Abstract
Early detection of viral invasion by pattern recognition receptors (PRR) is crucial for the induction of a rapid and efficient immune response. Cytosolic DNA sensors are the most recently described class of PRR, and induce transcription of type I interferons (IFN) and proinflammatory cytokines via the key adaptor protein stimulator of interferon genes (STING). Herpesviruses are a family of large DNA viruses widely known for their immense arsenal of proteins dedicated to manipulating and evading host immune responses. Tantamount to the significant role played by DNA sensors and STING in innate immune responses, herpesviruses have in turn evolved a range of mechanisms targeting virtually every step of this key signaling pathway. Strikingly, some herpesviruses also take advantage of this pathway to promote their own replication. In this review, we will summarize the current understanding of DNA sensing and subsequent induction of signaling and transcription, and showcase the close adaptation of herpesviruses to their host reflected by the myriad of viral proteins dedicated to modulating this critical innate immune pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Approaching the world of herpesviruses
Herpesviruses are a large virus family characterized by lifelong persistence in their host. Herpesviridae consist of three sub-families, namely α-herpesvirinae, β-herpesvirinae and γ-herpesvirinae, members of which are classified based on biological and sequence similarities [1]. As a family they infect a broad range of hosts, including mammals, reptiles, and avians, however, specific viruses have only a single or narrow host range.
Herpesviruses are enveloped viruses with linear double-stranded DNA genomes ranging from 125 to 290 kbp [2]. Primary infection is lytic and occurs in permissive cell types. Subsequently, herpesviruses become latent in their hosts, with periods of reactivation which can be triggered by a range of factors including stress, immunosuppression and other environmental changes. Viral progeny are produced upon primary infection or following reactivation. Pathology generally follows primary infection, reactivation from latency or due to oncogenic potential, the latter mostly restricted to γ-herpesviruses [3,4,5].
Between the three sub-families, there is little genetic similarity [1] and their target cell types and clinical manifestations differ. α-herpesviruses infect epithelial cells followed by entry to sensory nerve ganglia as the site of latency. Periodic reactivation of herpes simplex virus (HSV)-1 and HSV-2 leads to mucosal lesions such as oral or genital sores, respectively. Primary Varicella Zoster Virus (VZV) infection causes chickenpox, while reactivation from latency results in shingles. α-herpesviruses have important agricultural impact, being the etiologic agents of respiratory and neurological disorders in horses (Equine herpesvirus EHV-1 and -4) [6] and cattle (Bovine BoHV-1) [7], as well as Aujeszky’s disease in pigs (Pseudorabies virus) [8] and immunosuppression and T cell lymphomas in chicken (oncogenic Marek’s disease virus) [9].
β-Herpesviruses infect and establish latency in myeloid cells, lymphocytes and epithelial cells. The prototype virus for this sub-family is cytomegalovirus (CMV). Human CMV (HCMV), while asymptomatic in healthy individuals, causes a range of diseases, such as retinitis and hepatitis, in immunocompromised individuals including transplant recipients and HIV-1/AIDS patients [5]. HCMV is underappreciated as the leading viral cause of congenital birth defects. Due to restricted host specificity, MCMV in mice became a well-established disease model of HCMV infection as it shares biological and sequence similarities with HCMV [10]. Aside from CMV, human herpesvirus 6 (HHV-6) is also a member of the β-herpesvirus family. There are two variants, HHV-6A and HHV-6B, which are considered as two distinct herpesvirus species due to their distinctive biological properties [11]. Almost all humans are infected with HHV-6B, usually in early childhood. Infection can result in fever, diarrhea, and sometimes a rash known as roseola. Although rare, this initial HHV-6B infection can also cause seizures or encephalitis. HHV-6A has not yet been clearly linked to any disease [12]. The fourth human member of the β-herpesviridae, HHV-7, is closely related to HHV-6 and infects up to 80% of children in infancy. At this time, there is no link between HHV-7 and any specific disease.
γ-Herpesvirus infection is restricted to lymphocytes, B cells and endothelial cells. This sub-family has the distinction of containing two oncogenic human viruses. Kaposi’s sarcoma-associated herpesvirus (KSHV) is the cause of the endothelial tumor Kaposi’s sarcoma (KS) and the lymphoproliferative diseases primary effusion lymphoma (PEL) and multicentric Castleman’s disease (MCD) [13]. Epstein-Barr virus (EBV) is the causative agent of mononucleosis but infection can also cause the cancers Burkitt’s lymphoma, nasopharyngeal carcinoma, and Hodgkin’s lymphoma [14]. Murid herpesvirus 4 strain 68 (here referred to as MHV68) has emerged as a small animal model for the human γ-herpesviruses KSHV and EBV [15, 16].
Antiviral therapy against herpesviruses is limited. The majority of the currently licensed drugs ultimately antagonize the viral DNA polymerase. These include the nucleoside analogues acyclovir and gancyclovir, and the more recently developed drugs foscarnet and cidofovir. Letermovir (Prevymis®), which inhibits the viral terminase complex [17, 18], has been approved since 2017 for use in patients after allogeneic hematopoietic stem cell transplantation to combat CMV. Limitations to antiviral therapy include poor bioavailability, toxicity and the emergence of drug-resistant strains, which have been reported in immunocompromised individuals such as transplant recipients and HIV-infected patients [reviewed in 19]. Currently, the only licensed vaccine against a herpesvirus is targeted at VZV. Introduction of varicella vaccination has resulted in a decline in rates of infection and morbidity, proving to be protective against 85% cases of chickenpox and 95% cases of shingles [20]. Given the efficacy of the varicella vaccine, there is potential for successful vaccines against other herpesvirus, which demands further studies to identify suitable antigens for vaccine candidates.
