Abstract
Purpose
Resistance to chemotherapy drugs remains a difficult problem in bladder cancer treatment. Protein expression is an important factor underlying multidrug resistance (MDR) in bladder cancer. The aim of the study was to explore differentially expressed proteins responsible for MDR between an adriamycin-resistant human bladder cancer cell line (pumc-91/ADM) and its parental cell line (pumc-91).
Methods
Two-dimensional gel electrophoresis (2-DE) combining image analysis was used to screen the differentially expressed protein spots between the pumc-91/ADM and pumc-91 cell lines. Then, the protein spots were identified using MALDI-TOF/TOF mass spectrometry. Among the identified proteins, annexin A2 (ANXA2) and nucleophosmin (NPM1) were then further verified using RT-PCR and Western blot analysis.
Results
A total of 30 proteins, including 19 up-regulated and 11 down-regulated proteins, were successfully identified in pumc-91/ADM. According to their different functions, these 30 proteins were classified into 12 categories. Annexin A2 (ANXA2) and nucleophosmin (NPM1) were up-regulated in pumc-91/ADM compared with pumc-91.
Conclusion
The proteins identified may have an important clinical significance in MDR, and ANXA2 and NPM1 may take part in mechanism of MDR in bladder cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bladder cancer is one of the most common malignancies worldwide. It is the second most common cancer affecting the genitourinary tract and is also the second most common genitourinary tract cancer lead to mortality (Ichimi et al. 2009). The biological characteristics of bladder cancer render it prone to drug resistance and recurrences. Despite the application of various therapies, approximately 35–45 % of bladder tumors will recur, and 10 % of patients will progress to muscle-invasive disease (Gingrich 2011). Chemotherapy is an important tool for bladder cancer treatment: systemic chemotherapy is effective for advanced tumors; intravesical chemotherapy is a primary therapy for superficial tumors and can help prevent recurrence. Adriamycin is an important drug used for systemic and intravesical chemotherapy against bladder cancer. However, owing to multidrug resistance (MDR), treatment with Adriamycin or other agents often fails. “MDR is a phenomenon whereby resistance to 1 anticancer drug is accompanied by resistance to drugs whose structures and mechanisms of action may be completely different (Baguley 2010).” Several mechanisms had been implicated in the development of MDR, such as “extrusion of the drug by cell membrane pumps, enhanced drug detoxification, increased DNA damage repair, redistribution of intracellular accumulation of drugs, modification of drug target molecules, suppression of drug-induced apoptosis, up-regulation of lipids” and other biochemical changes (Hong et al. 2005). Resistance to anticancer drugs remains a major obstacle to overcome, and further studies on MDR in bladder cancer are required. We have previously established a human bladder cancer cell line is resistant to adriamycin (pumc-91/ADM). In order to assess whether the pumc-91/ADM resistant to adriamycin, MTT and flow cytometry analysis were performed. The results showed that pumc-91/ADM exhibited a tenfold resistance to adriamycin and were also cross-resistance to methotrexate, cisplatin, vincristine, and epirubicin. The pumc-91/ADM was a good model for further research (Zhang et al. 2009a, b).
In addition, protein expression is an important factor underlying MDR in bladder cancer. Two classical proteins related to MDR, P-glycoprotein (P-gp) and the multidrug resistance protein 1 (MRP1), were over-expressed in bladder cancer cells resistant to adriamycin and patients after chemotherapeutic treatment (Hasegawa et al. 1995; Nakagawa et al. 1997; Tada et al. 2000). Few other meaningful proteins associated with MDR were identified. In this study, we attempted to reveal new markers related to MDR in bladder cancer by examining protein expression changes between pumc-91/ADM and pumc-91.
Materials and methods
Reagents
Adriamycin (Sigma), IPG dry strips (pI 3–10, 24 cm), thiourea, urea, CHAPS, DTT, tris-base, SDS, TEMED, coomassie brilliant blue G-250, trizol, the enhanced HRP-DAB chromogenic kit. Horseradish-peroxidase-conjugated goat anti-mouse, goat anti-rabbit IgG (Santa Cruz), polyclonal antibodies against annexin2 (ANXA2, ab41803) and monoclonal antibodies against nucleophosmin (NPM1, ab10530) (Abcam).
