Abatract
Purpose
To explore the expression of ribosomal protein L7A (RPL7A) in osteosarcoma and its correlation with clinical features.
Methods
Ribosomal protein L7A mRNA expression was quantified by real-time reverse transcription (RT)-polymerase chain reaction (PCR) in 47 specimens from osteosarcoma, 8 from normal bone tissues and 12 from benign bone lesion tissues. Expression of RPL7A mRNA was also detected in human osteosarcoma cell line MG-63. The relationship between RPL7A mRNA expression and clinicopathological factors was statistically analyzed. The immunoblotting pattern of RPL7A was also analyzed in 20 osteosarcomas, 8 normal bone and 8 benign bone tissues.
Results
Ribosomal protein L7A mRNA expression in osteosarcoma samples was significantly down-regulated compared with that in samples from normal bone (P < 0.001) and benign bone lesion tissues (P < 0.001). Low expression of RPL7A mRNA was also found in osteosarcoma cell line MG-63. Low expression of RPL7A mRNA was significantly associated with increased serum level of alkaline phosphatase (ALP, P = 0.008), but was not correlated with other clinicopathological parameters including sex, age, serum lactate dehydrogenase (LDH), tumor location, histological type, histological grading, lung metastasis and overall survival. Interestingly, survival analysis suggested low RPL7A mRNA expression was a significant poor prognostic indicator for overall survival in patients with high grade lesion developed lung metastasis at the time of diagnosis of the primary osteosarcoma (P < 0.05). On western blot, reduced expression of RPL7A protein was observed in osteosarcoma samples (n = 20) compared with normal bone (n = 8) (P < 0.001) and benign bone tissues (n = 8) (P < 0.001).
Conclusion
Under-expression of RPL7A may be involved in the carcinogenesis of osteosarcoma. Loss of RPL7A expression may be associated with poor survival of osteosarcoma patients with lung metastasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma is the most common malignant tumor of bone in adolescents and young adults. Despite advances in multimodality treatments, including aggressive adjuvant chemotherapy and wide resection of the tumor, patients still have a poor prognosis. Five-year survival for patients with metastatic osteogenic sarcoma remains only 20% three decades ago (Mankin et al. 1996; La Quaglia 1998). Moreover, in China, the majority of families have only one child at present, and so it is a big hit for the families with children suffering from osteosarcoma. The identification of biomarkers linked to development and/or prognosis of this disease is crucial for the diagnosis and treatment of these patients.
Altered gene expression caused either by mutations or by loss of heterozygosity has been reported in osteosarcoma. Such alterations can occur in tumor suppressor genes, such as retinoblastoma tumor suppressor (RB1) (Yamaguchi et al. 1992; Wadayama et al. 1994; Miller et al. 1996) and tumor protein 53 (TP53) (Miller et al. 1990). In addition, serum alkaline phosphatase (ALP) and lactate dehydrogenase (LDH) have been reported to be over-expressed in osteosarcoma and have an influence on prognosis of osteosarcoma (Bacci et al. 1993; Bacci et al. 2004). However, these genetic changes do not precisely reflect the biologic nature or clinical characteristics of all osteosarcomas.
Ribosomal proteins are a major component of ribosomes that catalyze protein biosynthesis in the cytoplasm of cells. In humans, 80 ribosomal proteins in males and 79 ribosomal proteins in females have been identified and characterized at the molecular level (Warner and Nierras 1998). However, their precise function and tissue expression have not been fully understood. Progress in ribosome research provides growing evidence that ribosomal proteins can also function during various cellular processes such as replication, transcription, RNA processing, DNA repair, and even inflammation besides their functional involvement in the protein biosynthesis (Wool 1996; Yamamoto 2000). In addition, recent data have demonstrated that defects in some ribosomal proteins contribute to carcinogenesis (Draptchinskaia et al. 1999; Amsterdam et al. 2004; Gazda et al. 2006; Choesmel et al. 2008; Ebert et al. 2008). However, little is known about the expression of these genes and their clinical significance in osteosarcoma. Ribosomal protein L7A, a component of the 60S ribosomal subunit located on chromosome 9q34, the chromosome frequently lost in osteosarcoma, is also believed to be involved in cell growth and differentiation. RPL7A specifically interacts with human thyroid hormone receptor (THR) and retinoic acid receptor (RAR) which in turn inhibits transactivation of the two nuclear hormone receptors (Burris et al. 1995). Thyroid hormone (TH) and retinoic acid (RA) are known to play an important role in regulation of cell proliferation, differentiation and death. While RPL7A was over-expressed in some types of cancer cells (Zhu et al. 2001; Vaarala et al. 1998; Kroes et al. 2000; Wang et al. 2000). However, there were no or only a small number of clinical samples in most studies and the specific clinical significance of RPL7A expression was not fully reported previously.
