Abstract
Purpose
To discover new molecular targets for cancer therapy and diagnosis, we surveyed signal transducers and activators of transcription 3 (Stat3)-regulated genes, because constitutive activation of Stat3 is associated with a wide variety of human malignancies.
Methods
We investigated the Stat3-regulated genes in 293 cells with cDNA microarray analysis and found that Nicotinamide N-methyltransferase (NNMT) was induced on stimulation of the cells with leukemia inhibitory factor. We examined the expression of NNMT in several types of cancer cells by real-time quantitative RT-PCR. To examine the role of Stat3, Hep-G2 hepatocellular carcinoma cells were transfected with NNMT promoter-luciferase reporter construct together with conditionally active Stat3 (Stat3ER) or dominant-negative Stat3 expression vector and NNMT promoter activity was determined. The expression of NNMT and activated Stat3 in 88 colon cancer tissues and 17 normal colon tissues was examined with immunohistochemical analysis.
Results
In Hep-G2 cells and SW480 colon cancer cells, NNMT expression increased on stimulation of the cells with interleukin 6. NNMT promoter activity in Hep-G2 cells was dependent on the activation of Stat3. MDA-MB-468 breast cancer cells and HT29 colon cancer cells expressed constitutively a high level of NNMT. Treatment of these cells with Stat3 siRNA or curcumin, which inhibited Stat3 phosphorylation, resulted in reduction of the NNMT level. We found a correlation between the expression of NNMT and activated Stat3 (P < 0.001) in the colon cancer tissues.
Conclusion
NNMT is a novel Stat3-regulated gene. Its expression is enhanced with the activation of Stat3 in colon cancer tissues. NNMT may be a potential candidate for a tumor marker of various kinds of cancers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Signal transducers and activators of transcription 3 (Stat3) were identified originally as key components of cytokine [such as interleukin (IL)-6 and leukemia inhibitory factor (LIF)] signaling pathways (Akira et al. 1994; Zhong et al. 1994). It was also shown that non-receptor and receptor tyrosine kinases, such as Src and EGF receptor, activate Stat3 (Zhong et al. 1994; Yu et al. 1995). Constitutive activation of Stat3 has been detected in a wide variety of human tumor cell lines and primary human tumors including breast cancer (Watson and Miller 1995), lung cancer (Fernandes et al. 1999; Song et al. 2003; Yeh et al. 2006), pancreatic cancer (DeArmond et al. 2003; Wei et al. 2003), gastric cancer (Kanda et al. 2004), and colon cancer (Ma et al. 2004; Corvinus et al. 2005; Kusaba et al. 2005; Lin et al. 2005; Kawada et al. 2006). Furthermore, a constitutively active mutant form of Stat3 (Stat3-C) in immortalized fibroblasts causes cellular transformation and tumor formation in nude mice (Bromberg et al. 1999). Stat3 up-regulates cyclin D1, c-Myc, survivin and Bcl-x, and causes stimulation of the proliferation and survival of cells (Yu and Jove 2004). It also stimulates tumor angiogenesis through activation of vascular endothelial growth factor and hypoxia-inducible factor 1 genes. We previously showed that Stat3 activated constitutively by autocrine LIF and/or IL-6 is responsible for hepatocyte growth factor (HGF) expression in human cancer cells and normal fibroblasts (Tomida and Saito 2004a, b). HGF is involved in tumor invasion and metastasis by stimulating the motility of cancer cells and by stimulating angiogenesis (Jiang et al. 2005; Saito and Tomida 2005). These results support the idea that Stat3 is a molecular target for cancer therapy (Yu and Jove 2004) and suggest that many Stat3-regulated genes are associated with cancer.
In the present study, we surveyed new Stat3-regulated genes and found that expression of the nicotinamide N-methyltransferase (NNMT) gene is associated with activation of Stat3. NNMT catalyzes the N-methylation of nicotinamide and other structural analogs, and is involved in the biotransformation of many drugs and xenobiotic compounds (Aksoy et al. 1994, 1995). NNMT is highly expressed in the liver and weakly expressed in other tissues, such as lung, colon, kidney, and thyroid (Yan et al. 1999). Recently, Roessler et al. (2005) found that NNMT was expressed at markedly higher levels in malignant tissues compared with normal colonic epithelium and they proposed that the NNMT serum level has significance as to the early detection of patients with colorectal cancer. We examined the expression of NNMT and activated Stat3 in colon cancer tissues and normal colon tissues, and found a correlation between the expression of NNMT and activated Stat3 in the colon cancer tissues.
