Abstract
Purpose: Epstein-Barr virus (EBV) is associated with the development of several lymphoid and epithelial malignancies, including Burkitt’s lymphoma. The EBV latent protein, EBV Nuclear Antigen 1 (EBNA1), is detectable in almost all types of EBV-associated tumors and is essential for replication and maintenance of the latent episome of EBV. We here examined whether the RNA interference (RNAi) technique could be employed to suppress expression of EBNA1 in EBV-positive Burkitt’s lymphoma cells. Methods: A Raji cell line expressing small hairpin RNAs (shRNAs) against EBNA1 was established and EBNA1 mRNA level was determined by real-time RT-PCR analysis. We investigated the effects of EBNA1 silence on lymphoma cell growth and cell cycle progression. Results: Transfection of an EBNA1 RNAi plasmid resulted in substantial loss of EBNA1 mRNA and significantly inhibited proliferation of Raji cells relative to the control plasmid case. Suppression of EBNA1 was also associated with downregulation of EBV oncogene EBNA2, a decreased PCNA labeling index and increased G0/G1 fraction in cell cycle analysis. Conclusions: These findings point to potential therapeutic applications for vector-mediated siRNA delivery to control EBV-associated malignant disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Epstein-Barr virus (EBV) is a ubiquitous human herpesvirus implicated in the pathogenesis of several lymphoid and epithelial malignancies, including Burkitt’s lymphoma and nasopharyngeal carcinoma, as well as Hodgkin’s disease, lymphomas occurring in immune-deficient patients and gastric carcinomas (Young and Murray 2003). Infection with EBV in vitro easily transforms resting B cells from human peripheral blood to permanently growing lymphoblastoid cell lines (LCLs) (Sugimoto et al. 2004; Middeldorp et al. 2003). Most EBV diseases are associated with EBV latency, during which the viral gene expression is limited to the six members of the EBNA family of nuclear proteins, two membrane proteins, and two small polymerase III transcripts (Kieff 1996). EBV can establish three types of latency (I, II, and III), each of which is characterized by differential expression of a group of latency proteins.
During latency, the EBV genome exists predominantly as a multicopy circular episome that is replicated once per cell cycle and partitioned at mitosis (Adams 1987; Yates and Guan 1991). Dimers of EBNA1 bind with high affinity to two elements within the origin of plasmid replication (OriP), namely, the family of repeats (FR) and the dyad symmetry (DS) (Yates et al. 1984, 1985; Reisman et al. 1985; Chen et al. 1993). At the same time EBNA1 associates with host-cell chromosomes through its chromosome-binding domains (Marechal et al. 1999). The exact mechanisms by which EBNA1 facilitates replication and maintains viral episome are incompletely understood but possible functions include the recruitment of cellular replication machinery during S phase (Schepers et al. 2001; Zhang et al. 1998) and segregation of episomes to daughter cells during mitosis (Hung et al. 2001).
EBNA1 furthermore participates in regulation of latency gene transcription. Binding of EBNA1 to a site downstream of the BamHI Q promoter (Qp) represses Qp activity, but this repression can be overcome by E2F that displaces EBNA1 (Sung et al. 1994). EBNA1 has also been shown to upregulate two key EBV latency promoters, the BamHI C promoter (Cp) (Sugden and Warren 1989) and the LMP1 promotor (Gahn and Sugden 1995). Although EBNA1 is exogenous to the host, EBV-infected cells expressing EBNA1 are barely recognized by cytotoxic T lymphocytes (CTLs) and the Gly-Ala repeat domain of EBNA1 has been shown to prevent the presentation of CTL epitopes via the Major Histocompatibility Complex (MHC) class I pathway by blocking ubiquitin/proteasome-dependent processing (Levitskaya et al. 1995). This evasion mechanism has been implicated in the pathogenesis of EBV-associated diseases.
