Abstract
Small terminal or interstitial deletions involving bands 4q34 and 4q35 have been described in several patients with a relatively mild phenotype such as mild to moderate intellectual disability and minor dysmorphic features. We present a boy born from unrelated parents with a de novo 4q34.1–q35.2 deletion and clinical features resembling 22q11.2 deletion syndrome. To the best of our knowledge, this is the first reported patient with 4q34–q35 deletion and phenotype resembling 22q11.2 deletion syndrome without fifth finger anomalies as a specific feature of 4q- syndrome. G-banding karyotyping disclosed the deletion, which was further delineated by microarray comparative genomic hybridization. Fluorescence in situ hybridization and multiplex ligation-dependent probe amplification analyses did not reveal rearrangements of 22q11.2 region. MLPA confirmed the deletion within the 4q35.2 region. Conclusion: Given the considerable clinical overlaps between the 22q11.2 deletion syndrome and clinical manifestation of the patient described in this study, we propose that region 4q34.1–q35.2 should be considered as another region associated with phenotype resembling 22q11.2 deletion syndrome. We also propose that distal 4q deletions should be considered in the evaluation of patients with phenotypic manifestations resembling 22q11.2 deletion syndrome in whom no 22q11.2 microdeletion was detected, even in the absence of distinctive fifth finger anomalies. Additionally, we underline the importance of applying array CGH that enables simultaneous genome-wide detection and delineation of copy number changes (e.g., deletions and duplications).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microscopically visible interstitial and terminal deletions of the long arm of chromosome 4 often cause a recognizable pattern of malformations known as 4q- syndrome [28, 30, 32]. This syndrome is usually associated with growth retardation, mild to moderate mental retardation, craniofacial anomalies (microcephaly, low-set and malformed pinnae, short nose with a low nasal bridge and micrognathia), palatal, skeletal and fifth finger anomalies (clinodactyly, ulnar ray defects, stiff fifth finger with hypoplastic or tapering distal phalanx, hooked or volar nail), as well as urogenital and cardiovascular malformations (mostly atrial or ventricular septal defects) [1, 11, 17, 30]. Several authors have suggested that the critical region for most of the 4q- common characteristics might be assigned to the 4q31–q34 interval [16, 26, 28, 29]. Smaller terminal or interstitial deletions involving bands 4q34–q35 have also been described in several cases [2, 5–7, 10, 17, 24, 33–35]. These smaller deletions usually do not lead to major anomalies, but rather give mild to moderate mental retardation and minor dysmorphic features such as coarse face, ear malformations and fifth finger anomalies [6]. Major anomalies have been described in a few cases associated with smaller deletions and the most common are congenital heart defects, genitourinary anomalies and cleft palate [2, 33]. An association of the 4q34.2–qter deletions and the phenotype of velocardiofacial syndrome has been suggested by several authors [5, 7, 10, 25, 33]. So far, no consistent clinical phenotype for 4q34–q35 deletions could be delineated [2]. It has, however, been suggested that the severity of the phenotype correlates with the size of deletion [17].
22q11.2 deletion syndrome (MIM# 188400) is the most common deletion disorder in humans with an incidence of approximately 1/4,000 per live births [9]. This syndrome is characterized by a broad spectrum of features including conotruncal heart anomalies, craniofacial dysmorphism (long face with malar flattening and mandibular retrusion, narrow palpebral fissures, small and malformed pinnae, prominent nasal bridge and small mouth), cleft palate, absent or small thymus with T lymphocyte dysfunction and hypoparathyroidism [11, 14].