Materials and methods
Cells and cell culture
Human embryo kidney 293 cells were cultured in Dulbecco’s minimum essential medium supplemented with 10% fetal bovine serum (FBS). Human breast carcinoma cell line MDA-MB-468, and human colon carcinoma cell lines HT-29 and SW480 were obtained from the American Type Culture Collection, Rockville, MD, USA. These cell lines, human hepatocellular carcinoma cell line Hep-G2 and breast carcinoma cell line MCF-7 were grown in RPMI 1640 medium supplemented with 10% FBS.
Materials
Recombinant human LIF was produced in Chinese hamster ovary (CHO) cells and purified to homogeneity as described previously (Lowe et al. 1989). Recombinant human IL-6 was kindly provided by Ajinomoto Co., Kawasaki, Japan. Chicken anti-NNMT antibodies were obtained from GenWay Biotech Inc., San Diego, CA, USA. Anti-phospho Stat3 (Tyrosine 705) antibodies, no. 9131 and sc-7993, were obtained from Cell Signaling Technology Inc., Beverly, MA, USA, and Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA, respectively. Curcumin was purchased from LKT Laboratories, St. Paul, MN, USA, and was prepared as a 20 mM solution in DMSO and then further diluted in cell culture medium. A synthetic (Z)-4-hydroxytamoxifen (4HT), purchased from Sigma-Aldrich, Fremont, CA, USA, was dissolved in ethanol (2 mM stock).
Microarray analysis
The 293 cells were stimulated with or without 10 ng/ml of recombinant human LIF for 10 h. Total RNAs were extracted from the cells using guanidium thiocyanate and then subjected to Human Expression CHIP (Takara Biotechnology Co., Kusatsu, Japan) analysis.
Plasmid construction
The NNMT promoter sequence from −2,568 to −105 relative to the translation start site was isolated, because transcription initiation occurred 105–109 nucleotides 5′-upstream from the translation initiation codon in human liver (Aksoy et al. 1995). The DNA was amplified by PCR using a forward primer, 5′-TAGGTACCATAACATGGATGGAACTGGAGGTC-3′, and a reverse primer, 5′-TACTCGAGCAGCATTCCTTCCATCAGTTCAGC-3′, with human blood cell DNA (Clontech, Palo Alto, CA, USA), and then subcloned into the Kpn I-Xho I sites of luciferase reporter plasmid pGL2-basic (Promega, Madison, WI, USA). Deletion construct NNMT-329 was generated with restriction enzymes, Sac I/Hind III, and then subcloned into the Sac I-Hind III sites of the pGL2 vector.
A conditionally active form of Stat3 was constructed by fusing the entire coding region of mouse Stat3 to the modified ligand binding domain (G525R) of mouse estrogen receptor, as described previously (Matsuda et al. 1999). This construct was cloned into pCAGGS vector. The dominant-negative Stat3 expression vector (pRc/CMV-ΔStat3B) was derived from pME18S-ΔStat3B (O’Farrell et al. 1998; Tomida et al. 1999).
Luciferase reporter assays
Hep-G2 and 293 cells (105) were inoculated into 24-well plates 1 day before transfection. The cells were transfected with the HGF promoter constructs (0.5 μg/well) together with Stat3 expression vector (0.5 μg/well) and the pRL-TK vector (0.05 μg), using 3 μl of FuGENE 6 reagent (Roche Diagnostic Corporation, Indianapolis, IN, USA). After 24 h, the cells were treated overnight with LIF (10 ng/ml) or IL-6 (25 ng/ml) or 4HT (2 μM). Cell lysates were prepared at 38-h post-transfection. Luciferase activity was determined with a luminometer and a Dual-Luciferase Reporter Assay System (Promega). The activity was normalized as to transfection efficiency using the Renilla luciferase activity of pRL-TK.