Prompted by these, in the current study, we investigated RPL7A expression level in specimens from osteosarcoma as well as human osteosarcoma cell line MG-63 compared with that from normal and benign bone lesion tissues and further evaluated the correlation between RPL7A mRNA expression and the clinicopathological parameters in osteosarcoma for the first time to our knowledge.
Materials and methods
Patients and tissue samples
A total of 47 patients were studied. All samples were snap-frozen fresh pretreatment samples obtained by biopsy in the Department of Orthopaedic Surgery, Shanghai 6th People’s Hospital, Shanghai Jiaotong University, during the period from 2002 to 2006. The mean age was 19 years (range: 5~56 years). Most of patients range from 12 to 20 years in age (32/47). Thirty patients were males and 17 females. Fifteen patients with G3 grade lesion at diagnosis developed lung metastasis. Diagnosis of lung metastasis was made by computed axial tomographic scans and by progression always showed as nodules in lung in computed tomographic scans. Following the initial biopsy, 41 patients received 2 cycles of multi-agent neoadjuvant chemotherapy. Patients underwent limb salvage 38 (38/47) cases, amputation 9 (9/47) cases. Each postoperative case received 3~6 cycles of adjuvant chemotherapy of equal cycles. The cytotoxic drugs used as each equal cycle were high-dose methotrexate, cis-diammine dichloroplatinum, adriamycin, and iphosphamide. Patients with recurrence obtained additional operation therapy if possible. In the duration of follow-up 15 cases were detected as lung relapse, 4 cases underwent local recurrence and 16 cases died of cancer (Table 1). Eight normal bone tissues were taken from healthy donors who had undergone total knee arthroplasty. Twelve benign specimens were derived from patients with bone cysts (n = 4), giant cell tumor (n = 3), osteofibrous dysplasia (n = 3) and chondroma (n = 2). Informed consent was obtained from all subjects prior to conducting the study.
Cell culture
The human osteosarcoma cell line MG-63 was originally derived from the American Tissue Culture Collection (ATCC, Rockville, MD). Cells were routinely cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin under standard tissue culture conditions. Cells were harvested and washed with PBS before RNA extraction for real-time PCR.
RNA extraction and cDNA synthesis
Total RNA was extracted with TRIZOL Reagent (Invitrogen, Carlsbad, CA, USA) and the concentration of RNA was determined by the absorbance at 260 nm, and the purity was determined by the 260/280 ratio with a BioPhotometer (Eppendorf, Hamburg, Germany). 1 μg RNA was reverse-transcribed with random primer by ReverTra Ace (Toyobo, Osaka, Japan) at 42°C for 1 h.
Real-time quantitative polymerase chain reaction
The mRNA expression of RPL7A and internal control gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were quantified by real-time PCR with iCycler iQ (Bio-Rad, Hercules, CA, USA). Genebank ID for RPL7A is NM_000972.2 and for GAPDH is NM_002046.3. The gene-specific exon-spanning PCR primer pair for RPL7A was 5′-ACCGCCAAACAGCTACTCAG-3′, 5′-GCAGGCAAGAAGACAACCAG-3′, and for GAPDH was 5′-ACGGATTTGGTCGTATTGGG-3′, 5′-ATCTCGCTCCTGGAAGATGG-3′. The sequences of the primers for RPL7a and GAPDH for RT-PCR were checked by Nucleotide BLAST for specific gene amplification. PCR product size for RPL7A is 254 bp and for GAPDH is 218 bp. Omission of cDNA template was used as a negative control. PCR amplification reaction conditions for RPL7A and GAPDH were 40 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s and finally elongated at 72°C for 5 min. All PCR were performed in duplicate and data of mean were used. For relative quantification of RPL7A gene expression level, standard curves were built for both genes by considering at least four points of a tenfold dilution series of cDNA in water; the slopes observed were −3.46 for RPL7A gene and -3.49 for GAPDH. The correlation coefficient of amplification was 0.992 and 0.996, respectively. Relative gene expression data are given as the n-fold change in transcription of the RPL7A genes normalized to the endogenous control in the same sample.