Cell lines
The human bladder cancer cell line pumc-91 was provided from the Cell Laboratory of Beijing Union Medical College Hospital. When grew to the logarithmic phase, the pumc-91 cell was prepared into single cell suspension and incubated with adriamycin. We induced and established the multidrug-resistant bladder cancer cell lines pumc-91/ADM by increasing use of adriamycin. To preserve the biologic characteristics of MDR, pumc-91/ADM was cultured in RPMI1640 medium with 15 % fetal calf serum and 1.0 mg/L adriamycin.
Protein extraction
Pumc-91/ADM and pumc-91 cells were collected and washed three times with PBS, the cells were lysed in lysis buffer (Triscl 30 mM, thiourea 2 mol/L, urea 7 mol/L, 1 % DTT, 4 % CHAPS, 1 % Pharmalyte, 1 g/L DNase, pH adjusted to 8.5 with HCL) followed by sonication treatment (30 × 1 s, total 60 s) placed in a beaker with ice water mixture. The cell lysate was placed 30 min at room temperature and then centrifuged at 15,000 rpm for 30 min at 4 °C to obtain the supernatant. Protein concentrations were measured with the Bradford assay (Bradford 1976), and the extracted protein was packed and stored at −80 °C.
Two-dimensional gel electrophoresis (2-DE)
Two-dimensional gel electrophoresis (2-DE) was performed as described by the manufacturer’s suggested protocols (Bio-Rad). The pumc-91/ADM and pumc-91 were run on 3 gels, respectively, to generate 6 2-DE maps for analysis. For each gel, the protein sample extracted was mixed with rehydration buffer (thiourea 2 mol/L, urea 7 mol/L, 1 % DTT, 4 % CHAPS, 1 % ampholyte and trace bromophenol blue), and the total volume was adjusted to 450 µL (1 mg of proteins). The IPG strip placed side down in a 24-cm focusing tray containing the protein sample solutions and then covered with mineral oil. Isoelectric focusing was carried out on PROTEAN IEF Cells (Bio-Rad). After rehydrating for 13 h at 50 V, the focusing was conducted successively according to the following conditions: 2 h at 250 V, 2 h at 500 V, 2 h at 1,000 V, and 5 h at 10,000 V to give a total of 120 kVh. First-dimension IPG strip was then equilibrated for 15 min in Equilibration Buffer I (6 M urea, 20 % glycerol, 2 % SDS, 0.375 M Tris–HCl pH 8.8, 0.1 g DTT) and transferred to the Equilibration Buffer II (6 M urea, 20 % glycerol, 2 % SDS, 0.375 M Tris–HCl pH 8.8, 0.4 g iodoacetamide) for 15 min. After equilibration, second-dimension sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was carried out in 12 % polyacrylamide gels and used the PROTEAN-II XI Electrophoresis Cell (Bio-Rad) at 60 v/gel for 1 h and then at 160 v/gel for 20 h. Gels were then fixed in 10 % methanol and 7 % acetic acid for 1 h, stained with Coomassie blue G-250 (Bio-Rad) overnight followed by being destained with deionized water. Each stained 2-DE gels were scanned on Image scanner and analyzed using PDQuest software (Bio-Rad).
Image analysis
The scanned six 2-DE images (3 images of pumc-91/ADM, 3 images of pumc-91) were analyzed using PDQuest software (v.7.1.0). Protein spots of differential abundance were screened between the pumc-91/ADM and pumc-91 2-DE images. In order to reduce experimental variations, three separate gels were run on for each sample at the same time. The six images were normalized prior to analysis to eliminate the background interference. An automatic match-set of all 6 2-DE maps was conducted after a master gel had been constructed, followed by manually matched, and quantitative analysis of differential protein abundance through statistical analysis was performed. Abundance of matched protein spots of the two images (pumc-91/ADM image and pumc-91 image) was defined as differential if the observed fold change was greater than 2 with p values less than 0.05.