Real time RT-PCR
Total RNA was extracted from the cells using guanidium thiocyanate. The RNA (0.5 μg) was reverse-transcribed using random nonamer primers. cDNA samples (Human Colon Matched cDNA Pair Panel) synthesized using total RNA from colon adenocarcinoma and corresponding normal tissues from individual patients were obtained from Clontech. Normalization of Matched Pair Panels was performed using β-actin and 23 kDa highly basic protein as standards. PCRs were performed with a LightCycler-FastStart DNA Master SYBR Green I kit (Roche Diagnostic Corporation) and a Control Kit DNA (Roche) according to the manufacturer’s instruction. The primer sequences were as follows: for NNMT, 5′-GAATCAGGCTTCACCTCCAA-3′ (forward), 5′-TCACACCGTCTAGGCAGAAT-3′ (reverse); for Stat3, 5′-CAGGATGGCCCAATGGAATC-3′ (forward), 5′-CCCAGGAGATTATGAAACACC-3′ (reverse); for Stat1, 5′-GCAGGATGTCTCAGTGGTAC-3′ (forward), 5′-GATCATCCAGCTGTGACAGG-3′ (reverse); for GAPDH, 5′-AGCTGAACGGGAAGCTCACT-3′ (forward), 5′-ATGAGGTCCACCACCCTGTT-3′ (reverse); for IL-6, 5′-AGCCAGAGCTGTGCAGATGA-3′ (forward), 5′-ACCAGAAGAAGGAATGCCCA-3′ (reverse); for LIF, 5′-CAACCTCATGAACCAGATCAG-3′ (forward), 5′-TCTGGAAGACATCCTTACCC-3′ (reverse).
Stat3 siRNA transfection
A double-stranded siRNA oligonucleotide against Stat3 (5′-CAUCUGCCUAGAUCGGCUAdTdT-3′ and 3′-dTdTGUAGACGGAUCUAGCCGAU-5′) was synthesized by Dharmacon Inc., Lafayette, CO, USA (Konnikova et al. 2003; Yuan et al. 2004). Control siRNA (sc-37007) was obtained from Santa Cruz Biotechnology Inc. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used as the transfection reagent according to the manufacturer’s directions with 200 nmol of siRNA per 60 mm dish. siRNA transfected cells were incubated for 48 h.
Western blot analysis
Cells (107) were lysed in 100 μl of lysis buffer (CelLytic™-M Cell Lysis Reagent, Sigma-Aldrich). Supernatants (10 μl) obtained on centrifugation were resolved on SDS-polyacrylamide gel electrophoresis, followed by transfer to Immobilon P (Millipore, Bedford, MA, USA) and then immunoblotting with anti-NNMT antibodies (1:1,000 dilution) and horseradish peroxdase-conjugated rabbit anti-chicken IgY (Chemicon International Inc., Temecula, CA, USA). The immune complex was detected using an enhanced chemiluminescence system (Amersham Biosciences, Little Chalfont, UK).
Tissue array analysis
Human colon cancer tissue array slides were obtained from Chemicon International Inc. and Super Bio Chips, Seoul, Korea. Immunohistochemical staining was performed by the standard method (Bankfalvi et al. 1996; Kobayashi et al. 2004). For phospho-Stat3 (pStat3) staining, slides were autoclaved for 20 min in 10 mM citrate buffer (pH 6.0) for antigen retrieval, and the primary antibody was used at 1:100 dilution. For NNMT staining, the autoclaving was omitted and the primary antibody was used at 1:200 dilution. Detection of the immune complex was performed by the streptavidin-biotin-horseradish peroxidase method. The immune complex was visualized with 3, 3′-diaminobenzidine tetrahydrochloride (DAB) and hydrogen peroxide.
Statistical analysis
The association between pStat3 and NNMT expression in cancer tissues and normal colon tissues was analyzed by the χ 2-test.
Results
Stimulation by LIF of NNMT expression in 293 cells
Although Stat3 in 293 cells was extremely activated by LIF, the proliferation and morphology of the cells were little affected (Tomida and Saito 2004a). Therefore, we expected observation of primary effects of Stat3. After stimulation with LIF for 10 h, the cells were harvested. Total RNA was extracted and subjected to human cDNA array analysis. Of the 16,600 genes examined, the expression of eight was increased or decreased more than twofold by LIF. Among these genes, only suppressor of cytokine signaling 3 (SOCS3) is known as a target of Stat3. The microarray data showed that NNMT was increased 3.3-fold.
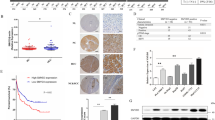
We chose five genes including NNMT as candidate Stat3-regulated genes. The promoter region (1–3 kb upstream of the genes) of each gene was cloned and fused to a luciferase reporter vector. These constructs were transfected transiently into 293 cells. LIF was subsequently added to the 293 culture. The promoter activity of a NNMT construct (NNMT-2568) was stimulated by LIF (Fig. 1). The consensus sequences for STATx binding elements (−1,625, −1,305, and −195), NF-IL6 binding elements (−1,422 and −204), and a hepatocyte nuclear factor-1β (HNF-1β) binding element (−154) are present in the NNMT-2568 construct. Removal of the sequence including distal 2 STATx binding elements and a NF-IL6 binding element to position −329 decreased the LIF responsiveness. This indicates that the sequence between −2,568 and −329 contains important regulatory elements for LIF responsiveness, and suggests that these elements are probably the STATx binding elements and/or the NF-IL6 binding element. The promoter activities of the other 4 genes did not change on the addition of LIF.