RNA interference is a new powerful tool for selective suppression of gene expression at the post-transcriptional level (Dykxhoorn et al. 2003). Small interfering RNAs (siRNAs) are short, double stranded RNAs (dsRNAs) that target mRNA with complementary sequences for endonucleolytic degradation. The Dicer family of RNase III enzymes cleaves long dsRNA to produce siRNAs, which become incorporated into a multiprotein RNA-inducing silencing complex (RISC). Introduction of chemically synthesized 21-nucleotide siRNAs is sufficient to initiate RNA interference in mammalian cells (Elbashir et al. 2001) and DNA vectors expressing small hairpin RNAs (shRNAs) using RNA polymerase III (RNA pol III) promoters have been developed for effective and persistent gene silencing (Brummelkamp et al. 2002; Tomar et al. 2003). This approach is finding increasing employment for analysis of gene function and gene therapy (Hall and Alexander 2003; Butz et al. 2003; Li et al. 2004).
We here aimed to suppress EBNA1 expression in Raji Burkitt’s lymphoma cells using the RNA interference strategy. A plasmid expressing shRNAs against EBNA1 was therefore constructed and transfected to Raji cells. Effective suppression at the mRNA level could be confirmed by real-time RT-PCR analysis post transient transfection. One month after antibiotics selection, the EBNA1 shRNA expressing Raji cells displayed a significantly lower cell growth as compared with control cells. Furthermore, we observed decreased expression of the EBV oncogenic protein EBV Nuclear Antigen 2 (EBNA2) as well as a lower labeling index for proliferating cell nuclear antigen (PCNA). Finally, we provided evidence that the growth inhibitive effect is associated with elevated frequency of G0–G1 phase cells but not increased apoptosis. Our findings support the possibility that RNA interference could be an efficacious novel therapy for EBV-related diseases.
Materials and methods
Cell culture
Raji is an EBV-positive human Burkitt’s lymphoma cell line whose cells contain 50–60 copies of EBV genome per cell (Adams 1987; Polack et al. 1984; Hatfull et al. 1988). Raji cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin, in a humidified atmosphere of 5% CO2 at 37°C.
Plasmid construction
The sequence of EBNA1 hairpin was designed by iGENE Therapeutics (Ibaraki, Japan). The shRNA-encoding DNA template comprised the 19-base sense sequence followed by the hairpin loop and antisense sequences and five consecutive Ts as an RNA pol III terminator sequence. This template was subcloned downstream of the human U6 promotor in the RNAi-Ready pSIREN-RetroQ vector (BD Biosciences Clontech), between the BamH I and EcoR I restriction sites. A control vector was generated by utilizing the Negative Control siRNA Annealed Oligonucleotide supplied with the vector.
Transfection
Transfection was performed with NucleofectorTM Technology (Amaxa), with 2×106 logarithmically growing cells nucleofected using a Cell Line NucleofectorTM Kit T, program O-17 and 2 μg of plasmid. Raji cells transfected with EBNA1 RNAi or control plasmid were plated in 100-mm dishes and 24–48 hours post transfection, puromycin (BD Biosciences Clontech) was added at a concentration of 5 μg/ml. The cells were maintained under these conditions with medium change twice a week.
RNA extraction and reverse transcription (RT)
Total cellular RNA was isolated using an RNeasy Midi Kit (QIAGEN) as recommended by the supplier. Samples were digested with an RNase-Free DNase Set (QIAGEN) during purification to remove residual DNA. Twenty micro litres of cDNA was synthesized using 200 ng of total RNA as the template and random hexamers as primers, as described in the manufacturer’s protocol for Taqman Reverse Transcription Reagents (Applied Biosystems).