Protein identification by MALDI-TOF/TOF
Differentially expressed protein spots confirmed by PDQuest software were excised from the gels, destained with 100 mmol/L NH4HCO3 in 50 % acetonitrile. After dried in a vacuum centrifuge, the gel particles were then digested with the 0.01 µg/µl trypsin and 25 mmol/L NH4HCO3 overnight at 37 °C. The next day, peptides were firstly extracted using extraction buffer I (5 % trifluoroacetic acid), followed by the second in the extraction buffer II (50 % acetonitrile, 2.5 % trifluoroacetic acid). The resulting peptides were subjected to the MS analysis dissolved in 7 mg/ml CHCC matrix in 50 % acetonitrile, 0.1 % trifluoroacetic acid.
MS and MS/MS spectra of the differentially expressed proteins were obtained by the ABI 4700 Proteomics Analyzer MALDI-TOF/TOF (Applied Biosystems) with a mass range of 700–4,000 m/z. Before analysis, internal and external calibration was performed in MS and MS/MS mode. The peptides with signal-to-noise ratio above 100 at the MS were selected for the MS/MS. For MS/MS analysis, a minimum signal-to-noise ratio of 50 was set to be picked from each spot. The MS combining with MS/MS spectra was searched against the IPI human database using the MASCOT (Matrix Science) and GPS Explorer (Applied Biosystems). The searching parameters were set up as follows: homo sapiens, trypsin cleavage, the missed cleavage sites were allowed up to 1, carbamidomethylation was the fixed modification (cysteine); oxidation of methionine was the variable modification; the mass tolerance set to ±1.0 Dalton; the MS/MS tolerance set to ±0.5 Dalton.
Reverse transcription (RT)-PCR
Total RNA was extracted from the pumc-91/ADM and pumc-91 cells using the Trizol method according to manufacturer’s protocol. Three micrograms of total RNA from each sample was reverse transcribed to obtain single-stranded cDNA. PCR assays were carried out in a PCR amplification instrument (Eppendorf). The gene anxa2, npm1, and β-actin were amplified using the specific primers (Table 1). The PCR conditions were 30–34 cycles, an annealing temperature of 55 °C was used for the anxa2 and npm1 primers, and 58 °C was used for the β-actin primers. All primers were annealed for 1 min and denaturated at 94 °C for 40 s, elongated at 72 °C for 1 min. The PCR products were detected on a 1.5 % agarose gel with ethidium bromide staining. Using the Laned 1D gel analysis software (Sage Creation, China), the integrated optical density (IOD) of each band was analyzed. Every experiment was repeated at least three times. To assess the differential mRNA level of anxa2 and npm1 between the two cell lines, the average IOD of the gen in the pumc-91/ADM was calculated relative to which in the pumc-91, after normalization against a β-actin standard. The Histogram for relative fold expression change of the gen was drawn using SPSS (v 16.0).
Western blotting
Western blotting was performed with proteins extracted from the two cell lines prepared for 2-DE. The process was done as previously described (Pastorelli et al. 2007). Briefly, total 30 µg proteins were separated by 10 % SDS-PAGE and transferred 1.5 h to a PVDF membrane (Bio-Rad). The membrane was blocked with 5 % nonfat dry milk for 2 h at room temperature and then incubated with primary annexin A2, nucleophosmin, and β-actin antibodies overnight at 4 °C. Horseradish-peroxidase-conjugated goat anti-mouse, goat anti-rabbit IgG were used as secondary antibodies. The results were detected by the enhanced HRP-DAB chromogenic kit.
Statistical analysis
The significant difference of proteins and relative mRNA expression levels of ANXA2, NPM1 between the pumc-91/ADM and pumc-91 cell lines was evaluated using Student’s t-test. p values for the variance of protein and mRNA were determined with p < 0.05. Statistical analysis was done by using SPSS (v16.0).