Stimulation by LIF of NNMT promoter activity in 293 cells. A schematic representation of NNMT promoter constructs is shown in the upper panel. The consensus binding elements for transcription factors of Stats (STATx), NF-IL6, and HNF-1β are depicted. The 293 cells were transfected with the NNMT promoter constructs. After 24 h, half of the cultures was treated overnight with 10 ng/ml of LIF. Luciferase activity was determined at 38-h post-transfection. Relative luciferase activity was normalized using pRL-TK as an internal control. The values are mean + SE for duplicate transfections. These experiments were repeated twice with similar results
Effect of dominant-negative Stat3 and conditionally active Stat3 on NNMT promoter activity
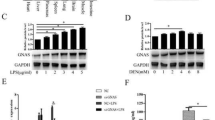
Next, we examined expression of NNMT in several types of cancer cells by real-time quantitative RT-PCR using a LightCycler. Mammary carcinoma MDA-MB-468 cells and colon cancer HT29 cells express constitutively high levels of NNMT. The expression of NNMT in Hep-G2 hepatoma cells and SW480 colon cancer cells was low. However, IL-6, but not LIF, stimulated NNMT expression in both cell lines (Fig. 2). Activation of Stat3 was examined by Western blot analysis and it was found that IL-6, but not LIF, activated Stat3 in these cells (data not shown). It is likely that expression of LIF receptor is too low in Hep-G2 and SW480 cells, but exact reasons for lack of responsiveness to LIF in these cells are presently unknown.
Induction by IL-6 of NNMT mRNA in Hep-G2 and SW480 cells. The cells were stimulated with 30 ng/ml of IL-6 or LIF for 13 h. The mRNA level was determined with a LightCycler. The NNMT mRNA level is expressed in arbitrary units and standardized as to the level of GAPDH mRNA. The values are mean + SE for duplicate samples. Similar results were obtained in two additional experiments
The promoter activity of the NNMT gene in Hep-G2 cells was examined (Fig. 3). The activity of the NNMT-2568 construct was drastically induced by IL-6. However, the truncated NNMT promoter construct was less active (data not shown). This suggests that the distal STATx binding elements and NF-IL6 binding element are important for IL-6 responsiveness. To examine the role of Stat3, a truncated Stat3 (ΔStat3), which lacks the transactivation domain and acts as dominant negative manner was generated (O’Farrell et al. 1998). A conditionally active Stat3 (Stat3ER), a fusion protein composed of Stat3 and the modified ligand binding domain of estrogen receptor, was also constructed (Matsuda et al. 1999). The modified ligand binding domain binds with the synthetic steroid ligand 4-hydroxytamoxifen (4HT) but not with 17β-estradiol. Hep-G2 cells were transfected with the NNMT-2568 promoter construct together with each Stat3 expression vector. Expression of dominant-negative Stat3 abolished the NNMT promoter activity stimulated with IL-6. On the other hand, activation of Stat3ER by 4HT resulted in a significant induction of the promoter activity. These results suggest that Stat3 plays major role in the induction of NNMT mRNA in Hep-G2 cells stimulated with IL-6.
Effect of dominant-negative (dn) Stat3 and conditionally active Stat3 (Stat3ER) on NNMT promoter activity in Hep-G2 cells. The Hep-G2 cells were transfected with the NNMT-2568 promoter construct together with dnStat3 or Stat3ER expression vector or an empty expression vector (mock). After 24 h, the cells were treated for 13 h with IL-6 (25 ng/ml) or 4HT (2 μM). Luciferase activity was determined at 38-h post-transfection. Relative luciferase activity was normalized using pRL-TK as an internal control. The values are mean + SE for duplicate transfections. These experiments were repeated three times with similar results
Suppression by curcumin or Stat3 siRNA of NNMT expression
Recently, it was reported that curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible Stat3 phosphorylation in human multiple myeloma cells (Bharti et al. 2003). We confirmed previously that more than 50 μM curcumin suppressed LIF-inducible Stat3 phosphorylation in 293 cells (Tomida and Saito 2004b). We examined the effect of curcumin on NNMT gene expression. Treatment of MDA-MB-468 cells and HT29 cells with 50 μM curcumin for 14 h caused suppression of NNMT expression in these cells, but did not affect expression of the control GAPDH gene (Fig. 4). Next, the effect of Stat3 siRNA was examined. Treatment of MDA-MB-468 cells with Stat3 siRNA for 2 days decreased the expression of Stat3 and NNMT, but did not affect Stat1 expression (Fig. 5). HT29 cells were not transfected with the siRNA in the presence of various kinds of transfection reagents.