Real-time PCR
PCR was carried out using 1×of TaqMan Universal PCR Master Mix (Applied Biosystems), 900 nM of each primer, 250 nM of TaqMan probe and cDNA template in a 25 μl reaction mixture. The PCR primers used for the quantitation of EBNA1 mRNA were 5′-CGCATCATAGACCGCCAGTA-3′ (forward) and 5′-CTGGCCCCTCGTCAGACAT-3′ (reverse). A TaqMan probe (5′-[FAM]-CCGCGGCCGTCTCCTTTAAGTGTG-[TAMRA]-3′) located between the PCR primers was synthesized by Nippon EGT (Toyama, Japan). The Pre-Developed TaqMan Assay Reagent for GAPDH (20×primers and probe mixture) was obtained from Applied Biosystems. The reaction conditions were as follows: 50°C for 2 min (activation of the AmpErase uracil N-glycosylase [UNG]) and then 95°C for 10 min (activation of the AmpliTaq Gold DNA polymerase), followed by 50 cycles of 15 s at 95°C and 1 min at 60°C. All reactions were performed in duplicate. A standard curve for threshold cycle (CT) values was run in parallel using serial twofold dilutions of 50 ng cDNA of untreated Raji cells. Amplification data were collected using an ABI PRISM 7700 Sequence Detector system (Applied Biosystems) and analyzed using Sequence Detector software version 1.7 (Applied Biosystems).
Western blot analysis
Total cellular protein was prepared as described in previous report (Davenport and Pagano 1999). Briefly, cells were washed once with 1×phosphate-buffered saline (PBS) and then resuspended in lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 5 mM dithiothreitol [DTT], 10 mM NaF, protease inhibitor cocktail [Sigma]). After freezing and thawing three times, debris was spun down at 4°C for 30 min and the protein concentration was determined by the Bradford protein assay. Twenty microgram aliquots of lysates were separated on 8% sodium dodecyl sulfate—polyacrylamide gels (SDS-PAGE) and proteins were transferred to Immobilon-P (Millipore). Expression of EBNA1 and EBNA2 proteins was examined using mouse monoclonal antibodies to EBNA1 (Advanced Biotechnologies) and PE2 (Dako), respectively. Signals were detected with either anti-mouse or anti-rabbit immunoglobulin (Ig)—horseradish peroxidase (Daco) and Western Lightening Plus Chemiluminescence Reagent (PerkinElmer Life Sciences).
Immunocytochemistry
Immunocytochemical analysis was performed on 4 μm thick sections obtained from formalin-fixed and paraffin-embedded fibrin-clot preparations as described previously (Kumada et al. 2004). After antigen-retrieval using microwave heating, sections were inactivated for endogenous peroxidase in 3% H2O2 and then blocked with 5% BSA. Subsequently, slides were stained with mouse monoclonal antibodies against Ki-67 (Clone MIB-1, 1:50 dilution, Dako) or PCNA (Clone PC10, 1:100 dilution, Dako) followed by treatment with EnVision+, Peroxidase, Mouse (Dako). Finally, Peroxidase activity was determined with diaminobenzidine (DAB) solution and counterstaining was achieved with hematoxylin.
Cell proliferation and apoptosis assays
For growth curve analysis, stably transfected Raji cells cultured to mid-log phase were diluted in standard growth medium with 5 μg/ml puromycin to 0.25×105/ml and seeded at 2 ml/well in 6-well plates. Cell numbers of control or EBNA1 RNAi plasmid-transfected cells were determined on four consecutive days by trypan blue exclusion. The cell cycle distribution and numbers of apoptotic cells were determined using a BrdU Flow Kit (BD Biosciences) according to the manufacturer’s protocol. Logarithmically growing Raji cells were pulsed with 10 μM bromodeoxyuridine (BrdU) for 30 min and stained with FITC-conjugated anti-BrdU antibody in combination with a DNA-specific dye, 7-amino-actinomycin D (7-AAD). Two-color analyses were then conducted on a flow cytometer FACSCaliber (Becton Dickinson).
Results
Choice of RNAi target for EBNA1 silencing in Raji cells
Suppression of gene expression by RNA interference is highly target sequence-specific so that only one base pair mutation will greatly disturb the inhibition efficiency (Elbashir et al. 2001). The size of EBNA1 protein varies greatly among latently infected cell lines due to the different lengths of the Gly-Ala repeat (Heller et al. 1982; Grasser et al. 1994). Besides, although EBNA1 does not display any obvious polymorphism between the two major types of EBV, types 1 and 2, a degree of interstrain sequence variation, especially in the region between residues 466–527, has been reported from the analysis of EBV-associated tumors and samples from asymptomatic carriers (Habeshaw et al. 1999).