Results
Identification of differentially expressed proteins
In order to compare global protein expression profiles between the drug-resistant cell line pumc-91/ADM and its parental cell line pumc-91, 2-DE analysis was performed. Protein spots of differential abundance between the 2 cell lines were screened through the PDQuest software for further analysis. The mean number of protein spots detected in pumc-91/ADM and pumc-91 2-DE maps was 1,772 ± 57 and 1,739 ± 59, respectively. Thirty-four protein spots with a more than twofold expressed change between the two cell lines (Fig. 1) were selected and submitted to MALDI-TOF/TOF for identification. A total of 30 proteins, including 19 up-regulated and 11 down-regulated proteins, differentially expressed in pumc-91/ADM (but not in pumc-91) were successfully identified. The identification information of these proteins is shown in Table 2.
Of the 30 proteins, we choose ANXA2 and NPM1 for further verification by RT-PCR and Western blot. Peptide mass fingerprinting (PMF) and matched amino acid sequences of these proteins are illustrated in Fig. 2. Representative MS/MS spectra of ANXA2 (peptide sequence: AYTNFDAER) and NPM1 (peptide sequence: MSVQPTVSLGGFEITPP VVLR) are shown in Fig. 3, respectively.
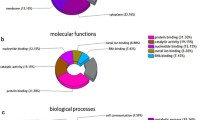
The 30 proteins, which were differentially expressed between the pumc-91/ADM and pumc-91 cell lines, were further classified into 13 functional categories (Fig. 4). The categories were as follows: transferase (20 %), oxidoreductase (13.33 %), transporter (10 %), transcription factor (10 %), calcium ion binding (10 %), nucleic acid binding (6.67 %), enzyme modulator (6.67 %), cytoskeletal protein (6.67 %), signaling molecule (3.33 %), ligase (3.33 %), hydrolase (3.33 %), lyase (3.33 %), and storage protein (3.33 %).
Validation of differentially expressed proteins
To further verify the identified proteins, semi-quantitative RT-PCR analysis was performed to determine the mRNA level of anxa2 and npm1 in the pumc-91/ADM and pumc-91 cell lines. Figure 5a, c showed the PCR products detected by agarose electrophoresis; the relative mRNA expression levels of the gens were up-regulated in pumc-91/ADM compared to the pumc-91 cell lines after normalization to a β-actin standard, and this finding was consistent with the results obtained by 2-DE. The histograms for relative mRNA fold expression change are shown in Fig. 5b, d.
Western blot analysis was also performed to determine the protein expression level of ANXA2 and NPM1 in the pumc-91/ADM and pumc-91 cell lines. As seen in Fig. 6, the relative expression levels of the proteins were up-regulated in the pumc-91/ADM compared to the pumc-91 cell line.
Discussion
Differential protein expression between the adriamycin-resistant human bladder cancer cell line pumc-91/ADM and its parental cell line pumc-91 was assessed using a classical 2-DE method and MALDI-TOF/TOF. A total of 30 proteins differentially expressed were discovered. Two of these proteins—ANXA2 and NPM1—that were further verified by semi-quantitative RT-PCR, and Western blot analysis was over-expressed in the pumc-91/ADM cell line. These results suggested that various drug-resistant mechanisms may work together to induce chemotherapy resistance in bladder cancer.
