the same treatments stated in a–e and g–k. Data are presented as the mean ± standard deviations of at least three independent experiments
The miR-23a-mediated migration and invasion of NSCLC cells can be antagonized by IRS-1
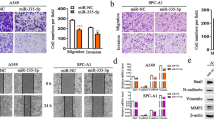
The interaction between IRS-1 and miR-23a on the migration and invasion of NSCLC cells was determined by transwell assay. First of all, the viability of the A549 cells transfected with miRNAs or plasmids was evaluated. There is no significant difference among the viabilities of the cells with various treatments (Fig. 2a). Western blotting showed that IRS-1 was highly expressed in the cells transfected with plasmid p-EZ-M02-IRS-1, while the IRS-1 expression was knocked down in the cells with Si-IRS-1. Interestingly, IRS-1 expression was somehow suppressed in the cells with miR-23a mimics. The suppression could be reversed by anti-miR-23a (Fig. 2b, c).
miR-23a-mediated migration and invasion of NSCLC cells can be antagonized by IRS-1. a MTT assay for the viability of A549 cells. There is no significant difference among the viabilities of A549 cells with various treatments at 48 h. b and c The expression of IRS-1 in the cells with various treatments was detected by Western blotting. The blank cells were treated with control mimics, p-EZ-M02 vector and negative control for anti-miR-23a, respectively. d–q The motility ability of the cells was detected by transwell assay. d–i The representative images of migration assay. k–p The representative images of invasion assay. d and k Blank control; e and l miR-23a-treated. f and m IRS-1-treated; G and N: treated with miR-23a and IRS-1; h and o: Si-IRS-1-treated; i and p treated with anti-miR-23a and Si-IRS-1. j and q illustrated the migration and invasion ability, respectively. Data are mean ± standard deviations of at least three independent experiments
Transwell assay showed that miR-23a could markedly promote the migration of A549 cells compared with untreated controls (P < 0.05, Fig. 2d, e, j). In the cells co-transfected with miR-23a mimics and p-EZ-M02-IRS-1, the increase of the miR-23a-mediated migration was significantly reduced (P = 0.027, Fig. 2f, g, j). The addition of miR-23a in the presence of exogenous IRS-1 noticeably promoted the migration of A549 cells (P = 0.026, Fig. 2f, g, j). Moreover, anti-miR-23a did not affect the migration cell number, while endogenous IRS-1 was blocked (P = 0.739, Fig. 2h, i, j). Similar patterns were observed in the invasion of A549 cells (Fig. 2k–q). These findings implied that there was an antagonistic effect between IRS-1 and miR-23a in modulating the migration and invasion of A549 cells.
miR-23a counteracts IRS-1 by targeting its mRNA
Above detection demonstrated that miR-23a could suppress IRS-1 expression (Fig. 2b, c). IRS-1 has been reported to block TGF-β-induced EMT (Shi et al. 2009), while we previously showed that miR-23a can be directly induced by the TGF-β/Smad pathway during the EMT (Cao et al. 2012). In this study, RT-PCR confirmed that TGF-β markedly stimulated the expression of miR-23a in A549 cells (P = 0.0028, Fig. 3a). Western blotting showed that TGF-β markedly suppressed IRS-1 expression, while anti-miR-23a apparently upregulated IRS-1 expression in A549 cells. The upregulation of IRS-1 expression by miR-23a was attenuated by TGF-β. These data suggest a negative correlation between miR-23a and IRS-1 (Fig. 3b). Therefore, we estimated that IRS-1 might be a direct target of miR-23a.