In order to block EBNA1 expression selectively in Raji cells by RNA interference, four potential 19-nt sequences within the EBNA1 open reading frame (ORF) were chosen as siRNA targets. Blast searching against the human genome was performed to ensure that no host cell gene was targeted. The corresponding oligonucleotides were then introduced into the pSIREN-RetroQ vector, which contains a puromycin resistance gene for selection of stable transfectants. Treatment with one plasmid targeting 5′-GTTCCAACCCGAAATTTGA-3′ (+1418 to +1436 of the EBNA1 ORF) reproducibly suppressed EBNA1 expression with high efficiency and was used in all subsequent experiments. The predicted secondary structure of the shRNA against EBNA1 is indicated in Fig. 1a.
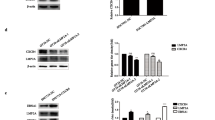
Designs of the EBNA1 shRNA template and real-time PCR Primers. a The predicted secondary structure of the shRNA against EBNA1. Since hairpin sequences are difficult to sequence, three nucleotides in the target sense sequence were substituted to facilitate insert verification. b Model for transcription and splicing of EBNA1 mRNAs. The top bar illustrates the linear form of the EBV genome and the positions of four different promoters (bent arrows). The lower panel shows three classes of EBNA1 transcripts and their corresponding infection types. Coding regions are indicated by boxes and excised intron sequences by thin lines. c Partial cDNA sequence of the 5′ end of the EBNA1 mRNA (GenBank accession number M13941). The sequences of the PCR primers and TaqMan probe used for EBNA1 quantitation are underlined. The splice site between exons U and K is indicated with an arrowhead. The start codon of EBNA1 ORF is shown translated. d Specificity and efficiency of PCR primers designed for real-time PCR analysis. GAPDH is amplified as an internal standard. Negative water blanks were included in each analysis
Quantitation of EBNA1 transcripts by real-time RT-PCR
Previous reports indicated that EBNA1 protein has a long half-life in excess of 36–48 h, which is in agreement with the finding that the Gly-Ala repeat inhibits processing by the ubiquitin/proteasome pathway (Davenport and Pagano 1999; Levitskaya et al. 1997). In order to investigate the effect of candidate RNAi sequences on EBNA1 transcription, the mRNA level was analyzed post transient transfection by performing real-time RT-PCR assays. Since TaqMan RT-PCR analysis with a low-abundance target is sensitive to very small amounts of DNA, we designed primers that anneal at intron splice junctions so that genomic DNA would not be amplified.
Although EBNA1 expression is common to the three types of latency, the promoter used for its expression differs (Fig. 1b). The open reading frame for EBNA1 is located in BamHI K exon at the 3′ end of the message and is preceded by a long 5′ untranslated region (5′ UTR) derived from several short exons (Fig. 1b). The U leader exon is common to all four transcripts and spliced directly to the K exon (Isaksson et al. 2003; Sample et al. 1986, 1991). Raji cells exhibit the type III latency program in which long primary transcripts, containing coding sequences for EBNA1-6, are alternatively spliced into bi- or monocistronic transcripts (Davenport and Pagano 1999).
For amplification of the EBNA1 transcript, primers flanking the U/K intron splice junctions were utilized (Fig. 1c). Figure 1d shows that EBNA1 primers amplified an 88 bp fragment corresponding to the EBNA1 coding region, nucleotides −69 to +19. No DNA contamination was detected following control RT-PCR experiments in which no reverse transcriptase was added to the RT step.
RNAi selectively reduces expression of the EBNA1 viral gene
To determine efficient siRNA sequence for EBNA1 targeting, Raji cells were transiently transfected with siRNA-generating plasmids and selected for successfully transfected cells by puromycin resistance (de la Luna and Ortin 1992). After 2 days of exposure to puromycin, total cellular RNA was prepared and EBNA1 transcripts were assessed by real-time RT-PCR. We observed marked reduction in the level of EBNA1 mRNA in cells transfected with one RNAi vector compared with that in control cells, when the levels were normalized to GAPDH expression (Fig. 2a). In two separate experiments the mRNA levels were consistently diminished by no less than 50% relative to the control (51.3 and 75.0%).