The early birds: pattern recognition receptors
The innate immune system recognizes pathogens during the initial stages of infection via germline-encoded pattern recognition receptors (PRR). Several classes of PRR have been proposed, including Toll-like receptors (TLR), NOD-like receptors (NLR), RIG-I-like receptors (RLR) and cytosolic DNA sensors, which can recognize molecular structures pinpointing pathogenic infection, so-called pathogen-associated molecular patterns (PAMPs). Upon sensing of viruses or other intracellular pathogens, PRR activate downstream signaling cascades leading to the secretion of type I interferons (IFN) and proinflammatory cytokines. This response is launched within the first hours of infection and eliminates in most cases the pathogen before it can establish a foothold in the host.
Within the last few years, research in the field of PRR has brought the importance of cytosolic DNA sensing into the forefront. Concurrently, the discovery of antagonists of these pathways encoded by pathogens contributes to our knowledge of how the immune system combats infections. Given their immense coding capacity and the ability to establish lifelong infection, it follows that herpesviruses have evolved various strategies to counteract the host machinery and target critical checkpoints involved in DNA driven immune responses.
Step by step: the cGAS-STING signaling pathway
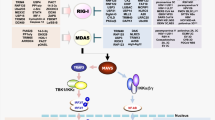
Upon herpesviral infection, the DNA sensor cyclic GMP-AMP (cGAMP) synthase (cGAS) recognizes viral DNA or aberrantly localized cellular DNA and catalyzes the formation of the second messenger 2′3′-cGAMP [21,22,23,24,25] (Fig. 1). While cGAS was described as a predominantly cytosolic DNA sensor, newer insights provide evidence that cGAS also localizes in the nucleus [26, 27]. The second messenger cGAMP binds to the endoplasmic reticulum (ER)-resident adaptor protein stimulator of interferon genes (STING) leading to its dimerization and activation [28]. cGAMP can also rapidly amplify the intruder alarm via direct transfer through gap junctions to neighbouring cells leading to a heightened antiviral state [29]. Two recent studies also reported that cGAMP can be delivered within viral particles to neighbouring cells [30, 31]. Upon activation by cGAMP, STING translocates from the ER to the Golgi apparatus [32]. Here, the E3 ubiquitin ligases tripartite motif family 56 (TRIM56) and TRIM32 catalyze K63-linked poly-ubiquitination [33, 34], whereas autocrine motility factor receptor (AMFR) catalyzes K27-linked poly-ubiquitination at different lysine residues on STING [35]. These modifications on STING serve as an anchor for the recruitment of Tank-binding kinase 1 (TBK1), which in turn phosphorylates STING at S366 [28]. This phosphorylation event leads to a negatively charged surface on STING, which attracts the positively charged interferon regulatory factor 3 (IRF3). Once in proximity of the STING-TBK1 complex, IRF3 is likewise phosphorylated by TBK1, triggering IRF3 activation and dimerization [28]. IRF3 dimers translocate into the nucleus, leading to the expression of type I IFN. After the activation of TBK1 and IRF3, STING degradation is mediated by p62/Sequestosome 1 (SQSTM1)-dependent autophagy [36]. Notably, the trafficking step from the ER to the Golgi apparatus is crucial for the induction of type I IFN transcription by STING [37].
Step by step: the cGAS-STING signaling pathway. The DNA sensor cGAS detects pathogenic or aberrantly localized DNA and produces the second messenger 2′3′-cGAMP which binds to STING. IFI16 also senses DNA and signals via STING or enhances cGAS activity. Activated STING dimerizes and translocates from the ER to the Golgi apparatus. Then, STING is ubiquitinated which serves as an anchor for TBK1. This in turn triggers IRF3 activation and homodimer formation. Activated IRF3 then translocates into the nucleus and induce the expression of type I IFN. Prior to its translocation to the Golgi, activated STING induces NF-κB-dependent production of proinflammatory cytokines
While the focus has predominantly been on its role in IRF3-dependent signaling, STING also activates NF-κB-mediated signaling. This has been suggested to occur via TBK1 and TNF receptor-associated factor 6 (TRAF6) [38]. We have since shown, via siRNA knockdown of TBK1 coupled to reporter assays, that STING-mediated NF-κB activation is independent of TBK1 and further that STING activates the NF-κB mediated proinflammatory cytokine response prior to its translocation to the Golgi compartment [39] (Fig. 1).
DNA sensing beyond cGAS: IFI16, AIM2, DDX41, DAI, and DNA-PK
Besides cGAS, several other putative cytosolic DNA sensors have been identified. Gamma-interferon-inducible protein 16 (IFI16) [40] belongs to the Pyrin and HIN domain (PYHIN) protein family. It contains one N-terminal Pyrin (protein–protein interaction) domain and two C-terminal HIN (DNA-binding) domains and localizes to the nucleus in steady-state [40]. Upon herpesviral infection, IFI16 can translocate to the cytoplasm and induce STING-mediated signaling [41] (Fig. 1). Moreover, IFI16 interacts with cGAS in a Pyrin-domain dependent manner to potentiate cGAS-mediated responses [42, 43]. Two recent studies reported that IFI16 synergizes with cGAS as a DNA co-sensor [44, 45]. However, while IFI16 was shown to enhance cGAMP production by interacting with cGAS in macrophages [44], cGAMP production was not affected in keratinocytes [45], indicating that the role of IFI16 may be cell type dependent.
IFN-inducible protein absent in melanoma 2 (AIM2) also detects aberrantly localized DNA; however, currently, it is proposed to be connected with activation of the inflammasome [46]. Other reported DNA sensors like DEAD-Box Helicase 41 (DDX41) [47], DNA-dependent activator of IFN-regulatory factors (DAI) [48], and DNA-dependent protein kinase (DNA-PK) [49] require further investigation to clarify their role during viral infection and if they act redundantly, cooperatively or in a cell-type dependent manner. Moreover, unlike cGAS and IFI16, none of these have so far been shown to be implicated in the evasion strategies of herpesviruses or to restrict viral growth.