Protein extraction and Western blot
A total of 100 mg of frozen tissue was ground in liquid nitrogen before lysing in 1 mL RIPA Lysis Buffer (Santa Cruz Biotechnology, Santa Cruz, CA, USA) by sonication on ice. The lysates were then centrifuged at 14,000g for 20 min at 4°C. Subsequently, the protein concentration of the supernatants was determined by the BCA Protein Assay Kit (Sangon, Shanghai, China). After being heated to 99°C for 5 min in sample buffer, aliquots of protein extract (50 μg) were run on a 12% sodium dodecylsulfate–polyacrylamide gel and transferred onto a Nitrocellulose Blotting Membranes (Millipore, MA, USA). The transferred membrane, which was blocked with TBS-Tween 20 (TBST) containing 5% non-fat dry milk for 2 h at room temperature, was incubated overnight at 4°C with rabbit polyclonal antibody against human RPL7A (Cell Signaling Technology Inc., MA, USA) at 1:1,000, and GAPDH (Santa Cruz) at 1:4,000 dilution, followed by incubating with peroxidase-conjugated goat antirabbit immunoglobulin (Santa Cruz) diluted 1:10,000 in TBST for 1 h. Signals were developed using enhanced chemiluminescent reagent (Pierce Biotechnology, Rockford, IL, USA). GAPDH is used as the internal loading control. The band intensity was analyzed using Quantity One. Relative expression was calculated as the intensity ratio of RPL7A to that of GAPDH.
Statistical analysis
All statistical analyses were performed using the SPSS 12.0 software package for Windows (SPSS Inc., Chicago, IL, USA). We performed statistical analysis using either the Mann–Whitney U test or the Kruskal–Wallis test for comparison of RPL7A gene expression between normal bone tissues, benign bone lesion tissues and osteosarcoma samples, and for analysis the relationship between RPL7A mRNA expression and clinicopathological parameters. Overall survival (OS) was defined as the time from the start of the treatment to death due to any causes or date of the latest follow-up. Survival curves were plotted by the Kaplan–Meier method; statistical differences were analyzed using the log-rank test. A probability value less than 0.05 was considered to indicate a statistically significant difference.
Results
Down-regulation of RPL7A mRNA in osteosarcoma samples and MG-63 cell line
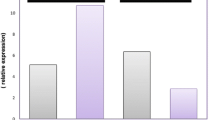
To investigate RPL7A mRNA expression level in osteosarcoma, 47 osteosarcoma samples were evaluated as distinct group versus normal bone and benign bone lesion tissues. Compared with the high expression in normal bone and benign bone lesion tissues, the RPL7A mRNA expression in osteosarcoma tissues showed significantly decreased levels (P < 0.001 and P < 0.001, respectively, Mann-Whitney U test). Low expression levels of RPL7A mRNA were also detected in a human osteosarcoma cell line MG-63 (Fig. 1). There was no significant difference in RPL7A mRNA expression level between normal bone tissues and benign bone lesion tissues (P = 0.186).
Differences in the expression of RPL7A mRNA among normal bone tissues (Normal), benign bone lesion tissues (Benign), osteosarcoma samples (Osteosarcoma) and MG-63 cell line (MG-63). There was a significant difference between the RPL7A mRNA levels in normal bone tissues and osteosarcoma (P < 0.001) and between benign bone lesion tissues and osteosarcoma (P < 0.001). There was no significant difference in RPL7A mRNA expression level between normal bone tissues and benign bone lesion tissues (P = 0.186). The line corresponds to the median
Correlation between RPL7A mRNA expression level and clinicopathologic parameters
Our data indicated that the low expression of RPL7A was significantly associated with increased serum level of ALP (P = 0.008). However, there were no statistically significant relationships with other clinicopathological parameters including sex, age, serum LDH, tumor location, histological type, histological grading and lung metastasis (Table 2). Further, the comparison among histological type and histological grading cannot be said to be entirely reliable due to the small number of patients involved in dilated blood vessels type and non-G3 grading.