Suppression of EBNA1 expression in Raji cells by RNA interference. a Quantitation of EBNA1 mRNA by real-time RT-PCR analysis. Values represent the mean ± standard deviation (SD) of two separate experiments performed in duplicate. **P<0.01 versus the control cells. Significance was determined using the Student’s t-test. b Cells stably transfected with EBNA1 RNAi or control vector were extracted and analyzed by Western blotting for EBNA1 and actin as a loading control. c Relative amounts of EBNA1 protein in Fig. 2b as determined by densitometry. Data are mean ± SD from three separate experiments. **P<0.01 versus the control cells. Significance was determined by using Student’s t-test
After 1 month’s puromycin selection, two Raji cell populations, stably transfected with either EBNA1 RNAi or control plasmid, were obtained. To test the effects of siRNA delivery on expression of the EBNA1 protein, we separated equal amounts of protein by SDS-PAGE and hybridized the blots with antibodies to EBNA1 and β-actin, respectively. As shown in Fig. 2b and c, Western blots confirmed partial (about 60%) decrease in EBNA1 expression in the cells stably transfected with the RNAi plasmid. Importantly, β-actin protein levels appeared unaffected, suggesting that RNAi against EBNA1 did not activate global suppression of protein expression.
RNA interference against EBNA1 inhibits proliferation of Raji cells
Having demonstrated the selective silencing of the viral gene, we next studied the cellular consequences of EBNA1 depletion. In cell growth curve assays, as illustrated in Fig. 3a, the EBNA1 shRNA-expressing Raji cells displayed a significantly lower growth rate as compared with control cells.
We also observed substantial reduction in the level of EBNA2 protein in Raji cells transfected with EBNA1 RNAi vector relative to the control cells (Fig. 3b). EBNA2 transcriptionally activates certain cellular proto-oncogenes as well as viral oncogenes and is involved in G0–G1 transition (Middeldorp et al. 2003). This finding suggested the possibility that suppression of EBNA1 might attenuate proliferation through down-regulating the expression of other EBV transforming genes.
Effects of EBNA1 RNAi on proliferation of Raji cells. a Cell growth curve assays. EBNA1-targeted RNA interference significantly inhibited the proliferation of Raji cells over four days (time×treatment interaction: P=0.0001). Data are mean ± SD from triplicate assays and are representative of two independent experiments. Significance was determined using repeated ANOVA (analysis of variance). b Downregulation of EBNA2 protein. Cells stably transfected with EBNA1 RNAi or control vector were extracted and analyzed by Western blotting for EBNA2 and α-Tubulin as a loading control. c Ki-67 immunoreactivity and PCNA labeling index determined by immunocytochemistry. Raji cells in three random fields of each slide were counted and the percentage of PCNA-positive cells was calculated and expressed as the mean ± SD. ***P<0.001. Significance was determined using the Student’s t-test
The inhibitory effect on cell growth was further corroborated by immunostaining studies of proliferative antigens. PCNA is an auxiliary protein of DNA polymerase δ, whose synthesis reaches a peak during the S phase of the cell cycle (Mathews et al. 1984). Immunocytochemistry revealed a marked decrease of PCNA-positive Raji cells stably transfected with EBNA1 RNAi vector as compared with those transfected with the control vector (Fig. 3c). In contrast, no apparent changes were detected in the immunoreactivity of Ki-67, which is known to be preferentially expressed during all active phases of cell cycle (G1, S, G2 and M phases) but absent in resting cells (G0 phase) (Gerdes et al. 1984).
To further elucidate whether the inhibited cell growth is associated with cell cycle arrest, log-phase Raji cells were analyzed using a BrdU Flow Kit. Compared with the control cells, the frequency of G0–G1 phase cells significantly increased in EBNA1 RNAi vector-transfected cells, while there was a marked reduction of the frequencies of S and G2/M phase cells (Fig. 4). Monitoring cell death showed that inhibition of cell proliferation by EBNA1 RNAi was not due to increased apoptosis under standard growth conditions (Fig. 4).