Here, we report a patient initially suspected of having 22q11.2 deletion syndrome because of facial dysmorphism, tetralogy of Fallot, right aortic arch and normal growth and development. Routine cytogenetic analysis, however, identified a microscopically visible distal 4q deletion. In order to delineate this chromosomal deletion, we applied microarray comparative genomic hybridization (array CGH). Array CGH confirmed the distal deletion of the long arm of chromosome 4. The deletion is approximately 17.4 Mb in size and extends from 4q34.1 to the subtelomeric region of 4q35.2. The 22q11.2 microdeletion was not detectable by fluorescence in situ hybridization (FISH) and multiplex ligation-dependent probe amplification (MLPA). Additionally, MLPA confirmed the deletion within the 4q35.2 region. Considering that this patient with clinical features of 22q11.2 deletion syndrome has a large 4q deletion and no fifth finger anomalies as specific feature of 4q- syndrome, we propose that 4q34.1–q35.2 region should be considered as another region associated with the phenotype resembling 22q11.2 deletion syndrome.
Case report
A 6-month-old male infant was referred for genetic counseling because of tetralogy of Fallot, right aortic arch and facial dysmorphism. He was the first child of a young, healthy and unrelated couple. Family history as well as the histories of pregnancy and delivery were unremarkable, without consanguinity or exposure to teratogenic factors.
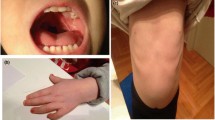
Physical examination revealed a small mouth and prominent ears. Height and weight were between the 25th and 50th centile and head circumference at 50th centile. Heart auscultation disclosed a grade 4/6 systolic murmur at upper left sternal border, accompanied with central cyanosis. He was mildly hypotonic, with otherwise normal motor and cognitive development.
At the age of 8 months, a complete correction of tetralogy of Fallot was undertaken. Postoperative course was complicated by junctional ectopic tachycardia, which was successfully treated with amiodarone. The motor milestones of the patient were within wider range of normal. He was able to sit unassisted at 9–10 months, shortly after surgical intervention, and walked at the age of 15–16 months.
Presently, at the age of 2 years and 2 months, previously observed facial dysmorphism persists. Growth and psychomotor development are normal, with height 90.5 cm (75th centile), weight 14.3 kg (between 75th and 90th centile) and head circumference 52 cm (between 75th and 90th centile). He speaks short sentences and smiles occasionally during examination. The hair, as well as, skull form is normal. The palpebral fissures and distance between eyes also appear normal. The outer canthal distance is 7.8 cm (75th centile), the inner canthal distance is 2.9 cm (between 75th and 90th centile), the interpupillary distance is 5.1 cm (75th centile) and the length of palpebral fissures is 2.5 cm (between 50th and 75th centile). The nasal root and bridge, as well as, length and form of the filtrum are normal. The ears are anteverted with otherwise normal morphology and dimensions. The mouth appears small and the chin pointed. The boy is not suffering any disturbance of heart rhythm, having only a mild residual pulmonary stenosis detected on the most recent echocardiographic examination.
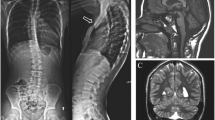
G-banding cytogenetic analysis of the patient identified a distal deletion within the long arm of one chromosome 4, with the breakpoints assigned between 4q34 and 4q35 (data not shown). Parental chromosomes were normal, indicating that deletion in the patient had arisen de novo (data not shown). The array CGH analysis provided precise delineation of the chromosomal rearrangement, showing an interstitial deletion approximately 17.4 Mb in size in the distal part of chromosome 4 (chr4: 172,977,872–190,351,861; genome build 18), extending from 4q34.1 to the subtelomeric region of 4q35.2 (Fig. 1). Copy number profiling was performed on an Agilent Human Genome CGH Microarray 60K (AMADID# 021924, hg18, NCBI build 36.1, March 2006) as previously described [4]. Microarray slides were scanned using an Agilent microarray scanner, quantified with Feature Extraction software 9.5.1 and further analyzed with arrayCGHbase (http://medgen.ugent.be/arraycghbase/) [20]. No other copy number variations and cryptic chromosomal rearrangements could be detected by array CGH. In addition, FISH with TUPLE1 probe and MLPA (kit P250-A1 DiGeorge; MRC-Holland) analyses were performed. Neither of these two analyses revealed rearrangements of 22q11.2 region. MLPA confirmed the deletion of the KLKB1 probe within the 4q35.2 region and rearrangements in other four regions at chromosomes 8, 9, 10 and 17 were not detectable (data not shown).