DNA sensors: crucial sentinels of herpesviral infections
Herpesviral infection strongly triggers the DNA sensing pathway. For the α-herpesvirus HSV-1 it has been reported that infection of cGAS-deficient mice drastically increases viral titers in vivo [24] and mice lacking STING succumb to infection due to the uncontrollable spread of the virus to the central nervous system [50]. IFI16 was shown to directly associate with the HSV-1 genome and restrict viral replication by preventing the association of viral transcriptional activators [51]. In addition, HSV-1 genome bound IFI16 is acetylated and shuttles to the cytoplasm, where it subsequently activates STING-dependent type I IFN induction [52]. While IFI16 was shown to be a restriction factor for infection with the β-herpesvirus HCMV [53], others have revealed that IFI16 is not essential for the type I IFN response to HCMV infection [54, 55]. However, cGAS is a key sensor for type I IFN production upon HCMV infection in monocyte-derived dendritic cells and macrophages [56]. Moreover, STING is critical for the primary detection and initial burst of type I IFN against MCMV infection in vivo [39, 57]. In the case of γ-herpesviruses, it was reported that mice lacking cGAS show significantly higher titers upon infection with MHV68 [58], and siRNA-mediated knockdown of cGAS and STING increases KSHV reactivation [59, 60]. Hence, DNA sensors and STING play a crucial role in controlling the establishment as well as reactivation of herpesviral infections in the host.
Raise the veil: herpesviral antagonism of the DNA sensing pathway
Herpesviruses, with their large coding capacity and close coevolution with their respective host species, remain a plentiful resource for enriching our understanding into host immune responses. Evasion of host immune responses is an ongoing aspect of every stage of the viral life cycle, from primary infection to establishing latency to reactivation. In this review, we will focus on the most recently discovered class of PRR: the DNA sensors cGAS and IFI16, and the key adaptor protein STING. We will outline all currently identified herpesviral antagonists that modulate this key defense pathway and demonstrate the resourcefulness of herpesviruses to target virtually every checkpoint downstream of DNA sensing. Strikingly, at least one antagonist, and in many cases multiple, has been identified for each level of the DNA sensing pathway, and the viral strategies range from directly inhibiting PRR binding to DNA to skewing STING function to disruption of the transcription factors IRF3 and NF-κB (Table 1).
We would like to point out that detailed mechanistic insight is missing for many of the antagonists mentioned in this review, and for the majority we lack confirmation of the phenotype in vivo due to the fact that many herpesviruses are strictly species specific and therefore cannot be studied in small animal models. Especially for VZV, experimental models to study the implications of immune evasion in the context of infection are limited [98] and only recent developments allow the investigation of VZV infection in vitro [99]. Fortunately, animal models of infection with the murine herpesviruses MCMV and MHV68 are available, which allows for verification of phenotypes in vivo for the human herpesviruses HCMV, EBV and KSHV.
From the top: herpesviruses paralyze the sensors
Since the discovery of the key adaptor protein STING in 2008 [100,101,102,103] further research has yielded the identification of multiple cytosolic DNA sensors and with it, a plethora of viral proteins which inhibit them. Figure 2 gives an overview of the identified proteins encoded by herpesviruses that directly antagonize the DNA sensors cGAS and IFI16. So far, antagonists of HSV-1, HCMV and the γ-herpesviruses KSHV, EBV, MHV68, and rhesus monkey rhadinovirus (RRV) have been described as dedicated inhibitors of these sensors.
From the top: herpesviruses paralyze the sensors. α-, β- and γ-herpesviral proteins that target the activation of IFI16 or cGAS are shown in red, blue or green, respectively. The black blind-ended lines indicate inhibition. The black arrow-headed lines indicate degradation mediated by the herpesviral protein. Solid black lines represent studies with mechanistic insights, while dashed black lines indicate findings from descriptive studies
HSV-1
The cGAS-STING signaling pathway is activated at very early stages after infection. Therefore, many of the herpesviral antagonists identified so far are tegument proteins or viral proteins expressed at early time points post infection. The highly abundant tegument protein VP22 of HSV-1 regulates the localization and expression of various viral and cellular proteins [104]. It was recently shown that overexpressed VP22 colocalizes and interacts with cGAS in 293T cells [61]. Infection of human foreskin fibroblasts (HFF) with a virus lacking VP22 led to reduced production of the second messenger cGAMP, resulting in lower levels of IRF3 dimers. The HSV-1 tegument protein, UL37, has previously been characterized to deamidate the RNA sensor RIG-I [105], and was more recently shown to also deamidate cGAS upon HSV-1 infection, thereby antagonizing cGAMP production [62]. HSV-1 UL41 is proposed to selectively degrade host mRNA with AU-rich element (ARE) core motifs in the 3′-untranslated region (3′-UTR) [106] which are present in cGAS, but not STING or IRF3, mRNA [63]. As a result, the presence of UL41 decreases cGAS, but not STING or IRF3, protein levels and eventually diminishes cGAS-mediated signaling in HFF upon HSV-1 infection [63].
HSV-1 likewise targets DNA sensing by IFI16. The tegument protein and E3 ubiquitin ligase ICP0 was proposed to specifically target IFI16 for proteasomal degradation in HFF [66, 107]. However, in contrast, Cuchet-Lourenco and colleagues showed that ICP0 expression alone is neither sufficient nor necessary for IFI16 degradation, since infection with an ICP0-null mutant still resulted in IFI16 degradation [67]. This was also observed by another study [108], which suggests that other viral proteins are involved in the process of IFI16 degradation. However, these contrasting results may be explained by the usage of different cell types and virus strains (Table 1). Further studies are necessary to fully elucidate the relationship between ICP0 and IFI16 and the mechanistic basis behind this immune evasion strategy.
HCMV
The HCMV tegument protein UL31 interacts with cytoplasmic and nuclear cGAS in 293T and HFF cells during infection. In 293T cells this interaction leads to disassociation of DNA from cGAS, which results in reduced production of cGAMP and subsequent dampening transcription of type I IFN [64].