ANXA2 is a 36-kDa protein belonging to the annexin superfamily of Ca2+-dependent phospholipid-binding proteins. More than 60 different annexins have been identified, all of which play important roles in membrane traffic and organization, signal transduction, cell proliferation, differentiation, and apoptosis. More than ten annexin isoforms are expressed in human (Flood and Hajjar 2011; Hayes and Moss 2004; Gerke and Moss 2002). ANXA2 is also involved in angiogenesis, positive regulation of vesicle fusion, fibrinolysis, and other biological processes. It can regulate cell proliferation and is over-expressed in many tumors, such as pancreatic (Vishwanatha et al. 1993), gastric (Emoto et al. 2001a, b), colorectal (Emoto et al. 2001a, b), and breast cancer (Sharma et al. 2006). ANXA2 can promote cell growth and thus may contribute to drug resistance. Studies had found that ANXA2 was up-regulated in some chemotherapeutic drug-resistance cell lines (Zhang et al. 2009a, b; Cole et al. 1992), which was in concordance with the results of the current study. Zhang et al. (2009a, b) discovered that down-regulation of ANXA2 decreases cell proliferation in a drug-resistant MCF-7/ADM cell line. However, in prostate cancer, it had been reported that reduced or lost ANXA2 may lead to the development and progression of prostate cancer, and ANXA2 expression had been indicated to suppress cell migration (Liu et al. 2003). It is therefore apparent that the role of ANXA2 in different tumor types may rely on tumor origin and tissue specificity (Yao et al. 2009). We should study the function of ANXA2 and its relationship with MDR in different tumors, respectively.
ANXA2 is also associated with metastasis and prognosis. It is a receptor for plasminogen and tissue-type plasminogen activator (tPA). When ANXA2 binds to plasminogen, the latter gets converted to plasmin. The plasmin can facilitate the degradation of the extracellular matrix to facilitate cell invasion and migration (Schulz et al. 2009). Sharma et al. reported that ANXA2 was expressed in invasive/metastatic MDA-MB231 cells but not in noninvasive/non-metastatic MCF-7 cells (Sharma et al. 2006). ANXA2 had been suggested to promote the invasion and metastasis of cancer cells. Moreover, drug resistance was possibly associated with cancer invasion and metastasis. Certain tumor cells selected for drug resistance were found to be more invasive and metastatic compared to nonresistant parental cells, and in some cases, tumors with increased metastatic potential were found to be more resistant to drugs than their primary counterparts (Liang et al. 2002). It is reasonable that ANXA2 may contribute to MDR by helping cancer cells invade and migrate. Zhang et al. also found that ANXA2 may enhance cell invasion of the drug-resistant MCF-7/ADM cells line (Zhang et al. 2009a, b). In summary, the over-expression of ANXA2 in the bladder cancer pumc-91/ADM cell line may promote cell growth, enhanced cell invasion and metastasis; thus, it may be a factor in drug resistance.
Nucleophosmin (NPM 1), also known as B23, is a 37-kDa/pI 5.1 protein (Yung et al. 1985). It is originally identified as a non-ribosomal nucleolar phosphoprotein found at high levels in the granular regions of the nucleolus (Lim and Wang 2006). The protein is multifunctional and involved in cell proliferation and differentiation (Liu et al. 2007), nucleosome assembly (Herrera et al. 1995), P53 regulation (Colombo et al. 2002), transportation (Borer et al. 1989), centrosome duplication (Tokuyama et al. 2001), and many other biological processes. NPM1 was highly up-regulated in proliferative cells, and it promoted cell growth by inhibiting tumor suppressors (Ye 2005). NPM1 was over-expressed in many tumors and had been considered a marker for colon (Nozawa et al. 1996), gastric (Tanaka et al. 1992), and prostate carcinomas (Subong et al. 1999) and thyroid tumors (Pianta et al. 2010). Additionally, NPM1 had been found to be mutated in acute myeloid leukemia (AML; Falini et al. 2005; Verhaak et al. 2005). Over-expression of NPM1 may be associated with tumor recurrence and progression and indicated a poor prognosis in bladder cancer and Ewing’s sarcoma (Tsui et al. 2004, 2008; Kikuta et al. 2009). There are multiple mechanisms associated with the anti-apoptotic process of NPM1. It can interact with several transcription factors to promote cell growth, such as NF-κB, E2F1, pRB (Dhar et al. 2004; Lin et al. 2006), and p53 (Colombo et al. 2002). NPM1 is a factor in the ARF-p53 pathway, and it can compete with Mdm2 to bind with the tumor suppressor ARF and inhibit its function. Mutation and/or ectopic localization of the protein may be a key step in malignant transformation (Lim and Wang 2006; Korgaonkar et al. 2005).