Discussion
It has been proposed that distal deletions of chromosome 4q involving 4q34-qter are usually characterized with relatively mild phenotypic abnormalities, such as mild to moderate mental retardation and minor dysmorphic features [6, 33]. We have identified a de novo interstitial deletion, spanning the 4q34.1–q35.2 region, in a boy with a phenotype resembling the 22q11.2 deletion syndrome. The approximate size of 17.4 Mb and breakpoints of the deletion have been determined by array CGH.
Based on literature analysis, only a few authors have postulated that distal 4q deletions might be associated with a phenotype resembling the 22q11.2 deletion syndrome [10, 25, 33]. Tsai et al. described a boy with 4q34.2-qter deletion, having velocardiofacial syndrome-like phenotype accompanied by typical 4q- syndrome anomaly, i.e., dysplastic and duplicated nail of the fifth finger [33]. A second report describes two patients with distal 4q deletions, initially suspected of having 22q11.2 deletion syndrome [10]. The first patient was presented with intellectual disability, congenital heart defect, cutaneous syndactyly of fingers, omphalocele and deletion of the 4q33-qter region. The second reported patient had a milder phenotype and haploinsufficiency of both 4q35-qter and 4p16 regions. A third report by Ravnan et al. described a patient with suspected 22q11.2 deletion syndrome and a deletion of the 4q31–q33 region, located more proximal comparing to deletion of the patient presented in this study [25]. Furthermore, certain craniofacial characteristics of velocardiofacial syndrome were described in four patients with 4q34 deletions [5, 7].
The patient presented here has normal growth as well as normal motor and cognitive development, with no major extracardiac malformations. In association with tetralogy of Fallot, right aortic arch, small mouth appearance and protuberant ears, such characteristic clinical findings initially suggested the 22q11.2 deletion syndrome (Table 1). To the best of our knowledge, this is the first reported case having 4q34–q35 deletion and phenotype resembling 22q11.2 deletion syndrome without fifth finger anomalies as a specific feature of 4q- syndrome. Given the considerable clinical overlap between the 22q11.2 deletion syndrome and clinical manifestation of the patient described in this study (Table 1), we propose that region 4q34.1–q35.2 should be considered as another region associated with a phenotype resembling 22q11.2 deletion syndrome.
Additionally, a phenotypically normal patient with a deletion approximately 10.7 Mb in size spanning the 4q34.1–q34.3 region (chr4: 172,142,000–182,840,000) has been described recently [1]. The deletion in this patient overlaps with the one described in our patient. By comparing the breakpoints in the asymptomatic patient having a 4q34.1–q34.3 deletion [1] and the patient presented in this study, we hypothesize that the deletion of the distal 4q34.3–q35.2 region might be responsible for the heart defects and phenotypic manifestation resembling the 22q11.2 deletion syndrome.
HAND2 (heart and neural crest derivatives expressed 2) gene, which is previously reported to play an essential role in cardiac morphogenesis [22], is among the genes affected by both deletions (4q34.1–q34.3 and 4q34.1–q35.2). The absence of any cardiac abnormality in the patient with 4q34.1–q34.3 deletion [1] further supports the observation that haploinsufficiency of the HAND2 gene is either non-causative or shows incomplete penetrance [13, 15, 36]. The absence of cardiac features has been described in several patients having 4q deletions involving the HAND2 gene [1, 13, 15, 36]. Accordingly, it has been suggested that other genes residing in the distal 4q interval, including microRNA has-mir-1305, might be involved in cardiac morphogenesis [27, 36].