IRS-1 mRNA is a direct target of miR-23a. a A549 cells were treated with TGF-β1 (5 ng/ml) and miR-23a levels were examined by qRT-PCR. TGF-β1 induced a 58.00 % increase of miR-23a compared with control. b A549 cells were treated with miR-23a inhibitors in the presence or absence of TGF-β1 (5 ng/ml). IRS-1 expression was examined by Western blotting. TGF-β1 significantly suppressed IRS-1 expression. c The sequence of miR-23a and the corresponding sequence in the 3′-UTR of IRS-1 mRNA. d A549 cells were transfected with appropriate vectors and miR-23a mimics (pmirGLO, pmiGLO + miR-23a, pmirGLO-IRS-1, pmirGLO-IRS-1 + miR-23a, pmirGLO-IRS-1 + control mimics, and pmirGLO-mut-IRS-1 + miR-23a), and the luciferase activities were determined. miR-23a induced a 42.59 % reduction of luciferase activity compared to cell treated with miRNA control. Data are showed as the mean ± standard deviations. At least three independent experiments were performed
Indeed, upon the bioinformatic analysis using web-based tools for miRNA target prediction, we found that the 3′-UTR of IRS-1 mRNA contained putative target sites of miR-23a that were complementary to the seed sequence of miR-23a (Fig. 3c). Using a reporter plasmid in which the 3′ UTR of luciferase was replaced by the 3′ UTR of IRS-1, luciferase assay showed that, compared to control mimics, miR-23a mimics caused a marked reduction in luciferase activities of A549 cells co-transfected with the reporter plasmid (P < 0.05, Fig. 3d), supporting that miR-23a directly targets the IRS-1 gene.
IRS-1 expression is variable in NSCLCs and correlates with NSCLC patient survival
To determine whether IRS-1 expression is dysregulated in NSCLC patients and whether it correlates with clinicopathologic characteristics of NSCLC patients, we examined the expression of IRS-1 in 105 surgically resected NSCLC specimens by immunohistochemistry. Twenty-eight (26.67 %) patients showed low IRS-1 expression, while 73.33 % of the patients showed high IRS-1 expression (Fig. 4a–c). We found no significant difference in clinicopathologic characteristics, including patient age, gender, tumor size, node status, pathologic type, differentiation, and smoking status between patients with low and high IRS-1 expression except in EGFR expression (low vs. high EGFR expression, P = 0.038, Table 1).
Immunohistochemistry detection of IRS-1 in NSCLC tissues and the survival of NSCLC patients stratified by IRS-1 expression. a and a′ The NSCLC tissue with high IRS-1 expression. b and b′ The NSCLC tissue with low IRS-1 expression. c and c′ The NSCLC tissue without IRS-1 expression. a–c ×100, a′–c′ ×400. d Kaplan–Meier overall survival (OS) curve and (E) disease-free survival (DFS) stratified by IRS-1 expression
Kaplan–Meier analysis revealed that the median OS of NSCLC patients with high IRS-1 expression was markedly longer than that of the patients with low IRS-1 expression (31.7 months vs. NA, P = 0.002, Fig. 4d). Similarly, we observed a significantly longer median DFS in NSCLC patients with high IRS-1 expression than that of those with low IRS-1 expression (24.0 vs. 54.4 months, P < 0.001, Fig. 4e). Univariate analysis and multivariate Cox regression analysis showed that IRS-1 expression was a prognostic factor for the OS of NSCLC patients, independent of other tested prognostic factors such as smoking status, histology type, lymph node involvement, differential degree, and histological grade (RR 0.413, CI 0.238–0.718, P = 0.002, Table 2).
Discussion
Migration and invasion are two important hallmarks of cancer and are responsible for the majority of cancer deaths (Xia et al. 2013; Mattie et al. 2006). Despite recent advances in chemotherapies and molecular-targeted therapies, resectable NSCLC only sees modest improvement and treatment failure ensues with eventual development of distant metastasis (Saintigny and Burger 2012). Multiple signaling pathways are involved in regulating tumor cell proliferation, migration, and invasion. IRS-1, a major substrate of insulin, insulin-like growth factors and cytokine signaling, plays an important role in mediating apoptosis, cell differentiation, and cell transformation. Downregulation of IRS-1 expression has been correlated with the progression of breast cancer (Schnarr et al. 2000). The loss of IRS-1 enhances metastasis of head and neck cancer (Luo et al. 2012). A study of 94 cases of stage I NSCLC found that loss of IRS-1 was more frequent in stage 1B than 1A NSCLC, suggesting that IRS-1 loss could be an early event in NSCLC development (Han et al. 2006). We found that IRS-1 was variably expressed in NSCLC, but observed no statistically significant difference in clinicopathologic characteristics, including age, gender, tumor size, node status, pathologic type, differentiation and smoking status between patients with low and high IRS-1 expression except in EGFR expression. EGFR was found to interact with and recruit IRS-1 in tamoxifen-resistant MCF-7 (Tam-R) breast cancer cells and EGFR-selective tyrosine kinase inhibitor Gefitinib could reduce this association (Knowlden et al. 2008). A recent study revealed that EGF dramatically upregulated IRS-1 in EGFR-positive breast cancer cells (Cui et al. 2006). Han et al. (2006) found that IRS-1 expression did not correlate with OS and DFS; however, their study cohort was limited to stage I NSCLC. We found that high IRS-1 expression correlated with markedly longer OS and DFS despite of different stages of NSCLC. Therefore, we suggest that IRS-1 could be a useful predictor for NSCLC survival time.