Suppression by curcumin of NNMT mRNA in HT29 and MDA-MB-468 cells. The cells were treated with 50 μM of curcumin for 14 h. The mRNA level was determined with a LightCycler. The NNMT and GAPDH mRNA levels are expressed in arbitrary units. The values are mean + SE for duplicate samples. Similar results were obtained in two additional experiments
Suppression by Stat3 siRNA of NNMT mRNA in MDA-MB-468 cells. The cells were transfected with a Stat3 siRNA or a control siRNA. After incubation for 48 h, the mRNA level was determined with a LightCycler. The NNMT and Stat mRNA levels are expressed in arbitrary units. The values are mean + SE for duplicate samples. Similar results were obtained in two additional experiments
Enhanced expression of NNMT and activated Stat3 in colorectal cancer tissues
We further examined expression of NNMT and activation of Stat3 in colon cancer tissues and normal colon tissues. NNMT mRNA was examined by RT-PCR analysis using matched normal and cancer cDNA pairs. Expression of IL-6 and LIF mRNA was also examined, because these cytokines activate Stat3 and secretion of the cytokines from many cancer cell lines was known. Expression of NNMT in colon adenocarcinoma tissue from all five patients was higher than that in normal tissue (Fig. 6). In three cases, expression of NNMT in cancer tissue was more than twofold compared with the normal level. Metastases were seen in lymph nodes of patients B and D. The other three patients were without metastasis. Level of IL-6 mRNA in cancer tissue from patients B and E was significantly higher than that in normal tissues. LIF mRNA was hardly detected in these samples.
NNMT and IL-6 mRNA expression in colon cancer and normal tissues. mRNA levels are determined with cDNA samples synthesized using total RNA from tumor and corresponding normal tissues from individual patients (A–E). The values are mean + SE for duplicate assays. These experiments were repeated twice with similar results
Next, we examined expression of NNMT and pStat3 by immunohistochemical analysis using tissue array slides. The NNMT antibody used in this study was a chicken polyclonal antibody to human NNMT. To test its specificity for NNMT, Western blot analysis was performed (Fig. 7). Lysates of MDA-MB-468 cells and HT29 cells each gave a clear single band corresponding to the molecular weight of NNMT (29,600 Da), while lysates of non-stimulated 293 cells and MCF-7 cells did not give this specific band. These results are accord with results of RT-PCR analysis of NNMT mRNA. We observed that the enhanced expression of NNMT in cytoplasm of 76 (86%) colon adenocarcinoma tissues among 88 evaluable samples (Fig. 8, Table 1). On the other hand, 16 of 17 normal colon tissues were weakly stained with NNMT antibody by immunohistochemistry. The low-grade staining of NNMT was assigned as negative in Table 1. Of the 88 cancer tissues, 66 (75%) were positive for phosphorylated Stat3 (tyrosine 705) antibody, whereas all normal tissues were almost negative. Of the 66 nuclear pStat3–positive cases, 61 (92%) were also strongly stained with anti-NNMT antibody. These results showed that enhanced expression of NNMT was correlated with activation of Stat3 (P < 0.001, χ 2 = 33.1).
Immunostaining of NNMT and phospho-Stat3 in colon cancer tissues. Colon tissue slides were incubated with anti-phospho-Stat3 or anti-NNMT antibodies. After overnight incubation at 4°C, the slides were incubated with secondary biotinylated antibodies and streptavidin/horseradish peroxidase complex and then visualized with DAB chromogen. Original magnification, ×400. Colon cancer 1, well differentiated adenocarcinoma. Colon cancer 2, poorly differentiated adenocarcinoma
Discussion
We examined the Stat3-regulated gene in 293 cells by cDNA array analysis and found that expression of NNMT was induced by LIF. There have been reports on pituitary gene expression profiles after treatment with LIF (Abbud et al. 2004), IL-6-dependent gene expression profiles in multiple myeloma INA-6 cells (Brocke-Heidrich et al. 2004), and Stat3-C-induced gene expression profiles in NIH3T3 cells (Paz et al. 2004). These reports showed that the expression of many genes was changed after activation of Stat3, but they did not mention NNMT. The cells used previously were induced to proliferate or differentiate through activation of Stat3, whereas the proliferation and morphology of the 293 cells were little affected by Stat3-activation. This is one reason why the degree of the change in gene-expression in our experiment was less than in previous experiments. Recently, Azare et al. (2007) examined the role of Stat3-C in prostate tumorigenesis and showed that NNMT expression was increased 2.64-fold in Stat3-C-expressing cells. This is in accord with our present results.