The tegument protein UL83 (also known as pp65) of HCMV, through its proposed effect on nuclear localized cGAS and IFI16, exemplifies the complex interplay between a viral protein and the targeted host proteins. While Browne and Shenk reported in 2003 that UL83 reduces nuclear translocation of NF-κB, but not IRF3 [109], Abate et al. observed reduced nuclear translocation of IRF3 but not NF-κB in the presence of UL83 [110]. Since both studies were performed in HFF using the AD169 strain of HCMV, there is no obvious explanation for these opposing observations, and moreover, a mechanism was lacking. A recent study provided fresh insight by showing that the N-terminal domain of UL83 interacts with nuclear cGAS and blocks its activation during HCMV infection. This leads to impairment of the cGAS-STING-IRF3 signaling pathway and results in significantly lower IFNβ levels [55]. Concurrently, UL83 inhibits the function of IFI16 upon HCMV infection [68, 111, 112]. The UL83-IFI16 interaction leads to a block of IFI16 oligomerization upon activation in the nucleus, thus dampening the transcriptional activation of cytokines [68]. Moreover, there is evidence that UL83 hijacks the DNA binding ability of IFI16 and forms a complex with the HCMV major immediate early promoter (MIEP) to kickstart viral transcription at early stages of infection [111, 112]. These results suggest that UL83 prevents the activation of the cGAS-STING and IFI16-STING axis, thus dampening the transcriptional activation of cytokines, while actively contributing to the expression of IE genes at early stages of HCMV infection.
KSHV
The KSHV tegument protein ORF52 is conserved in other members of the γ-herpesvirinae, namely EBV, MHV68, and RRV. KSHV ORF52 selectively interacts with cGAS, thereby blocking its binding to DNA [65]. This eventually results in reduced phosphorylation of IRF3 and transcriptional activation of IFNβ mRNA. The KSHV protein latency associated nuclear antigen (LANA) is localized exclusively to the nucleus in infected cells and is essential for maintenance of KSHV latency [113]. However, during reactivation a short isoform of LANA devoid of the N-terminal domain (LANA-Δ161) is expressed. This short isoform lacks the nuclear localization signal and localizes in the cytoplasm, where it interacts with cGAS in the PEL cell line BCBL-1 [60]. In HeLa cells, cytoplasmic LANA leads to reduced cGAMP production and promotes reactivation of KSHV from latency.
Cushion the STING: herpesviruses smother the key adaptor
Due to its instrumental role in the innate immune response, STING is consequently a prime target for herpesviruses (Fig. 3). Until now, viral proteins of HSV-1, HCMV, MCMV, KSHV, and MHV68 have been unveiled as antagonists of this crucial adaptor protein. On the other hand, parts of STING’s signaling activity can also be exploited by herpesviruses, as revealed by studies on the MCMV antagonist m152.
Cushion the STING: herpesviruses smother the key adaptor. Activity of the STING protein is manipulated by all members of the herpesvirus family (α: red, β: blue, γ: green). They either redirect it for degradation (black arrow-headed line), modulate the translocation to the Golgi apparatus or prevent its ubiquitination upon activation. The black blind-ended lines indicate inhibition. Solid black lines represent studies with mechanistic insights, while dashed black lines indicate findings from descriptive studies
Herpesvirus deubiquitinases
The tegument protein and deubiquitinase (DUB) ORF64 of MHV68 inhibits STING-dependent signaling in a DUB-dependent fashion. An MHV68 mutant lacking ORF64 induces higher STING-dependent responses in primary macrophages compared with wild-type (WT) MHV68. Notably, the presence of ORF64 is dispensable in STING−/− mice indicating that ORF64 acts on the level of or upstream of STING [74]. Assays to show or exclude an effect on cGAS were not performed and the detailed mechanism of ORF64 antagonism needs to be further investigated. However, a similar phenotype was obtained for the MHV68 ORF64 homologue KSHV ORF64 in the cancer monocytic cell line THP-1. In addition, expression of the HSV-1 and MCMV DUBs VP1-2 and M48, respectively, was shown to inhibit type I IFN secretion in primary macrophages to the same extent as MHV68 ORF64 [74]. Consistent with these observations, overexpression of HCMV UL48 was reported to inhibit signaling downstream of STING, TRAF6, TRAF3, IRAK1, IRF7 in a DUB-dependent manner [73].
HSV-1
The relationship between HSV-1 and STING is highly cell type- and context-dependent. It was reported that the STING protein is stabilized in cancer-derived HEP-2 and HeLa cells infected with HSV-1 [108]. This stabilization is dependent on the viral proteins ICP0, ICP4 and the US3 protein kinase, since infection with the respective HSV-1 mutants led to degradation of STING upon infection. However, in human embryonic lung fibroblasts (HEL), STING protein levels were unaffected by ICP0. Interestingly, HSV-1 infection of STING knockdown HEP-2 and HeLa cells resulted in significantly decreased viral titers. In contrast, depletion of STING in HEL cells did significantly increase the viral load after HSV-1 infection [108]. These results suggest a pivotal role of the STING protein for optimal cell-type specific replication of HSV-1, and that the HSV-1 antagonist ICP0 affects the stability of STING in a cell-type dependent manner.
The tegument protein VP11/12, encoded by UL46, was reported to mediate the degradation of STING in both HEL and HEP-2 cells [69]. This seems to be directed by an interaction of UL46 and STING and a yet unidentified mechanism. Consequently, an HSV-1 mutant lacking UL46 induces elevated transcript levels of IFNβ and ISG56 [69]. These combined findings implicate a complex interplay between HSV-1 proteins and the usurpation or inactivation of the STING protein.
HCMV and MCMV
The HCMV glycoprotein US9 localizes to the mitochondria and the ER upon overexpression and targets the RIG-I adaptor molecule mitochondrial antiviral signaling protein (MAVS) and STING [72]. Upon overexpression of US9 in 293T cells the membrane potential and membrane integrity of the mitochondria are disrupted, which are prerequisites for the induction of efficient MAVS signaling. Moreover, US9 prevents the dimerization of STING. Consistent with these observations, HCMV lacking the region US7-16 induces stronger MAVS- and STING-dependent responses [72].