Recent studies have demonstrated that specific miRNAs implicate in the processes of migration and invasion (Ortholan et al. 2009; Yanaihara et al. 2006; Hayashita et al. 2005). Huang et al. reported that miR-23a can function as a proliferation-promoting factor, and TGF-β can induce specific miRNA expression to escape from tumor-suppressive response in hepatocellular carcinoma (HCC) cells. Tang et al. suggest that miR-23a overexpression promotes the growth, invasion, and metastasis of colon carcinoma cells, and miR-23a may serve as important biological markers for the malignant phenotypes of colon cancer, such as invasion and metastasis (Tang et al. 2012; Huang et al. 2008). Shi et al. have shown that TGF-β1 induces EMT by modulating IRS-1 signaling (Shi et al. 2009), and we found that miR-23a promotes TGF-β-induced EMT in NSCLC cells (Cao et al. 2012). We speculated a close association between miR-23a and IRS-1 in NSCLC cells. Indeed, we found that IRS-1 was a direct target of miR-23a and overexpression of miR-23a was associated with significant IRS-1 downregulation. Transwell assay showed that miR-23a significantly promoted the migration and invasion of A549 cells, while it failed to boost this function when IRS-1 was downregulated. We also found that overexpression of IRS-1 aborted miR-23a-induced increase in the migration and invasion of A549 cells. Our findings suggest the existence of a regulatory loop of miR-23a and IRS-1 in NSCLC cells: miR-23a suppresses IRS-1 expression, while IRS-1 counteracts miR-23a activity. miR-23a functions as an oncogenic miRNA in various malignancies and is upregulated in lung cancer, breast cancer, hepatocellular carcinoma, and colon carcinoma (Cao et al. 2012; Mattie et al. 2006; Tang et al. 2012; Huang et al. 2008). More specifically, miR-23a promotes growth, invasion, and metastasis of colon cancer cells (Tang et al. 2012), and acts as a growth-promoting and anti-apoptotic factor in HCC cells (Huang et al. 2008). miR-23a is decreased in human prostate cancer, which is associated with increased c-Myc expression (Gao et al. 2009). Taken together, these findings suggest that miR-23a may act as an oncogene in multiple human malignancies. Previously, we reported that miR-23a regulated TGF-β-induced EMT by suppression of E-cadherin in lung cancer cells (Cao et al. 2012); this finding may explain the above observations that miR-23a functions as a metastasis driver.
However, the exact mechanisms underlying how NSCLC cells are regulated after the interaction of miR-23a and IRS-1 remains intriguing. Since IRS-1 conveys signals to PI-3K/Akt and ERK1/2 pathways (Gao et al. 2009). IRS-1/PI-3K/Akt signaling may serve as the downstream effects after interaction in part. Moreover, in breast cancer, action of IRS-1 siRNA induces the apoptotic response and reduces cell viability (Gao et al. 2009); it is possible that miR-23a/IRS-1 influence proliferation and apoptosis in this aspect leading to the OS prediction.
In conclusion, our study indicated that IRS-1 was variably expressed in NSCLC patients. IRS-1 expression correlated with NSCLC patient survival. miR-23a directly targeted and suppressed the expression of the IRS-1 gene, while IRS-1 counteracts miR-23a migration/invasion-promoting activity. IRS-1 may serve as a useful predictor of the outcome of NSCLC patients.