Interestingly, although the deletion of 4q approximately 17 Mb in size is delineated in our patient, no obvious cognitive impairment could be observed. Literature analysis of 20 patients reported with deletions located solely within 4q34–q35 region (Table 1) showed that 11 of them (55%) had normal or near normal cognitive development [2, 7, 31] while 13 (65%) had normal motor development [2, 7, 27, 31, 33]. Among patients with normal or near normal cognitive development, six patients were found to have large deletions of this region ranging from approximately 7–13 Mb [2, 31]. On the other hand, the deletions of such extent in other genomic regions have been described very rarely in individuals with a normal cognitive development [i.e., chromosomes 18q21.3-qter (17 Mb) and 21q11.2–q21.3 (14 Mb)] [18, 23, 37]. One of possible explanations for no cognitive delay in our patient may lie in the fact that such sizeable imbalance comprises one of the longest human gene deserts 6 Mb in size located within the region 4q34.3 [12]. Gene deserts are segments devoid of protein-coding genes or segments with low gene density and thus remain largely unknown regarding their function. Furthermore, the unusual clinical presentation of our patient could be a consequence of somatic mosaicism, as recently postulated by Mkrtchyan et al. [21].
The comparison of phenotypic characteristics of distal 4q34–q35 deletions and 22q11.2 deletion syndrome (Table 1) suggests that correct establishment of proper diagnosis could be challenging, especially in patients with normal or near normal development, conotruncal heart defects and the absence of digital anomalies.
Our findings support observations that the analysis of only one locus in patients with suspected 22q11.2 deletion syndrome or any other idiopathic malformation syndrome might be insufficient [3, 8, 19]. Thus, we underline the importance of applying array CGH that enables simultaneous genome-wide detection and delineation of copy number changes. Our findings further indicate that distal 4q deletion should be considered in patients with phenotypic manifestations resembling 22q11.2 deletion syndrome in whom no 22q11.2 microdeletion was detected [36]. We further suggest that distal 4q deletion should be considered in such patients even in the absence of distinctive fifth finger anomalies. The precise delineation of deletion breakpoints in cases of 4q deletions, together with studies focused on potential influence of yet unidentified modifying genes and epigenetic factors, should clarify which genes and pathways are responsible for the pathogenesis of heart defects and phenotype of patients with 4q deletions.
References
Bateman MS, Mehta SG, Willatt L, Selkirk E, Bedwell C, Zwolinski S, Sparnon L, Simonic I, Abbott K, Barber JC (2010) A de novo 4q34 interstitial deletion of at least 9.3 Mb with no discernible phenotypic effect. Am J Med Genet A 152A:1764–1769
Bendavid C, Pasquier L, Watrin T, Morcel K, Lucas J, Gicquel I, Dubourg C, Henry C, David V, Odent S, Leveque J, Pellerin I, Guerrier D (2007) Phenotypic variability of a 4q34–>qter inherited deletion: MRKH syndrome in the daughter, cardiac defect and Fallopian tube cancer in the mother. Eur J Med Genet 50:66–72
Brunet A, Armengol L, Heine D, Rosell J, Garcia-Aragones M, Gabau E, Estivill X, Guitart M (2009) BAC array CGH in patients with Velocardiofacial syndrome-like features reveals genomic aberrations on chromosome region 1q21.1. BMC Med Genet 10:144