The HCMV protein, IE86 (also known as IE2, encoded by UL122), was reported to negatively affect IFNβ mRNA transcription [114]. In a follow-up study using electrophoretic mobility shift assay (EMSA), stably expressed IE86 was shown to disrupt the binding of the transcription factor NF-κB to the promoter region of IFNβ, IL6, IL8 and RANTES [115]. Since IE86 is predominantly expressed in the nucleus, the authors suggested that IE86 blocks NF-κB mediated transcription [115]. Interestingly, a recent study showed that stably expressed IE86 led to reduced levels of STING protein, which was restored upon treatment with the proteasome inhibitor MG132 [70]. IE86 did not affect STING mRNA levels, suggesting that IE86 induces proteasomal degradation of STING. Accordingly, the authors observed that reduced protein levels of STING led to diminished activation of TBK1 and the transcription factors IRF3 and NF-κB. Nonetheless, an interaction between IE86 and STING in the context of HCMV infection could not be detected [70], and since IE86 is localized to the nucleus, a direct effect of IE86 on STING is unlikely. Taken altogether, only overexpression studies have been published for IE86 so far and the mechanisms by which IE86 induces proteasomal degradation of STING and how IE86 affects NF-κB mediated transcription need further investigation.
Another tegument protein of HCMV, UL82 (also known as pp71), localizes to the nucleus or the cytoplasm depending on its phosphorylation status at amino acid T223 [116]. Phosphorylated UL82 localizes to the cytoplasm while unphosphorylated UL82 is exclusively detected in the nucleus. A recent study showed that UL82 interacts with STING and eventually disrupts the translocation of STING to the Golgi apparatus in 293T cells [71]. Stable expression of UL82 in HFF leads to a complete block of STING translocation from the ER to the Golgi compartment. Expression of WT UL82, but not T223A UL82, impairs the activation of TBK1 and IRF3, resulting in dampened induction of type I IFN transcription in fibroblasts. Likewise, infection of HFF with HCMV lacking UL82 led to stronger induction of STING-dependent responses compared to WT HCMV [71].
MCMV is equipped with another manipulator of STING. The MCMV m152 protein, which is an ER-resident type I transmembrane protein, has been previously reported to efficiently thwart both NK- and T cell-dependent immune responses [117,118,119,120,121]. We recently showed that m152 targets yet another arm of the host’s immune response. At a very early time point after MCMV infection, m152 perturbs the translocation of activated STING from the ER to the Golgi compartment and thereby delays the type I IFN response to MCMV infection in macrophages and fibroblasts [39]. Interestingly, m152 has no effect on STING-mediated NF-κB activation, and through detailed analysis of m152 antagonism of STING we revealed that STING activates the NF-κB signaling from the ER prior to its translocation to the Golgi compartment. Strikingly, this STING-dependent NF-κB response is beneficial for the early stages of MCMV infection in fibroblasts. This study is a vivid example of how studies on viral proteins can lead to new insights into cellular mechanisms and responses. At the same time, it highlights the delicate balance between escape from host defense and the manipulation of said host responses to the advantage of the virus.
KSHV
The KSHV protein viral interferon regulatory factor 1 (vIRF1), encoded by ORF K9, binds directly to STING without affecting its translocation, but prevents its association with TBK1 upon reactivation of KSHV. This results in reduced phosphorylation of STING and, subsequently, lower IFNβ mRNA levels [59].
Take out the middleman-herpesviruses antagonize TBK1
TANK-binding kinase 1 (TBK1) is an ubiquitously expressed kinase of the IkB kinase family. It plays a central communication role between activated sensors and adaptor proteins and the transcription factor IRF3 [28, 122]. Only a few antagonists of TBK1 have been identified so far, among them several proteins of HSV-1 and a single MHV68 protein (Fig. 4).
Take out the middleman-herpesviruses antagonize TBK1. HSV-1 (red) and MHV68 (green) proteins inhibit the activation or activity of TBK1 to dampen the downstream signaling cascade (black blind-ended lines). Solid black lines represent studies with mechanistic insights, while dashed black lines indicate findings from descriptive studies
HSV-1
The γ134.5 protein of HSV-1 is known to facilitate viral replication by countering the translation arrest machinery upon infection [123, 124]. Ma et al. described that γ134.5 interacts via its N-terminal domain with TBK1 in an overexpression context. In addition, HSV-1 (F strain) lacking γ134.5 failed to replicate in the presence of TBK1, while HSV-1 expressing the N-terminus of γ134.5 could replicate and overcome the TBK1-mediated restriction of HSV-1 replication in vitro and in vivo [75]. A separate study in 2017 could confirm the interaction between γ134.5 and TBK1 upon overexpression; however, an interaction with endogenous TBK1 was not detected upon infection with neither the 17 + nor the F strain [76]. It is worth noting that this later study used human fibroblasts (HFF), compared to mouse fibroblasts (MEF) in the earlier study, and therefore, a cell-type-specific effect may explain the findings, or weak or transient interaction between TBK1 and γ134.5. In addition, the proposed N-terminal TBK1-binding domain of γ134.5 was dispensable for the impact on IRF3 phosphorylation and replication in vitro and in vivo, respectively [76]. However, while Ma et al. used Balb/c mice for their study, Manivanh and colleagues made use of B6J mice, which may explain the opposing observations in vivo. The authors postulated that the lack of γ134.5 also alters ICP0 protein levels upon infection of HFF, thus the effects seen on IRF3 activation are likely due to the inability to sustain ICP0 expression [76]. In conclusion, further studies are necessary to fully understand the impact of HSV-1 γ134.5 on the innate immune response.
The HSV-1 protein Us11 was shown to interact with heat shock protein 90 (Hsp90) in a yeast-two-hybrid screen [78]. In MEF, via its interaction with Hsp90, Us11 prevents the interaction between TBK1 and IRF3 in the context of HSV-1 infection, resulting in disruption of the TBK1 signalosome and abolishment of IRF3 phosphorylation. Consistently, the lack of Us11 led to lower viral titres in MEF [78]. Interestingly, the phenotype was only partially rescued in TBK1-deficient cells, indicating that Us11 may contribute to other immune evasion strategies of HSV-1.