NPM1 over-expression also prevents apoptosis induced by various pathways. For example, over-expression of NPM1 could not only reduce the susceptibility of human leukemia HL-60 cells to retinoic acid-induced apoptosis and differentiation (Hsu and Yung 2000), but it could also enhance the tolerance of NIH 3T3 cells and HeLa cells to ultraviolet radiation-induced cell death (Wu et al. 2002a, b). Yang et al. also reported that NPM1 was over-expressed in the gastric cancer drug-resistant SGC7901/ADM cell line, and that the suppression of NPM1 expression in the cell line enhanced adriamycin chemosensitivity (Yang et al. 2007). Recently, Hu et al. found that NPM1 may be associated with drug resistance and may aid in the prognostic assessment of leukemia cases (Hu et al. 2011). These results indicate that the over-expression of NPM1 may have important implications in the bladder cancer pumc-91/ADM cell line. This protein possibly decreases adriamycin-induced cell apoptosis. In addition, as a chaperone protein, NPM1 can shuttle between the nucleus and cytoplasm, and thus, it may have some molecular chaperone activities such as “the promotion of the renaturation of chemically denatured proteins or the inhibition of temperature-dependent and temperature-independent protein aggregation (Szebeni and Olson 1999).” Therefore, it is conceivable that NPM1 may play an important role in helping cells survive chemotherapy treatment.
In the 30 proteins differentially expressed identified in this study, 3 are involved in glycolysis: pyruvate kinase isozymes M1/M2 (PKM2), l-lactate dehydrogenase A chain (LDHA), and fructose-bisphosphate A (ALDOA). It is possible therefore that cancer cell glycolysis may have been enhanced by the addition of chemotherapeutic drugs in our experiment. The factors contributing to the enhanced glycolysis may be a result of mitochondrial metabolic defects that limit oxygen supply to tumor cells. The metabolic adaptation in response to these alterations is associated with reduced sensitivity to common anticancer agents (Xu et al. 2005). Xu et al. found that inhibiting glycolysis can induce apoptosis in multidrug-resistant cells (Xu et al. 2005), suggesting that inhibition of cellular glycolysis may be an effective way to overcome MDR. Subsequently, proteins involved in glycolysis may be new markers of MDR.
In addition to the previously described proteins, many other differentially expressed proteins with diverse functions, including transferase, oxidoreductase, transcription factors, and signal transduction factors, were identified. Our results suggest that a variety of mechanisms and proteins may contribute to the development of MDR, although further verification and research are required to elucidate their exact roles.
Conclusion
Using a proteomic method, 30 differentially expressed proteins between pumc-91/ADM and pumc-91 cell lines were identified. Association of ANXA2 and NPM1, two of the upregulated proteins in pumc-91/ADM, were verified by RT-PCR and Western blot. These findings may have important clinical significance associate with MDR. Additionally, future studies would focus on the other identified proteins with similar strategies. All of the proteins in this study, including ANXA2 and NPM1, may be new targets for overcoming multidrug-resistant of bladder cancer.