To evaluate the impact of expression of RPL7A gene on overall survival, the patients were divided into two groups: patients who had values above the median of expression are defined as high-expression of the gene and patients who had a value below the median are considered low-expression of RPL7A gene. Patients with high grade (including G3 and G4) osteosarcoma were included in this survival analysis. RPL7A mRNA expression did not significantly correlate with patients’ overall survival (P = 0.058, Log rank test). However, overall survival of 15 patients with G3 grade lesion at diagnosis developed lung metastasis were analyzed and found that high-expression RPL7A mRNA osteosarcoma patients had better overall survival than low-expression RPL7A mRNA osteosarcoma patients did (P = 0.039, Log rank test) (Fig. 2).
Downregulation of RPL7A protein expression in osteosarcoma tissues
The level of RPL7A protein in osteosarcoma tissues (n = 20) was lower than that in normal bone (n = 8) and benign bone tissues (n = 8) on western blot (RPL7A/GAPDH ratio ± SD: osteosarcoma tissue, 0.4915 ± 0.2605; normal bone tissue, 1.1463 ± 0.3185; benign bone tissue, 1.0863 ± 0.4312; P < 0.001 and P < 0.001, respectively; Mann–Whitney U test; Fig. 3).
RPL7A immunoblotting of benign bone lesion tissues (Benign), normal bone tissues (Normal) and osteosarcoma samples (Osteosarcoma). Western blot indicated reduced RPL7A protein expression in osteosarcoma samples when compared with normal bone and benign bone lesion tissues. Shown are two individual benign bone lesion tissues, three individual normal bone and four individual osteosarcoma tissue samples
Discussion
In the present study we analyzed RPL7A mRNA and protein expression in human osteosarcoma. Quantitative real-time RT-PCR indicated that RPL7A mRNA is under-expressed in primary osteosarcoma samples and in an aggressive human osteosarcoma cell line MG-63 compared with that in normal and benign bone tissues. Western blot further demonstrated the down-regulation of RPL7A protein in the test osteosarcoma samples. The findings suggest that RPL7A may be a predictor of osteogenic malignancy, and RPL7A may be involved in osteosarcoma development.
Several recent studies have demonstrated that defects in some ribosomal proteins through somatic chromosomal deletions or mutation cause tumors or associate with carcinogenesis. It was recently shown in zebra fish that 11 ribosomal protein genes act as haploinsufficient tumor suppressors; the haploinsufficiency for any of these genes caused malignant tumors of the peripheral nerve sheath (Amsterdam et al. 2004). In humans, germline heterozygous mutations for two ribosomal proteins-RPS19 and RPS24 have been described in the congenital disorder known as Diamond-Blackfan anemia (Draptchinskaia et al. 1999; Gazda et al. 2006; Choesmel et al. 2008), a disease with an increased risk of solid tumor and leukemia (Janov et al. 1996; van Dijken and Verwijs 1995; Lipton et al. 2001). More recently, the 5q- syndrome, a subtype of myelodysplastic syndrome with a propensity to progress to acute myeloid leukemia, was demonstrated to be caused by partial loss of function of RPS14 though the somatic deletion of one allele (Ebert et al. 2008). Partial and/or complete loss of chromosome 9 is also notable in osteosarcoma (Bridge et al. 1997; Sandberg and Bridge 2003). RPL7A is a component of the 60S ribosomal subunit located on chromosome 9q34. Data have been demonstrated that RPL7A specifically interacts with human thyroid hormone receptor (THR) and retinoic acid receptor (RAR), which in turn inhibits transactivation of the two nuclear hormone receptors (Burris et al. 1995). Thyroid hormone (TH) is known to play an important role in the regulation of cell proliferation, differentiation and death including osteosarcoma cells (Patricia et al. 2008).