Buysse K, Delle Chiaie B, Van Coster R, Loeys B, De Paepe A, Mortier G, Speleman F, Menten B (2009) Challenges for CNV interpretation in clinical molecular karyotyping: lessons learned from a 1001 sample experience. Eur J Med Genet 52:398–403
Caliebe A, Waltz S, Jenderny J (1997) Mild phenotypic manifestations of terminal deletion of the long arm of chromosome 4: clinical description of a new patient. Clin Genet 52:116–119
Cingoz S, Bisgaard AM, Bache I, Bryndorf T, Kirchoff M, Petersen W, Ropers HH, Maas N, Van Buggenhout G, Tommerup N, Tumer Z (2006) 4q35 deletion and 10p15 duplication associated with immunodeficiency. Am J Med Genet A 140:2231–2235
Descartes M, Keppler-Noreuil K, Knops J, Longshore JW, Finley WH, Carroll AJ (1996) Terminal deletion of the long arm of chromosome 4 in a mother and two sons. Clin Genet 50:538–540
Emanuel BS (2008) Molecular mechanisms and diagnosis of chromosome 22q11.2 rearrangements. Dev Disabil Res Rev 14:11–18
Fernandez L, Lapunzina P, Arjona D, Lopez Pajares I, Garcia-Guereta L, Elorza D, Burgueros M, De Torres ML, Mori MA, Palomares M, Garcia-Alix A, Delicado A (2005) Comparative study of three diagnostic approaches (FISH, STRs and MLPA) in 30 patients with 22q11.2 deletion syndrome. Clin Genet 68:373–378
Fernandez L, Lapunzina P, Pajares IL, Palomares M, Martinez I, Fernandez B, Quero J, Garcia-Guereta L, Garcia-Alix A, Burgueros M, Galan-Gomez E, Carbonell-Perez JM, Perez-Granero A, Torres-Juan L, Heine-Suner D, Rosell J, Delicado A (2008) Unrelated chromosomal anomalies found in patients with suspected 22q11.2 deletion. Am J Med Genet A 146A:1134–1141
Hennekam RCM, Allanson JE, Krantz ID, Gorlin RJ (2010) Gorlin's syndromes of the head and neck. Oxford University Press, Oxford
Hillier LW, Graves TA, Fulton RS, Fulton LA, Pepin KH, Minx P, Wagner-McPherson C et al (2005) Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature 434:724–731
Huang T, Lin AE, Cox GF, Golden WL, Feldman GL, Ute M, Schrander-Stumpel C, Kamisago M, Vermeulen SJ (2002) Cardiac phenotypes in chromosome 4q- syndrome with and without a deletion of the dHAND gene. Genet Med 4:464–467
Jones KL, Smith DW (2006) Smith's recognizable patterns of human malformation. Elsevier, Philadelphia
Kaalund SS, Moller RS, Teszas A, Miranda M, Kosztolanyi G, Ullmann R, Tommerup N, Tumer Z (2008) Investigation of 4q-deletion in two unrelated patients using array CGH. Am J Med Genet A 146A:2431–2434
Keeling SL, Lee-Jones L, Thompson P (2001) Interstitial deletion 4q32-34 with ulnar deficiency: 4q33 may be the critical region in 4q terminal deletion syndrome. Am J Med Genet 99:94–98
Lin AE, Garver KL, Diggans G, Clemens M, Wenger SL, Steele MW, Jones MC, Israel J (1988) Interstitial and terminal deletions of the long arm of chromosome 4: further delineation of phenotypes. Am J Med Genet 31:533–548
Lindstrand A, Malmgren H, Sahlen S, Schoumans J, Nordgren A, Ergander U, Holm E, Anderlid BM, Blennow E (2010) Detailed molecular and clinical characterization of three patients with 21q deletions. Clin Genet 77:145–154
Mantripragada KK, Tapia-Paez I, Blennow E, Nilsson P, Wedell A, Dumanski JP (2004) DNA copy-number analysis of the 22q11 deletion-syndrome region using array-CGH with genomic and PCR-based targets. Int J Mol Med 13:273–279
Menten B, Pattyn F, De Preter K, Robbrecht P, Michels E, Buysse K, Mortier G, De Paepe A, van Vooren S, Vermeesch J, Moreau Y, De Moor B, Vermeulen S, Speleman F, Vandesompele J (2005) arrayCGHbase: an analysis platform for comparative genomic hybridization microarrays. BMC Bioinforma 6:124