As mentioned above, HSV-1 VP11/12 (encoded by UL46) was shown to block STING signaling by mediating degradation of STING [69]. Using a GST-pulldown assay, they also showed that the C-terminal domain of UL46 interacts with TBK1 [69]. However, it is not yet known if this interaction also affects host signaling or if this interaction is present in the context of infection. The IE protein of HSV-1, ICP27, negatively regulates the type I IFN response by targeting TBK1-mediated IRF3 phosphorylation [77]. While ICP27 interacts with phosphorylated TBK1 in WT THP-1 cells, this interaction was abolished in STING−/− THP-1 cells. This led to the conclusion that ICP27 is recruited to the TBK1-activated STING signalosome, and this was dependent on the RGG motif in ICP27. However, ICP27 also inhibits signaling downstream of the RIG-I adaptor protein MAVS and the TLR adaptor protein TRIF [77], indicating that the effects seen are not exclusively STING-dependent. Thus, ICP27 interaction with activated TBK1 could potentially block signaling downstream of multiple PRR.
MHV68
The MHV68 tegument protein ORF11 was reported to be important for lytic replication of MHV68, but dispensable for the establishment of infection [125]. This observation may correlate with the report that stable expression of MHV68 ORF11 can inhibit IFNβ production in fibroblasts and macrophages by targeting signaling downstream of MAVS, RIG-I, STING and TRIF, but not downstream of IRF3 [79]. In agreement with these observations, ORF11 associates with the kinase domain of TBK1 upon MHV68 infection, thus blocking the phosphorylation and activation of the transcription factor IRF3 [79].
Seize the tail: herpesviruses converge on the transcription factors
The transcription factors IRF3 and NF-κB are master regulators of the innate immune response. STING-dependent signaling, RNA-dependent RLR signaling and TLR signaling converge on the activation of these transcription factors and their subsequent nuclear translocation and binding to promoter regions of cytokine genes. Not surprisingly, their major role in establishment of an antiviral state in the host has led to the evolution of a variety of herpesviral strategies to counteract these transcription factors (Fig. 5). So far, multiple antagonists for each of the herpesviruses HSV-1, VZV, HCMV, and KSHV have been uncovered, while only a single antagonist has been reported for MCMV (M35), HHV-6 (IE1), EBV (BGLF4), and MHV68 (ORF36).
Seize the tail: herpesviruses converge on the transcription factors. Herpesviruses (α: red, β: blue, γ: green) have evolved diverse strategies to prevent activation, nuclear translocation or transcriptional activity of the transcription factors IRF3 and NF-κB to suppress transcription of type I IFN and proinflammatory cytokines. The black blind-ended lines indicate inhibition. The black arrow-headed lines indicate degradation mediated by the herpesviral protein. Solid black lines represent studies with mechanistic insights, while dashed black lines indicate findings from descriptive studies
Conserved herpesviral protein kinases
One subgroup of herpesviral protein kinases is conserved throughout the α-, β- and γ-herpesviruses. HSV-1 UL13 is one representative of this group with homologues encoded in other herpesviruses: VZV ORF47, HCMV UL97, HHV-6 U69, EBV BGLF4, KSHV ORF36, and MHV68 ORF36. While HHV-6 U69 has not yet been reported to be involved in immune evasion, the other UL13 kinase family members target IRF3 in different manners. With a yeast-two-hybrid screen the EBV protein BGLF4 was identified to interact with the C-terminus of IRF3 [86]. Further analysis showed that this interaction is dependent on the kinase activity of BGLF4; however, BGLF4 does not affect phosphorylation and nuclear translocation of IRF3 nor the association of IRF3 with the CREB-binding protein (CBP), which is a prerequisite for transcriptional activation of IRF3. Instead, BGLF4 phosphorylates IRF3 at S123, S173 and T180, three residues of IRF3 within or close to the DNA binding domain of IRF3, thus preventing the promoter binding activity of IRF3. siRNA-mediated knockdown of BGLF4 in EBV-reactivated cells enhances IRF3-responsive reporter activities [86]. Simultaneously, Hwang et al. identified MHV68 ORF36 as a novel inhibitor of the type I IFN response using an in vivo screen in Balb/c mice with an MHV68 transposon mutant library [87]. Similar to EBV BGLF4, MHV68 ORF36 does not target activation or nuclear translocation of IRF3. However, in contrast to BGLF4, ORF36 interacts with active IRF3, eventually disrupting the interaction between IRF3 and CBP and thereby blocking IRF3-mediated transcription [87]. In addition, the authors used an EMSA to show that infection of NIH3T12 cells with an ORF36-deletion mutant enhances the recruitment of CBP and RNA Polymerase II to the IFNβ promoter. More importantly, using 293T and NIH3T3 cells, the authors could show that the MHV68 ORF36 homologues EBV BGLF4, KSHV ORF36, HCMV UL97, and HSV-1 UL13 inhibit IFNβ promoter induction to the same extent as MHV68 ORF36 [87]. VZV ORF47 was reported to target IRF3 as well [82]. In contrast to EBV BGLF4 and MHV68 ORF36, VZV ORF47 interacts with IRF3 to prevent its phosphorylation and homodimerization in a kinase-dependent manner upon infection of 293 cells. Taken together, the UL13 kinase family members EBV BGLF4, MHV68 ORF36 and VZV ORF47 target IRF3, but they use different mechanisms to achieve this goal. For the other homologues, HCMV UL97, HSV-1 UL13 and KSHV ORF36, which inhibit IFNβ promoter activation to the same extent as MHV68 ORF36, the mechanism of action merits further investigation.