References
Baguley BC (2010) Multi-drug resistance in cancer. Methods Mol Biol 596:1–9
Borer RA, Lehner CF, Eppenberger HM et al (1989) Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379–390
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cole SP, Pinkoski MJ, Bhardwaj G et al (1992) Elevated expression of annexin II (lipocortin II, p36) in a multidrug resistant small cell lung cancer cell line. Br J Cancer 65(4):498–502
Colombo E, Marine JC, Danovi D et al (2002) Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol 4:529–533
Dhar SK, Lynn BC, Daosukho C et al (2004) Identification of nucleophosmin as an NF-kappaB co-activator for the induction of the human SOD2 gene. J Biol Chem 279:28209–28219
Emoto K, Sawada H, Yamada Y et al (2001a) Annexin II overexpression is correlated with poor prognosis in human gastric carcinoma. Anticancer Res 21:1339–1345
Emoto K, Yamada Y, Sawada H et al (2001b) Annexin II overexpression correlates with stromal tenascin-C overexpression: a prognostic marker in colorectal carcinoma. Cancer 92:1419–1426
Falini B, Mecucci C, Tiacci E et al (2005) Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 352:254–266
Flood EC, Hajjar KA (2011) The annexin A2 system and vascular homeostasis. Vasc Pharmacol 54:59–67
Gerke V, Moss SE (2002) Annexins: from structure to function. Physiol Rev 82:331–371
Gingrich JR (2011) Chemohyperthermia for bladder cancer—clinically effective? Nat Rev Urol 8:414–416
Hasegawa S, Abe T, Naito S et al (1995) Expression of multidrug resistance-associated protein (MRP), MDR1 and DNA topoisomerase II in human multidrug-resistant bladder cancer cell lines. Br J Cancer 71:907–913
Hayes MJ, Moss SE (2004) Annexins and disease. Biochem Biophys Res Commun 322:1166–1170
Herrera JE, Savkur R, Olson MO (1995) The ribonuclease activity of nucleolar protein B23. Nucleic Acids Res 23:3974–3979
Hong L, Piao Y, Han Y et al (2005) Zinc ribbon domain containing 1 (ZNRD1) mediates multidrug resistance of leukemia cells through regulation of P-glycoprotein and Bcl-2. Mol Cancer Ther 4:1936–1942
Hsu CY, Yung BY (2000) Over-expression of nucleophosmin/B23 decreases the susceptibility of human leukemia HL-60 cells to retinoic acid-induced differentiation and apoptosis. Int J Cancer 88:392–400
Hu J, Lin M, Liu T et al (2011) DIGE-based proteomic analysis identifies nucleophosmin/B23 and nucleolin C23 as over-expressed proteins in relapsed/refractory acute leukemia. Leukemia Res 35:1087–1109
Ichimi T, Enokida H, Okuno Y et al (2009) Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 125:345–352
Kikuta K, Tochigi N, Shimoda T et al (2009) Nucleophosmin as a candidate prognostic biomarker of Ewing’s sarcoma revealed by proteomics. Clin Cancer Res 15:2885–2894
Korgaonkar C, Hagen J, Tompkins V et al (2005) Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol 25:1258–1271
Liang Y, McDonnell S, Clynes M (2002) Examining the relationship between cancer invasion/metastasis and drug resistance. Curr Cancer Drug Targets 2(3):257–277
Lim MJ, Wang XW (2006) Nucleophosmin and human cancer. Cancer Detect Prev 30:481–490
Lin CY, Liang YC, Yung Benjamin YM (2006) Nucleophosmin/B23 regulates transcriptional activation of E2F1 via modulating the promoter binding of NF-κB, E2F1 and pRB. Cell Signal 18:2041–2048
Liu JW, Shen JJ, Tanzillo-Swarts A et al (2003) Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene 22:1475–1485
Liu X, Liu ZX, Jang SW et al (2007) Sumoylation of nucleophosmin/B23 regulates its subcellular localization, mediating cell proliferation and survival. PNAS 104(23):9679–9684
Nakagawa M, Emoto A, Nasu N et al (1997) Clinical significance of multi-drug resistance associated protein and P-glycoprotein in patients with bladder cancer. J Urol 157:1260–1265
Nozawa Y, Van Belzen N, Vander Made AC et al (1996) Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J Pathol 178:48–52
Pastorelli R, Carpi D, Campagna R et al (2007) Proteome characterization of a human urothelial cell line resistant to the bladder carcinogen 4-aminobiphenyl. Proteome Sci 5:6–14