Mkrtchyan H, Gross M, Hinreiner S, Polytiko A, Manvelyan M, Mrasek K, Kosyakova N, Ewers E, Nelle H, Liehr T, Bhatt S, Thoma K, Gebhart E, Wilhelm S, Fahsold R, Volleth M, Weise A (2010) The human genome puzzle—the role of copy number variation in somatic mosaicism. Curr Genomics 11:426–431
Morikawa Y, Cserjesi P (2008) Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ Res 103:1422–1429
Netzer C, Helmstaedter C, Ehrbrecht A, Engels H, Schwanitz G, Urbach H, Schroder R, Weber RG, Kornblum C (2006) Global brain dysmyelination with above-average verbal skills in 18q- syndrome with a 17 Mb terminal deletion. Acta Neurol Scand 114:133–138
Pickard BS, Hollox EJ, Malloy MP, Porteous DJ, Blackwood DH, Armour JA, Muir WJ (2004) A 4q35.2 subtelomeric deletion identified in a screen of patients with co-morbid psychiatric illness and mental retardation. BMC Med Genet 5:21
Ravnan JB, Chen E, Golabi M, Lebo RV (1996) Chromosome 22q11.2 microdeletions in velocardiofacial syndrome patients with widely variable manifestations. Am J Med Genet 66:250–256
Robertson SP, O'Day K, Bankier A (1998) The 4q-syndrome: delineation of the minimal critical region to within band 4q31. Clin Genet 53:70–73
Rossi MR, DiMaio MS, Xiang B, Lu K, Kaymakcalan H, Seashore M, Mahoney MJ, Li P (2009) Clinical and genomic characterization of distal duplications and deletions of chromosome 4q: study of two cases and review of the literature. Am J Med Genet A 149A:2788–2794
Sarda P, Lefort G, Fryns JP, Humeau C, Rieu D (1992) Interstitial deletion of the distal long arm of chromosome 4. J Med Genet 29:259–261
Sensi A, Prontera P, Buldrini B, Palma S, Aiello V, Gruppioni R, Calzolari E, Volinia S, Martini A (2008) Cytogenetic and array CGH characterization of an intrachromosomal complex rearrangement of 4q in a patient with a 4q-phenotype. Am J Med Genet A 146A:110–115
Strehle EM, Bantock HM (2003) The phenotype of patients with 4q-syndrome. Genet Couns 14:195–205
Strehle EM, Middlemiss PM (2007) Children with 4q-syndrome: the parents' perspective. Genet Couns 18:189–199
Townes PL, White M, Di Marzo SV (1979) 4q-Syndrome. Am J Dis Child 133:383–385
Tsai CH, Van Dyke DL, Feldman GL (1999) Child with velocardiofacial syndrome and del (4)(q34.2): another critical region associated with a velocardiofacial syndrome-like phenotype. Am J Med Genet 82:336–339
Tupler R, Berardinelli A, Barbierato L, Frants R, Hewitt JE, Lanzi G, Maraschio P, Tiepolo L (1996) Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet 33:366–370
Van Buggenhout G, Maas NM, Fryns JP, Vermeesch JR (2004) A dysmorphic boy with 4qter deletion and 4q32.3-34.3 duplication: clinical, cytogenetic, and molecular findings. Am J Med Genet A 131:186–189
Vogt J, Ryan E, Tischkowitz MD, Reardon W, Brueton LA (2006) The tale of a nail sign in chromosome 4q34 deletion syndrome. Clin Dysmorphol 15:127–132
Wakui K, Toyoda A, Kubota T, Hidaka E, Ishikawa M, Katsuyama T, Sakaki Y, Hattori M, Fukushima Y (2002) Familial 14-Mb deletion at 21q11.2-q21.3 and variable phenotypic expression. J Hum Genet 47:511–516
Acknowledgments
This work was supported by the Ministry of Education and Science, Republic of Serbia (AK, DD and MS Grant Nos. 143028 and 173051).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cuturilo, G., Menten, B., Krstic, A. et al. 4q34.1–q35.2 deletion in a boy with phenotype resembling 22q11.2 deletion syndrome. Eur J Pediatr 170, 1465–1470 (2011). https://doi.org/10.1007/s00431-011-1533-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00431-011-1533-3