The NNMT promoter construct fused to the luciferase gene was transiently transfected into 293 and Hep-G2 cells, and it was found that the promoter activity was stimulated by LIF and IL-6, respectively. Deletion analysis suggests that distal STATx binding elements and/or a NF-IL6 binding element are important for the cytokine-responsiveness. We introduced a mutation into each element and then analyzed promoter activity. However, we could not identify a particular element, because each mutation reduced similarly the promoter activity. It is likely that these elements function coordinately. Activation of Stat3 alone caused fourfold induction in NNMT promoter activity (Fig. 3). However, the promoter activity is less than that stimulated with IL-6. These results suggest that NF-IL6, another mediator of IL-6, plays an additional role in NNMT expression. Xu et al. (2005) showed that HNF-1β functions as a transcription activator for NNMT gene expression in some thyroid cancer cells. The HNF-1β binding site is located between nucleotides −148 and −162 relative to the translation initiation codon. They also suggested that the sequence between −2,706 and −343 contains important positive regulatory elements for expression of NNMT in a papillary thyroid cancer cell line. If Stat3 in these cancer cells was constitutively activated, these elements would be the STATx binding element and NF-IL6 binding element.
Proteomics analysis or DNA microarray analysis showed that NNMT is expressed at markedly higher levels in several kinds of cancer, including pancreatic cancer (Iacobuzio-Donahue et al. 2002; Rogers et al. 2006), gastric cancer (Jang et al. 2004; Lim et al. 2006), papillary thyroid carcinoma (Xu et al. 2003, 2005), clear-cell renal carcinoma (Yao et al. 2005), and colorectal cancer (Roessler et al. 2005). We are interested in the relation between the expression of NNMT and activation of Stat3 in these cancers, and thus examined this relation in colon cancers using matched normal and cancer cDNA pair panels and tissue arrays. High levels of IL-6 and NNMT mRNAs were expressed in two colon adenocarcinomas (B and E). Autocrine IL-6 could be responsible for the activation of Stat3 and enhanced expression of NNMT in these carcinomas. In carcinoma D, NNMT mRNA was highly expressed, but IL-6 and LIF mRNAs were not. Although the true reason for this enhanced expression of NNMT is unknown, it is possible that Stat3 in the carcinoma was activated through a mechanism other than stimulation by IL-6 or LIF.
Recently, activation of Stat3 in colon carcinomas was analyzed by means of an immunohistochemical method (Ma et al. 2004; Kusaba et al. 2005; Lin et al. 2005; Kawada et al. 2006). These works showed that 58–73% carcinomas were stained with phosphor-Stat3 antibodies. We confirmed that 75% carcinomas were stained with the antibodies and that normal colon nuclei were hardly stained. We observed that the cytoplasm of 86% colon adenocarcinoma tissues was markedly stained with NNMT antibodies and most normal colon tissues were weakly stained. Of the 66 nuclear pStat3–positive cancer tissues, 61 (92%) were also strongly stained with anti-NNMT antibodies. These results showed that enhanced expression of NNMT was correlated with activation of Stat3 (P < 0.001). Because constitutive activation of Stat3 has been detected in a wide variety of primary human tumors (Turkson and Jove 2000; Yu and Jove 2004), the present work suggests that expression of NNMT is enhanced in various kinds of cancer tissue other than colon cancer tissues, and that NNMT may be a new marker for clinical diagnosis of these cancers. However, inflammation is also correlated with activation of Stat3. It is necessary to examine whether enhanced expression of NNMT is specific for cancer tissues. NNMT is not a secreted protein and usually localized in the cytoplasm of cells. Further studies are needed to evaluate whether NNMT is useful for serologic cancer biomarkers.