Suppression of EBNA1 expression in Raji cells results in increased frequency of G0–G1 phase cells. FACS analysis of the cell cycle distribution using a BrdU Flow Kit. Data are mean ± SD from three separate experiments. **P<0.01 versus the control cells. Significance was determined using the Student’s t-test
Discussion
Of the EBV-encoded products, EBNA1 is the only latent protein that is found to be consistently expressed in EBV-associated tumor cells. EBNA1 ensures episomal replication and genome maintenance to establish EBV latent infection efficiently. Furthermore, EBNA1 transactivates the enhancers of a number of other viral genes and is implicated in the evasion from host immune response. A previous report has also suggested that EBNA1 itself may have tumorigenic potential (Wilson et al. 1996). The present study demonstrated that the shRNA approach is effective for suppression of EBNA1 expression in Raji cells and that this is associated with decreased cell growth and increased frequency of G0–G1 phase cells.
A variety of different EBV-based therapies are currently being developed for the treatment of EBV-positive cancers, including prevention of viral oncogene expression, causing loss of the EBV episome, targeted induction of the lytic form of EBV infection, and enhancing host immune responses to virally encoded antigens (Israel and Kenney 2003). Several antisense-based strategies have previously been used in order to target EBV transcription (Roth et al. 1994; Mattia et al. 1997; Kenney et al. 1998) and studies have already shown that antisense oligodeoxynucleotides (Roth et al. 1994) or adenovirus-delivered ribozymes (Huang et al. 1997) directed to EBNA1 can suppress EBNA1 expression and inhibit proliferation of EBV-immortalized B lymphocytes. Anti-sense RNA, ribozymes and RNA interference all operate at the post-transcriptional level to suppress gene expression. However, the process of RNA interference is several orders of magnitude more efficient than anti-sense or ribozyme strategies (Jiang and Milner 2002). We therefore sought to down-regulate the EBNA1 gene by devising a vector-borne siRNA that, by binding to a target sequence on mRNA, would lead to the degradation of EBNA1 transcripts via endonucleolytic cleavage. We here screened several potential RNAi sequences directed against EBNA1 and established that one sequence significantly decreases the intracellular concentration of EBNA1 mRNA, and effectively inhibits cell proliferation of EBV-positive Raji Burkitt’s lymphoma cells.
However, the molecular mechanisms underlying inhibition of proliferation due to EBNA1 silencing remain obscure. Based on previous reports (Sugden and Warren 1989; Wilson et al. 1996; Huang et al. 1997), we propose that RNAi-mediated EBNA1 suppression inhibits cell growth through three potential mechanisms. First, given the indispensable role of EBNA1 in the replication of latent viral genome, it is possible that the viral episome might be eliminated over a number of cell divisions in the absence of EBNA1. Thus, suppression of EBNA1 expression would be expected to disrupt the latent infection and thereby reverse the transformed phenotype. This hypothesis is supported by a previous observation that suppression of EBNA1 by an adenovirus-delivered specific ribozyme reduced the number of EBV genomes in EBV-immortalized B lymphocytes (Huang et al. 1997). Second, suppression of EBNA1 may retard cell proliferation by down-regulating the expression of other viral genes directly involved in growth transformation, such as EBNA2. As inhibition of EBNA1 expression was shown to correlate with loss of EBV episomes (Huang et al. 1997), the lower level of EBNA2 detected in Western blotting may reflect the presence of cells that have reduced number of EBV plasmids. Reduction of EBNA2 protein level might also be explained by impaired transcription activation due to a possible role for EBNA1 as an enhancer protein (Sugden and Warren 1989). Finally, EBNA1 is the only viral protein expressed in the Burkitt’s lymphoma cells in vivo and in early passage in vitro, raising the possibility that EBNA1 may be directly linked to the oncogenic transformation of these cells.