To our knowledge, the present study demonstrates for the first time that RPL7A mRNA is down-regulated in the majority of osteosarcoma samples examined, compared with normal bone and benign bone lesion tissues. Similar results were observed when expression of RPL7A protein was detected on western blot. In particular, the current study first analyzed the prognostic value of RPL7A expression in osteosarcoma and for the first time found that low RPL7A mRNA expression was a significant poor prognostic indicator for overall survival in patients with high grade lesion at diagnosis developed lung metastasis although we failed to detect the significance of RPL7A expression in the assessment of prognosis in osteosarcoma. This suggests that RPL7A is not a cause of metastasis and will probably have no negative effect in the absence of metastasis. It is known that metastasis remains the primary cause of poor survival of the majority of tumor patients including osteosarcoma; however, survival of patients with metastatic tumor has a wide variation (Srivastava et al. 2006). These phenomenon of ‘‘metastatic heterogeneity’’ raises questions on the nature of the relationship between metastasis and patient survival. It has been reported that in the case of breast cancer, separate genes underlie metastasis and poor prognosis (Minn et al. 2005a, b; Kang et al. 2003). Recently, such gene has also been identified in the case of osteosarcoma (Srivastava et al. 2006). To our knowledge, RPL7A is the second such gene found to be associated with prognosis among osteosarcoma patients with lung metastases without being directly associated with metastasis.
The mechanism(s) by which ribosomal protein dysfunction is tumorigenic has yet to be determined. Some researchers favor the possibility that it is a shared, ribosome-associated function or defects in ribosomal biogenesis that allows them to be tumor suppressors, such as reduced protein synthesis leading to a decrease in the level of a critical tumor suppressor protein, or of a positive regulator of apoptosis or differentiation, and then a severe reduction in their expression below some critical, hypothetical threshold does favor tumor development (Amsterdam et al. 2004; Draptchinskaia et al. 1999; Gazda et al. 2006; Choesmel et al. 2008). Alternatively, other studies indicated that individual ribosomal protein gene has extra-ribosomal activities (Wool 1996; Frodin and Gammeltoft 1999; Noara 1999). Whether the diseases mainly associated with ribosomal protein genes due to a defect in ribosomal biogenesis, an abnormality in translation, and/or disruption of an extraribosomal function(s) of ribosome proteins important for cell differentiation, proliferation and death, remains a question. Data have shown that RPL7A specifically interacts with THR and RAR which in turn inhibits transactivation of the two nuclear hormone receptors (Burris et al. 1995). Hyperthyroidism status has been demonstrated to favor osteosarcoma cell growth (Patricia et al. 2008). In the present study we hypothesize that down-regulation of RPL7A may enhance the sensitivity of osteosarcoma cells to TH and thereby result in disruption of growth control. Furthermore, an inverse relationship was observed between RPL7A mRNA expression and serum ALP in our study. This finding is in agreement with Simsek et al. and Patricia et al., who reported an increased ALP activity in hyperthyroidism (Simsek et al. 2003; Patricia et al. 2008). Our data suggest a possible pathway by which RPL7A may be involved in osteosarcoma cell proliferation. However, previous studies have shown over-expression of RPL7A in some types of cancer cells, which were contrary to the present finding (Zhu et al. 2001; Vaarala et al. 1998; Kroes et al. 2000; Wang et al.2000). We explain that there may be different roles of RPL7A in distinct microenvironment in different types of cells. For example, in breast and prostate cancer cells, hyperthyroidism status has been proved to decreased cancer cell growth and over-expression of RPL7A may decrease the sensitivity of breast and prostate cancer cells to THR (Martinez et al. 2000), whereas it is the contrary in osteosarcoma cells (Patricia et al. 2008). However, future study will be required to determine the functional role and the exact molecular mechanism in osteosarcoma cells.