VZV
Since models to study VZV infection in vitro were developed only recently, most studies reported in this review utilize overexpression analysis of individual VZV proteins. Nonetheless, several VZV proteins were reported that target the innate immune response. The IE protein VZV ORF61, homologue of HSV-1 ICP0, was reported to target IRF3 [83]. Using luciferase assay and immunoblot analysis in 293T, the authors showed that overexpressed ORF61 specifically targets phosphorylated IRF3 in an ORF61 RING-finger-dependent manner and redirects it for proteasomal degradation. However, while the authors also observed by luciferase assay that ORF61 inhibited signaling when coexpressed with IκBα, its effect on IRF signaling was stronger and thus the authors focused on its impact on IRF3 [83]. Interestingly, another study highlighted that ORF61 targets NF-κB activation by inhibiting the ubiquitination of IκBα [90]. Infection with VZV (pOka strain) and further stimulation with TNFα showed that IκBα degradation is inhibited in an ORF61 RING-finger dependent manner. However, whether VZV ORF61 affects either IRF3 or NF-κB directly or if the observed phenotypes are indirect needs further investigation. In 2010, Sen et al. screened several VZV IE proteins for the capacity to modulate induction of the type I IFN response [81]. They reported that overexpression of IE62, a homologue of HSV-1 ICP4, blocked TBK1-mediated IRF3 phosphorylation in 293 cells, while signaling downstream of the constitutively active IRF3 mutant (IRF3-5D) was not inhibited. Using Serine/Threonine mutants of IRF3, the authors showed that IE62 blocks phosphorylation of IRF3 at positions S396, S398 and S402, which are crucial residues for IRF3 activation and homodimerization [81].
HSV-1
HSV-1 is reported to encode a variety of proteins targeting the transcription factors IRF3 and NF-κB. HSV-1 UL42 inhibits TNFα induced NF-κB activation [92]. During infection of 293T cells with HSV-1 (F strain), UL42 interacts with NF-κB and thereby prevents the nuclear translocation of activated NF-κB. Similarly, the UL24 protein interacts with NF-κB upon HSV-1 infection (KOS strain) and likewise prevents its nuclear translocation [91]. Analogous to VZV ORF61, HSV-1 UL36 was reported to diminish cGAS-STING mediated signaling by targeting IκBα [126]. Overexpression of UL36 in 293T cells prevented the ubiquitination of IκBα upon stimulation of the cGAS-STING signaling pathway, thereby blocking the activation and nuclear translocation of NF-κB. Using luciferase assays, the tegument protein VP16 was reported to downregulate IRF3 and NF-κB signaling [96]. Focusing on IRF3, the authors showed that overexpression of VP16 in 293T cells blocked the recruitment of CBP to IRF3, thus preventing IRF3-mediated transcription [96], which is reminiscent of MHV68 ORF36. The effect of VP16 on NF-κB was not further addressed in this study. While VP16 seems to prevent the association of IRF3 with CBP, the US3 kinase of HSV-1 significantly inhibits the activation of the type I IFN response by targeting IRF3 and NF-κB activation [94, 95]. Depending on its kinase activity, overexpressed US3 atypically hyperphosphorylates IRF3 and NF-κB in 293T cells, thus inhibiting their dimerization and nuclear translocation. In agreement, infection of 293T cells with a US3 K220M or D305A mutant, lacking the kinase activity of US3, induced more robust IFNβ and proinflammatory cytokine responses compared to WT HSV-1 [94, 95]. Moreover, the VP24 protein of HSV-1, encoded by UL26, inhibits IRF- but not NF-κB signaling downstream of cGAS-STING signaling [80]. The authors used luciferase-based assays and Co-IPs in 293T cells to show that overexpressed VP24 interacts with IRF3 to dampen its activation upon ISD stimulation.
β-herpesviruses (HHV-6, HCMV, MCMV)
Upon overexpression, the HHV-6 protein IE1 impairs activation and nuclear translocation of IRF3 in 293T cells, while NF-κB signaling remains intact [88], but the mechanism is unknown. The HCMV tegument protein UL26 blocks phosphorylation of the IKK complex, thereby inhibiting the activation of NF-κB, while IRF3 signaling is not affected. Infection of human lung fibroblasts with an HCMV mutant lacking UL26 leads to stronger induction of IL6 and TNFα transcription compared to infection with HCMV WT [93]. Following a luciferase-based screen with MCMV tegument and IE proteins in NIH3T3 cells, we identified the tegument protein M35 as an antagonist downstream of multiple PRR [97]. M35 is required for efficient replication of MCMV in macrophages, whereas replication in non-immune cells is not affected by its absence. This growth deficit was abolished in the absence of the interferon-α/β receptor IFNAR, indicating that the modulation of type I IFN responses by M35 is crucial for the ability of MCMV to replicate in macrophages. Further analysis showed that M35 is localized to the nucleus shortly after MCMV infection and inhibits transcription of type I IFN. In accordance, mice infected with an MCMV mutant lacking M35 respond with elevated levels of type I IFN and viral replication is significantly impaired in the spleen and salivary glands. These in vivo studies highlight the necessity for MCMV to counteract the type I IFN response to establish an infection. However, further investigation is required to understand the exact mechanism by which M35 is exerting this effect.
KSHV
The KSHV protein K-bZIP, encoded by ORF8, is expressed with IE kinetics [127], and, when overexpressed, it inhibits IFR3-mediated transcription [85]. Upon transfection of K-bZIP and infection with Sendai Virus to induce IRF3- and NF-κB activation, nuclear translocation of the transcription factors was not modulated. However, by EMSA it was shown that K-bZIP binds strongly to the IRF3 binding sites of the IFNβ promoter, thereby preventing association of IRF3, while NF-κB or ATF2 binding to the IFNβ promoter remained unaffected [85]. As mentioned above, the KSHV protein vIRF1 inhibits STING-dependent signaling by targeting the interaction of STING and TBK1, preventing the phosphorylation of STING and downstream signaling activation [59]. vIRF1 was also shown to inhibit IFNβ activation by blocking the recruitment of CBP to IRF [84]. Its effect on the STING-TBK1 axis and IRF3/CBP are proposed to be achieved by distinct mechanisms [59].