References
Abbud RA, Kelleher R, Melmed S (2004) Cell-specific pituitary gene expression profiles after treatment with leukemia inhibitory factor reveal novel modulators for proopiomelanocortin expression. Endocrinology 145:867–880
Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T (1994) Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77:63–71
Aksoy S, Brandriff BF, Ward A, Little PF, Weinshilboum RM (1995) Human nicotinamide N-methyltransferase gene: molecular cloning, structural characterization and chromosomal localization. Genomics 29:555–561
Aksoy S, Szumlanski CL, Weinshilboum RM (1994) Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J Biol Chem 269:14835–14840
Azare J, Leslie K, Al-Ahmadie H, Gerald W, Weinreb PH, Violette SM, Bromberg J (2007) Constitutively activated Stat3 induces tumorigenesis and enhances cell motility of prostate epithelial cells through integrin β6. Mol Cell Biol 27:4444–4453
Bankfalvi AA, Piffko J, Ofner D, Dreier R, Bocker W, Werner K (1996) Significance of wet autoclave pretreatment in immunohistochemistry. Pathol Oncol Res 2:71–77
Bharti AC, Donato N, Aggarwal BB (2003) Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol 171:3863–3871
Brocke-Heidrich K, Kretzschmar AK, Pfeifer G, Henze C, Loffler D, Koczan D, Thiesen HJ, Burger R, Gramatzki M, Horn F (2004) Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family independent survival pathway closely associated with Stat3 activation. Blood 103:242–251
Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr (1999) Stat3 as an oncogene. Cell 98:295–303
Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal K, Beug H, Ohlschlager P, Schutz A, Halbhuber KJ, Friedrich K (2005) Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia 7:545–555
DeArmond D, Brattain MG, Jessup JM, Kreisberg J, Malik S, Zhao S, Freeman JW (2003) Autocrine-mediated ErbB-2 kinase activation of STAT3 is required for growth factor independence of pancreatic cancer cell lines. Oncogene 22:7781–7795
Fernandes A, Hamburger AW, Gerwin BI (1999) ErbB-2 kinase is required for constitutive stat 3 activation in malignant human lung epithelial cells. Int J Cancer 83:564–570
Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH (2002) Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol 160:1239–1249
Jang JS, Cho HY, Lee YJ, Ha WS, Kim HW (2004) The differential proteome profile of stomach cancer: identification of the biomarker candidates. Oncol Res 14:491–499
Jiang WG, Martin TA, Parr C, Davies G, Matsumoto K, Nakamura T (2005) Hepatocyte growth factor, its receptor, and their potential value in cancer therapies. Crit Rev Oncol Hematol 53:35–69
Kanda N, Seno H, Konda Y, Marusawa H, Kanai M, Nakajima T, Kawashima T, Nanakin A, Sawabu T, Uenoyama Y, Sekikawa A, Kawada M, Suzuki K, Kayahara T, Fukui H, Sawada M, Chiba T (2004) STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene 23:4921–4929
Kawada M, Seno H, Uenoyama Y, Sawabu T, Kanda N, Fukui H, Shimahara Y, Chiba T (2006) Signal transducers and activators of transcription 3 activation is involved in nuclear accumulation of β-catenin in colorectal cancer. Cancer Res 66:2913–2917
Kobayashi Y, Tokuchi Y, Hashimoto T, Hayashi M, Nishimura H, Ishikawa Y, Nakagawa K, Sato Y, Takahashi A, Tsuchiya E (2004) Molecular markers for reinforcement of histological subclassification of neuroendocrine lung tumors. Cancer Sci 95:334–341
Konnikova L, Kotecki M, Kruger MM, Cochran BH (2003) Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer 3:23–31
Kusaba T, Nakayama T, Yamazumi K, Yakata Y, Yoshizaki A, Nagayasu T, Sekine I (2005) Expression of p-STAT3 in human colorectal adenocarcinoma and adenoma; correlation with clinicopathological factors. J Clin Pathol 58:833–838
Lim BH, Cho BI, Kim YN, Kim JW, Park ST, Lee CW (2006) Overexpression of nicotinamide N-methyltransferase in gastric cancer tissues and its potential post-translational modification. Exp Mol Med 38:455–465
Lin Q, Lai R, Chirieac LR, Li C, Thomazy VA, Grammatikakis I, Rassidakis GZ, Zhang W, Fujio Y, Kunisada K, Hamilton SR, Amin HM (2005) Constitutive activation of JAK3/STAT3 in colon carcinoma tumors and cell lines: inhibition of JAK3/STAT3 signaling induces apoptosis and cell cycle arrest of colon carcinoma cells. Am J Pathol 167:969–980
Lowe DG, Nunes W, Bombara M, McCabe S, Rangea GE, Henzel W, Tomida M, Yamamoto-Yamaguchi Y, Hozumi M, Goeddel DV (1989) Genomic cloning and heterologous expression of human differentiation-stimulating factor. DNA 8:351–359