The mechanisms involved in the downregulation of RPL7A expression remains to be unknown. It can be accomplished by loss of chromosomes (Bridge et al. 1997; Sandberg and Bridge 2003; Ebert et al. 2008). Previous studies have revealed that loss of chromosomes 9, 10, 13, and 17 is pronounced in osteosarcoma (Bridge et al. 1997; Sandberg and Bridge 2003). Several of these findings correspond with known molecular events in osteosarcoma such as loss or mutation of the RB1 and TP53 tumor suppressor genes located on chromosomes 13 and 17, respectively (Yamaguchi et al. 1992; Wadayama et al. 1994; Miller et al. 1990, 1996). Interestingly, the RPL7A gene is located on chromosomes 9q34. Moreover, data have shown that loss of chromosome 9 shared by the cell line MG-63 (Ozaki et al. 2003) but not shared by benign bone lesions. Therefore, detection of RPL7A may further make sense of distinguishing benign bone lesions such as benign bone cysts, giant cell tumor and osteofibrous dysplasia from osteosarcoma. Other mechanism(s), such as aberrant CpG methylation, transcriptional repressors and mutations also cannot be excluded while data have not been reported. Further investigation is needed to clarify the exact mechanism(s) involved in the decrease of RPL7A expression in osteosarcoma.
In conclusion, we analyzed RPL7A expression in osteosarcoma biopsy samples and found RPL7A expression is down-regulated in osteosarcoma. The present findings suggest the important role of RPL7A in the development of osteosarcoma. In particular, low RPL7A mRNA seems to be associated with very poor survival of patients with osteosarcoma metastasis. However, it should be emphasized that the present study is a retrospective study with limited samples. A multi-center prospective study with more clinical samples should be done before RPL7A can serve as a potential intervention target for osteosarcoma.
References
Amsterdam A, Sadler KC, Lai K et al (2004) Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2(E139):690–698. doi:10.1371/journal.pbio.0020139
Bacci G, Picci P, Ferrari S et al (1993) Prognostic significance of serum alkaline phosphatase measurements in patients with osteosarcoma treated with adjuvant or neoadjuvant chemotherapy. Cancer 71:1224–1230. doi:10.1002/1097-0142(19930215)71:4<1224::AID-CNCR2820710409>3.0.CO;2-M
Bacci G, Longhi A, Ferrari S et al (2004) Prognostic significance of serum lactate dehydrogenase in osteosarcoma of the extremity: experience at Rizzoli on 1,421 patients treated over the last 30 years. Tumori 90:478–484
Bridge JA, Nelson M, McComb E et al (1997) Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer Genet Cytogenet 95:74–87. doi:10.1016/S0165-4608(96)00306-8
Burris T, Nawaz Z, Tsal M-J et al (1995) A nuclear hormone receptor-associate protein that inhibits transactivation by the thyroid hormone and retinoic acid receptors. Proc Natl Acad Sci USA 92:9525–9529. doi:10.1073/pnas.92.21.9525
Choesmel V, Fribourg S, Aguissa-Touré AH et al (2008) Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet 17:1253–1263. doi:10.1093/hmg/ddn015
Draptchinskaia N, Gustavsson P, Andersson B et al (1999) The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet 21:169–175. doi:10.1038/5951
Ebert BL, Pretz J, Bosco J et al (2008) Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451:335–339. doi:10.1038/nature06494
Frodin M, Gammeltoft S (1999) Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol 151:65–77. doi:10.1016/S0303-7207(99)00061-1
Gazda HT, Grabowska A, Merida-Long LB et al (2006) Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet 79:1110–1118. doi:10.1086/510020
Janov AJ, Leong T, Nathan DG et al (1996) Diamond-Blackfan anemia. Natural history and sequelae of treatment. Medicine (Baltimore) 75:77–78. doi:10.1097/00005792-199603000-00004
Kang Y, Shu W, Siegel PM et al (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3:537–549. doi:10.1016/S1535-6108(03)00132-6
Kroes RA, Jastrow A, McLone MG et al (2000) The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett 156:191–198. doi:10.1016/S0304-3835(00)00462-6