Pianta A, Puppin C, Deganuto M et al (2010) Nucleophosmin is overexpressed in thyroid tumors. Biochem Biophys Res Commun 397:499–504
Schulz DM, llner CB, Thomas G et al (2009) Identification of differentially expressed proteins in triple-negative breast carcinomas using DIGE and mass spectrometry. J Proteome Res 8:3430–3438
Sharma MR, Koltowski L, Ownbey RT et al (2006) Angiogenesis-associated protein annexin II in breast cancer: selective expression in invasive breast cancer and contribution to tumor invasion and progression. Exp Mol Pathol 81:146–156
Subong EN, Shue MJ, Epstein JI et al (1999) Monoclonal antibody to prostate cancer nuclear matrix protein (PRO:4–216) recognizes nucleophosmin/B23. Prostate 39:298–304
Szebeni A, Olson MO (1999) Nucleolar protein B23 has molecular chaperone activities. Protein Sci 8:905–912
Tada Y, Wada M, Kuroiwa K et al (2000) MDR1 overexpression and altered degree of methylation at the promoter region in bladder cancer during chemotherapeutic treatment. Clin Cancer Res 6:4618–4627
Tanaka M, Sasaki H, Kino I et al (1992) Genes preferentially expressed in embryo stomach are predominantly expressed in gastric cancer. Cancer Res 52:3372–3377
Tokuyama Y, Horn HF, Kawamura K et al (2001) Specific phosphorylation of nucleophosmin on Thr(199) by cyclin dependent kinase 2-cyclin E and its role in centrosome duplication. J Biol Chem 276:21529–21537
Tsui KH, Cheng AJ, Chang PL et al (2004) Association of nucleophosmin/b23 mRNA expression with clinical outcome in patients with bladder carcinoma. Urology 64:839–844
Tusi KH, Juang HH, Lee TH et al (2008) Association of nucleophosmin/B23 with bladder cancer recurrence based on immunohistochemical assessment in clinical samples. Acta Pharmacol Sin 29(3):364–370
Verhaak RG, Goudswaard CS, van Putten W et al (2005) Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 106:3747–3754
Vishwanatha JK, Chiang Y, Kumble KD et al (1993) Enhanced expression of annexin II in human pancreatic carcinoma cells and primary pancreatic cancers. Carcinogenesis 14:2575–2579
Wu MH, Chang JH, Yung BY (2002a) Resistance to UV-induced cell-killing in nucleophosmin/B23 over-expressed NIH 3T3 fibroblasts: enhancement of DNA repair and up-regulation of PCNA in association with nucleophosmin/B23 over-expression. Carcinogenesis 23:93–100
Wu MH, Chang JH, Chou CC, Yung BY (2002b) Involvement of nucleophosmin/B23 in the response of HeLa cells to UV irradiation. Int J Cancer 97:297–305
Xu RH, Pelicano H, Zhou Y et al (2005) Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res 65:613–621
Yang YX, Hu HD, Zhang DZ et al (2007) Identification of proteins responsible for the development of adriamycin resistance in human gastric cancer cells using comparative proteomics analysis. J Biochem Mol Biol 40:853–860
Yao HX, Zhang ZQ, Xiao ZQ et al (2009) Identification of metastasis associated proteins in human lung squamous. carcinoma using two-dimensional difference gel electrophoresis and laser capture microdissection. Lung Cancer 65:41–48
Ye KQ (2005) Nucleophosmin/B23, a multifunctional protein that can regulate apoptosis. Cancer Biol Ther 4:918–923
Yung BY, Busch H, Chan PK (1985) Translocation of nucleolar phosphoprotein B23 (37 kDa/pI 5.1) induced by selective inhibitors of ribosome synthesis. Biochim Biophys Acta 826:167–173
Zhang M, Jin S, Zhang M (2009a) Establishing human multi-drug resistant bladder cancer cell lines Pumc-91/ADM and evaluating its biological characteristics. J Med Res 38(1):70–72
Zhang Fei, Zhang Lin, Zhang Bin et al (2009b) Anxa2 plays a critical role in enhanced invasiveness of the multidrug resistant human breast cancer cells. J Proteome Res 8:3430–3438
Acknowledgments
Research supported by Beijing Municipal Natural Science Foundation.
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meng, Q., Lei, T., Zhang, M. et al. Identification of proteins differentially expressed in adriamycin-resistant (pumc-91/ADM) and parental (pumc-91) human bladder cancer cell lines by proteome analysis. J Cancer Res Clin Oncol 139, 509–519 (2013). https://doi.org/10.1007/s00432-012-1350-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-012-1350-8