References
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193
Cao M, Seike M, Soeno C, Mizutani H, Kitamura K et al (2012) MiR-23a regulates TGF-beta-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol 41:869–875
Chekulaeva M, Filipowicz W (2009) Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 21:452–460
Cui X, Kim HJ, Kuiatse I, Kim H, Brown PH et al (2006) Epidermal growth factor induces insulin receptor substrate-2 in breast cancer cells via c-Jun NH(2)-terminal kinase/activator protein-1 signaling to regulate cell migration. Cancer Res 66:5304–5313
Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K et al (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458:762–765
Garzon R, Calin GA, Croce CM (2009) MicroRNAs in Cancer. Annu Rev Med 60:167–179
Gibson SL, Ma Z, Shaw LM (2007) Divergent roles for IRS-1 and IRS-2 in breast cancer metastasis. Cell Cycle 6:631–637
Gual P, Le Marchand-Brustel Y, Tanti JF (2005) Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie 87:99–109
Han CH, Cho JY, Moon JT, Kim HJ, Kim SK et al (2006) Clinical significance of insulin receptor substrate-I down-regulation in non-small cell lung cancer. Oncol Rep 16:1205–1210
Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K et al (2005) A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65:9628–9632
Huang S, He X, Ding J, Liang L, Zhao Y et al (2008) Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer 123:972–978
Jemal A, Bray F, Center MM, Ferlay J, Ward E et al (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R et al (2005) RAS is regulated by the let-7 microRNA family. Cell 120:635–647
Knowlden JM, Jones HE, Barrow D, Gee JM, Nicholson RI et al (2008) Insulin receptor substrate-1 involvement in epidermal growth factor receptor and insulin-like growth factor receptor signalling: implication for Gefitinib (‘Iressa’) response and resistance. Breast Cancer Res Treat 111:79–91
Luo X, Fan S, Huang W, Zhai S, Ma Z et al (2012) Downregulation of IRS-1 promotes metastasis of head and neck squamous cell carcinoma. Oncol Rep 28:659–667
Ma Z, Gibson SL, Byrne MA, Zhang J, White MF et al (2006) Suppression of insulin receptor substrate 1 (IRS-1) promotes mammary tumor metastasis. Mol Cell Biol 26:9338–9351
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S et al (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362:2380–2388
Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L et al (2006) Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 5:24
Moran C (2011) Importance of molecular features of non-small cell lung cancer for choice of treatment. Am J Pathol 178:1940–1948
Ohe Y, Ohashi Y, Kubota K, Tamura T, Nakagawa K et al (2007) Randomized phase III study of cisplatin plus irinotecan versus carboplatin plus paclitaxel, cisplatin plus gemcitabine, and cisplatin plus vinorelbine for advanced non-small-cell lung cancer: four-Arm Cooperative Study in Japan. Ann Oncol 18:317–323
Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B et al (2009) MicroRNAs and lung cancer: new oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr Med Chem 16:1047–1061
Reiss K, Wang JY, Romano G, Furnari FB, Cavenee WK et al (2000) IGF-I receptor signaling in a prostatic cancer cell line with a PTEN mutation. Oncogene 19:2687–2694
Saintigny P, Burger JA (2012) Recent advances in non-small cell lung cancer biology and clinical management. Discov Med 13:287–297
Schiller JH, Harrington D, Belani CP, Langer C, Sandler A et al (2002) Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 346:92–98
Schnarr B, Strunz K, Ohsam J, Benner A, Wacker J et al (2000) Down-regulation of insulin-like growth factor-I receptor and insulin receptor substrate-1 expression in advanced human breast cancer. Int J Cancer 89:506–513
Shi J, Wang DM, Wang CM, Hu Y, Liu AH et al (2009) Insulin receptor substrate-1 suppresses transforming growth factor-beta1-mediated epithelial-mesenchymal transition. Cancer Res 69:7180–7187
Tang HL, Deng M, Liao QJ, Zeng X, Zhou XT et al (2012) Expression and clinical significance of miR-23a and metastasis suppressor 1 in colon carcinoma. Zhonghua Bing Li Xue Za Zhi 41:28–32
Tsim S, O’Dowd CA, Milroy R, Davidson S (2010) Staging of non-small cell lung cancer (NSCLC): a review. Respir Med 104:1767–1774
White MF (2003) Insulin signaling in health and disease. Science 302:1710–1711
Xia H, Ooi LL, Hui KM (2013) MiR-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K et al (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189–198
Acknowledgments
Thanks to Professor Jin Xiaoming and Dr. Tong Dandan for providing pathologic evaluation. This work was supported by National Natural Science Foundation of China (30772540, 81172214 to L.C.).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Cao, M., Li, Y., Lu, H. et al. miR-23a-mediated migration/invasion is rescued by its target, IRS-1, in non-small cell lung cancer cells. J Cancer Res Clin Oncol 140, 1661–1670 (2014). https://doi.org/10.1007/s00432-014-1725-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-014-1725-0