Ma XT, Wang S, Ye YJ, Du RY, Cui ZR, Somsouk M (2004) Constitutive activation of Stat3 signaling pathway in human colorectal carcinoma. World J Gastroenterol 10:1569–1573
Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T (1999) STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J 18:4261–4269
O’Farrell AM, Liu Y, Moore KW, Mui AL (1998) IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J 17:1006–1018
Paz K, Socci ND, van Nimwegen E, Viale A, Darnell JE (2004) Transformation fingerprint: induced STAT3-C, v-Src and Ha-Ras cause small initial changes but similar established profiles in mRNA. Oncogene 23:8455–8463
Roessler M, Rollinger W, Palme S, Hagmann ML, Berndt P, Engel AM, Schneidinger B, Pfeffer M, Andres H, Karl J, Bodenmüller H, Rüschoff J, Henkel T, Rohr G, Rossol S, Rösch W, Langen H, Zolg W, Tacke M (2005) Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res 11:6550–6557
Rogers CD, Fukushima N, Sato N, Shi C, Prasad N, Hustinx SR, Matsubayashi H, Canto M, Eshleman JR, Hruban RH, Goggins M (2006) Differentiating pancreatic lesions by microarray and QPCR analysis of pancreatic juice RNAs. Cancer Biol Ther 5:1383–1389
Saito T, Tomida M (2005) Generation of inhibitory DNA aptamers against human hepatocyte growth factor. DNA Cell Biol 24:624–633
Song L, Turkson J, Karras JG, Jove R, Haura EB (2003) Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene 22:4150–4165
Tomida M, Heike T, Yokota T (1999) Cytoplasmic domains of the leukemia inhibitory factor receptor required for STAT3 activation, differentiation, and growth arrest of myeloid leukemic cells. Blood 93:1934–1941
Tomida M, Saito T (2004a) The human hepatocyte growth factor (HGF) gene is transcriptionally activated by leukemia inhibitory factor through the Stat binding element. Oncogene 23:679–686
Tomida M, Saito T (2004b) Stat3 activity stimulates HGF expression. In: Fenton MJ (ed) 5th Joint meeting of the international cytokine society and the international society for interferon and cytokine research. MEDIMOND, Bologna, pp 113–116
Turkson J, Jove R (2000) STAT proteins: novel molecular targets for cancer drug discovery. Oncogene 19:6613–6626
Watson CJ, Miller WR (1995) Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br J Cancer 71:840–844
Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL, Xie K (2003) Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene 22:319–329
Xu J, Capezzone M, Xu X, Hershman JM (2005) Activation of nicotinamide N-methyltransferase gene promoter by hepatocyte nuclear factor-1β in human papillary thyroid cancer cells. Mol Endocrinol 19:527–539
Xu J, Moatamed F, Caldwell JS, Walker JR, Kraiem Z, Taki K, Brent GA, Hershman JM (2003) Enhanced expression of nicotinamide N-methyltransferase in human papillary thyroid carcinoma cells. J Clin Endocrinol Metab 88:4990–4996
Yan L, Otterness DM, Weinshilboum RM (1999) Human nicotinamide N-methyltransferase pharmacogenetics: gene sequence analysis and promoter characterization. Pharmacogenetics 9:307–316
Yao M, Tabuchi H, Nagashima Y, Baba M, Nakaigawa N, Ishiguro H, Hamada K, Inayama Y, Kishida T, Hattori K, Yamada-Okabe H, Kubota Y (2005) Gene expression analysis of renal carcinoma: adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. J Pathol 205:377–387
Yeh HH, Lai WW, Chen HH, Liu HS, Su WC (2006) Autocrine IL-6-induced Stat3 activation contributes to the pathogenesis of lung adenocarcinoma and malignant pleural effusion. Oncogene 25:4300–4309
Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R (1995) Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 269:81–83
Yu H, Jove R (2004) The STATs of cancer–new molecular targets come of age. Nat Rev Cancer 4:97–105
Yuan ZL, Guan YJ, Wang L, Wei W, Kane AB, Chin YE (2004) Central role of the threonine residue within the p+1 loop of receptor tyrosine kinase in STAT3 constitutive phosphorylation in metastatic cancer cells. Mol Cell Biol 24:9390–9400
Zhong Z, Wen Z, Darnell JE Jr (1994) Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264:95–98
Acknowledgments
This work was supported in part by the Project, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, JST. We thank A. Kodaka and T. Tanaka for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tomida, M., Ohtake, H., Yokota, T. et al. Stat3 up-regulates expression of nicotinamide N-methyltransferase in human cancer cells. J Cancer Res Clin Oncol 134, 551–559 (2008). https://doi.org/10.1007/s00432-007-0318-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-007-0318-6