La Quaglia MP (1998) Osteosarcoma Specific tumor management and results. Chest Surg Clin N Am 8:77–95
Lipton JM, Federman N, Khabbaze Y et al (2001) Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol 23:39–44. doi:10.1097/00043426-200101000-00009
Mankin HJ, Gebhardt MC, Jennings LC et al (1996) Long-term results of allograft replacement in the management of bone tumors. Clin Orthop Relat Res 324:86–97. doi:10.1097/00003086-199603000-00011
Martinez MB, Ruan M, Fitzpatrick LA (2000) Altered response to thyroid hormones by prostate and breast cancer cells. Cancer Chemother Pharmacol 45:93–102. doi:10.1007/s002800050016
Miller CW, Aslo A, Tsay C et al (1990) Frequency and structure of p53 rearrangements in human osteosarcoma. Cancer Res 50:7950–7954
Miller CW, Aslo A, Campbell MJ et al (1996) Alterations of the p15, p16, and p18 genes in osteosarcoma. Cancer Genet Cytogenet 86:136–142. doi:10.1016/0165-4608(95)00216-2
Minn AJ, Kang Y, Serganova I et al (2005a) Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest 115:44–55
Minn AJ, Gupta GP, Siegel P et al (2005b) Genes that mediate breast cancer metastasis to lung. Nature 436:518–524. doi:10.1038/nature03799
Noara H (1999) Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol 77:197–205. doi:10.1046/j.1440-1711.1999.00816.x
Ozaki T, Neumann T, Wai D et al (2003) Chromosomal alterations in osteosarcoma cell lines revealed by comparative genomic hybridization and multicolor karyotyping. Cancer Genet Cytogenet 140:145–152. doi:10.1016/S0165-4608(02)00685-4
Patricia PP, Teixeira SS, Conde SJ et al (2008) The importance of hormone receptor analysis in osteosarcoma cells growth submitted to treatment with estrogen in association with thyroid hormone. Cell Biochem Funct 26:107–110. doi:10.1002/cbf.1408
Sandberg AA, Bridge JA (2003) Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet 145:1–30. doi:10.1016/S0165-4608(03)00105-5
Simsek G, Karter Y, Aydin S et al (2003) Osteoporotic cytokines and bone metabolism on rats with induced hyperthyroidism; changes as a result of reversal to euthyroidism. Chin J Physiol 46:181–186
Srivastava A, Fuchs B, Zhang K et al (2006) High WT1 expression is associated with very poor survival of patients with osteogenic sarcoma metastasis. Clin Cancer Res 12:4237–4243. doi:10.1158/1078-0432.CCR-05-2307
Vaarala MH, Porvari KS, Kyllönen AP et al (1998) Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int J Cancer 78:27–32. doi:10.1002/(SICI)1097-0215(19980925)78:1<27::AID-IJC6>3.0.CO;2-Z
van Dijken PJ, Verwijs W (1995) Diamond-Blackfan anemia and malignancy. A case report and a review of the literature. Cancer 76:517–520. doi:10.1002/1097-0142(19950801)76:3<517::AID-CNCR2820760324>3.0.CO;2-8
Wadayama B, Toguchida J, Shimizu T et al (1994) Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res 54:3042–3048
Wang Y, Cheong D, Chan S et al (2000) Ribosomal protein L7a gene is up-regulated but not fused to the tyrosine kinase receptor as chimeric trk oncogene in human colorectal carcinoma. Int J Oncol 16:757–762
Warner JR, Nierras CR (1998) Trapping human ribosomal protein genes. Genome Res 8:419–421
Wool IG (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem Sci 21:164–165
Yamamoto T (2000) Molecular mechanism of monocyte predominant infiltration in chronic inflammation: mediation by a novel monocyte chemotactic factor, S19 ribosomal protein dimer. Pathol Int 50:863–871. doi:10.1046/j.1440-1827.2000.01132.x
Yamaguchi T, Toguchida J, Yamamuro T et al (1992) Allelotype analysis in osteosarcomas: frequent allele loss on 3q, 13q, 17p, and 18q. Cancer Res 52:2419–2423
Zhu Y, Lin H, Li Z et al (2001) Modulation of expression of ribosomal protein L7a (rpL7a) by ethanol in human breast cancer cells. Breast Cancer Res Treat 69:29–38. doi:10.1023/A:1012293507534
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, Se., Yao, Y., Dong, Y. et al. Down-regulation of ribosomal protein L7A in human osteosarcoma. J Cancer Res Clin Oncol 135, 1025–1031 (2009). https://doi.org/10.1007/s00432-008-0538-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-008-0538-4