For the first time, we here selectively silenced EBV latent gene EBNA1 in human Burkitt’s lymphoma cells by exploiting RNAi technology. Our results indicate that targeting of EBNA1 by RNA interference represents a promising novel tool for fundamental research into the pathogenesis of EBV-induced diseases, and for the development of specific treatment strategies against EBV-positive cancers. Two key challenges in developing RNAi as a therapy are avoiding off-target effects and ensuring efficient delivery (Hannon and Rossi 2004). Transfection of cells with double-stranded (ds)RNAs can activate innate immune pathways, resulting in sequence-independent destruction of RNAs. In an in vivo study, hydrodynamically injected siRNAs targeting Fas protected mice from fulminant hepatitis (Randall et al. 2003). Despite the considerable obstacles to overcome, we anticipate the development of therapeutics based on RNA interference in the not-too-distant future.
References
Adams A (1987) Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol 61:1743–1746
Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553
Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F (2003) siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene 22:5938–5945
Chen MR, Middeldorp JM, Hayward SD (1993) Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J Virol 67:4875–4885
Davenport MG, Pagano JS (1999) Expression of EBNA-1 mRNA is regulated by cell cycle during Epstein-Barr virus type I latency. J Virol 73:3154–3161
de la Luna S, Ortin J (1992) pac gene as efficient dominant marker and reporter gene in mammalian cells. Methods Enzymol 216:376–385
Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4:457–467
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498
Gahn TA, Sugden B (1995) An EBNA-1-dependent enhancer acts from a distance of ten kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol 69:2633–2636
Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H (1984) Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 133:1710–1715
Grasser FA, Murray PG, Kremmer E, Klein K, Remberger K, Feiden W, Reynolds G, Niedobitek G, Young LS, Mueller-Lantzsch N (1994) Monoclonal antibodies directed against the Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin’s disease. Blood 84:3792–3798
Habeshaw G, Yao QY, Bell AI, Morton D, Rickinson AB (1999) Epstein-barr virus nuclear antigen 1 sequences in endemic and sporadic Burkitt’s lymphoma reflect virus strains prevalent in different geographic areas. J Virol 73:965–975
Hall AH, Alexander KA (2003) RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol 77:6066–6069
Hannon GJ, Rossi JJ (2004) Unlocking the potential of the human genome with RNA interference. Nature 431:371–378
Hatfull G, Bankier AT, Barrell BG, Farrell PJ (1988) Sequence analysis of Raji Epstein-Barr virus DNA. Virology 164:334–340
Heller M, van Santen V, Kieff E (1982) Simple repeat sequence in Epstein-Barr virus DNA is transcribed in latent and productive infections. J Virol 44:311–320
Huang S, Stupack D, Mathias P, Wang Y, Nemerow G (1997) Growth arrest of Epstein-Barr virus immortalized B lymphocytes by adenovirus-delivered ribozymes. Proc Natl Acad Sci USA 94:8156–8161
Hung SC, Kang MS, Kieff E (2001) Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci USA 98:1865–1870
Isaksson A, Berggren M, Ricksten A (2003) Epstein-Barr virus U leader exon contains an internal ribosome entry site. Oncogene 22:572–581
Israel BF, Kenney SC (2003) Virally targeted therapies for EBV-associated malignancies. Oncogene 22:5122–5130
Jiang M, Milner J (2002) Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21:6041–6048
Kenney JL, Guinness ME, Curiel T, Lacy J (1998) Antisense to the Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP-1) suppresses LMP-1 and bcl-2 expression and promotes apoptosis in EBV-immortalized B cells. Blood 92:1721–1727
Kieff E (1996) Epstein-Barr virus and its replication. In: Fields BN, Knipe DM, Howley PM (eds) Fields virology, 3rd edn, vol 2. Lippincott-Raven Publishers, Philadelphia, pp 2343–2396
Knudsen ES, Wang JY (1997) Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol 17:5771–5783
Kumada T, Tsuneyama K, Hatta H, Ishizawa S, Takano Y (2004) Improved 1-h rapid immunostaining method using intermittent microwave irradiation: practicability based on 5 years application in Toyama Medical and Pharmaceutical University Hospital. Mod Pathol 17:1141–1149
Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG (1995) Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375:685–688
Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG (1997) Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci USA 94:12616–12621
Li XP, Li G, Peng Y, Kung HF, Lin MC (2004) Suppression of Epstein-Barr virus-encoded latent membrane protein-1 by RNA interference inhibits the metastatic potential of nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 315:212–218
Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC (1999) Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol 73:4385–4392
Mathews MB, Bernstein RM, Franza BR Jr, Garrels JI (1984) Identity of the proliferating cell nuclear antigen and cyclin. Nature 309:374–376
Mattia E, Chichiarelli S, Hickish T, Gaeta A, Mancini C, Cunningham D, van Renswoude J (1997) Inhibition of in vitro proliferation of Epstein-Barr virus infected B cells by an antisense oligodeoxynucleotide targeted against EBV latent membrane protein LMP1. Oncogene 15:489–493
Middeldorp JM, Brink AA, van den Brule AJ, Meijer CJ (2003) Pathogenic roles for Epstein-Barr virus (EBV) gene products in EBV-associated proliferative disorders. Crit Rev Oncol Hematol 45:1–36
Polack A, Delius H, Zimber U, Bornkamm GW (1984) Two deletions in the Epstein-Barr virus genome of the Burkitt lymphoma nonproducer line Raji. Virology 133:146–157
Randall G, Grakoui A, Rice CM (2003) Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA 100:235–240
Reisman D, Yates J, Sugden B (1985) A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol 5:1822–1832
Roth G, Curiel T, Lacy J (1994) Epstein-Barr viral nuclear antigen 1 antisense oligodeoxynucleotide inhibits proliferation of Epstein-Barr virus-immortalized B cells. Blood 84:582–587
Sample J, Hummel M, Braun D, Birkenbach M, Kieff E (1986) Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc Natl Acad Sci USA 83:5096–5100
Sample J, Brooks L, Sample C, Young L, Rowe M, Gregory C, Rickinson A, Kieff E (1991) Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc Natl Acad Sci USA 88:6343–6347
Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W (2001) Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J 20:4588–4602
Sugden B, Warren N (1989) A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol 63:2644–2649
Sugimoto M, Tahara H, Ide T, Furuichi Y (2004) Steps involved in immortalization and tumorigenesis in human B-lymphoblastoid cell lines transformed by Epstein-Barr virus. Cancer Res 64:3361–3364
Sung NS, Wilson J, Davenport M, Sista ND, Pagano JS (1994) Reciprocal regulation of the Epstein-Barr virus BamHI-F promoter by EBNA-1 and an E2F transcription factor. Mol Cell Biol 14:7144–7152
Tomar RS, Matta H, Chaudhary PM (2003) Use of adeno-associated viral vectors for delivery of small interfering RNA. Oncogene 22:5712–5715
Wilson JB, Bell JL, Levine AJ (1996) Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J 15:3117–3126
Yates J, Warren N, Reisman D, Sugden B (1984) A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA 81:3806–3810
Yates JL, Guan N (1991) Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol 65:483–488
Yates JL, Warren N, Sugden B (1985) Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812–815
Young LS, Murray PG (2003) Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108–5121
Zhang D, Frappier L, Gibbs E, Hurwitz J, O’Donnell M (1998) Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res 26:631–637
Acknowledgements
We would like to thank Associate Professor Hiroyuki Kishi for advice on flow cytometry analysis of transfection efficiency and Tokimasa Kumada and Hideki Hatta for their expert technical support. We are also grateful to Associate Professor Hideto Yonekura and Dr Hui Li (Kanazawa University, Japan) for helpful discussions and Drs Fumihiro Tomoda and Hua-Chuan Zheng for their assistance with the statistical analysis. This work was supported in part by a Grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hong, M., Murai, Y., Kutsuna, T. et al. Suppression of Epstein-Barr nuclear antigen 1 (EBNA1) by RNA interference inhibits proliferation of EBV-positive Burkitt’s lymphoma cells. J Cancer Res Clin Oncol 132, 1–8 (2006). https://doi.org/10.1007/s00432-005-0036-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-005-0036-x