Concluding remarks
Due to their large genome and close association with their specific hosts, herpesviruses are renowned for their multilayered ability to evade host immune responses. A range of PRR have been described to recognize the very first stage of herpesviral infection and most recently the DNA sensing pathway, predominantly mediated by cGAS-STING-dependent signaling, has been at the forefront of efforts to understand how the host combats viral infection. In turn, herpesvirus proteomes have been systematically mined and in the last few years a plethora of viral proteins have been identified to specifically antagonize this key innate pathway. Remarkably, at least one antagonist, and in many cases multiple, has been characterized for each level of the signaling pathway, from directly inhibiting sensor binding to DNA to subverting STING function to disruption of transcription factor induction of type I IFN signaling. Since cytosolic DNA sensing itself is a relatively newly identified concept, the field of viral evasins specifically targeting this pathway is still in its infancy. Many of the studies we have described are the results of single studies, which lack verification from multiple groups, or demonstrate a highly cell and/or time and/or stimulation dependent phenotype. In addition, differences in virus strains can also contribute to opposing observations. Furthermore, mechanistic insight is missing for most of these antagonists and the overwhelming majority, acknowledged due to restriction by host specificity, lack confirmation of the phenotype in the host. We have endeavored to illustrate the context of these studies and encourage the reader to keep these in mind.
Notably, while so many viral antagonists of DNA sensing and innate signaling have been identified, induction of innate signaling upon viral infection is still evident. Does this then suggest that the activation of specific components of innate signaling are exploited by the virus to promote its replication in the host and ultimate dissemination throughout the population? There is emerging evidence to support this hypothesis, such as the finding that the HCMV UL83 protein exploits IFI16 to activate viral transcription, as well as our own studies with MCMV m152, which targets STING translocation to selectively shut down the antiviral type I IFN response while allowing the proviral NF-κB-mediated response to proceed. Alternatively, is this phenomenon merely indicative of the constant arms race between the herpesvirus and its host, a product of the close association from coevolution, where the virus wins some and the host wins some? Only time, and many more carefully executed studies, will tell.
References
Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E (2009) The order herpesvirales. Arch Virol 154(1):171–177
Pellett P, Roizman B (2007) Herpesviridae: a brief introduction. In: Howley P (ed) Fields virology, 5th edn. Lippincott, Philadelphia, pp 2480–2499
Kutok JL, Wang F (2006) Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 1:375–404
Roizman B, Taddeo B (2007) The strategy of herpes simplex virus replication and takeover of the host cell. In: Arvin A, Campadelli-Fiume G, Mocarski E et al (eds) Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, Chap. 13
Mocarski E (2007) Betaherpesvirus genes and their functions. In: Arvin A, Campadelli-Fiume G, Mocarski E et al (eds) Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, Chap. 15
Patel JR, Heldens J (2005) Equine herpesviruses 1 (EHV-1) and 4 (EHV-4)—epidemiology, disease and immunoprophylaxis: a brief review. Vet J 170(1):14–23
Yates WD (1982) A review of infectious bovine rhinotracheitis, shipping fever pneumonia and viral-bacterial synergism in respiratory disease of cattle. Can J Comp Med 46(3):225–263
Nauwynck HJ (1997) Functional aspects of Aujeszky’s disease (pseudorabies) viral proteins with relation to invasion, virulence and immunogenicity. Vet Microbiol 55(1–4):3–11
Nair V (2005) Evolution of Marek’s disease—a paradigm for incessant race between the pathogen and the host. Vet J (London, England: 1997) 170(2):175–183
Reddehase MJ, Lemmermann NAW (2018) Mouse model of cytomegalovirus disease and immunotherapy in the Immunocompromised host: predictions for medical translation that survived the “Test of Time”. Viruses 2018;10(12):693
Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T (2014) Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159(5):863–870
Mori Y, Yamanishi K (2007) HHV-6A, 6B, and 7: pathogenesis, host response, and clinical disease. In: Arvin A, Campadelli-Fiume G, Mocarski E et al (eds) Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, Chap. 46
Dittmer DP, Damania B (2013) Kaposi sarcoma associated herpesvirus pathogenesis (KSHV)—an update. Curr Opin Virol 3(3):238–244
Hjalgrim H, Smedby KE, Rostgaard K, Molin D, Hamilton-Dutoit S, Chang ET, Ralfkiaer E, Sundstrom C, Adami HO, Glimelius B, Melbye M (2007) Infectious mononucleosis, childhood social environment, and risk of Hodgkin lymphoma. Cancer Res 67(5):2382–2388
Barton E, Mandal P, Speck SH (2011) Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu Rev Immunol 29:351–397
Dittmer DP, Damania B, Sin SH (2015) Animal models of tumorigenic herpesviruses—an update. Curr Opin Virol 14:145–150
Goldner T, Hewlett G, Ettischer N, Ruebsamen-Schaeff H, Zimmermann H, Lischka P (2011) The novel anticytomegalovirus compound AIC246 (Letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J Virol 85(20):10884–10893
Lischka P, Hewlett G, Wunberg T, Baumeister J, Paulsen D, Goldner T, Ruebsamen-Schaeff H, Zimmermann H (2010) In vitro and in vivo activities of the novel anticytomegalovirus compound AIC246. Antimicrob Agents Chemother 54(3):1290–1297
Piret J, Drouot E, Boivin G (2017) Antiviral Drug Resistance in Herpesviruses. In: Gotte M, Berghuis A, Matlashewski G, Wainberg M, Sheppard D (eds) Handbook of antimicrobial resistance. Springer, New York, pp 1–32
Vazquez M, LaRussa PS, Gershon AA, Steinberg SP, Freudigman K, Shapiro ED (2001) The effectiveness of the varicella vaccine in clinical practice. N Engl J Med 344(13):955–960
Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ (2013) Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341(6148):903–906
Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339(6121):786–791
Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM (2014) The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J Immunol 193(5):2394–2404
Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V (2013) cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498(7454):380–384
West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS (2015) Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520(7548):553–557
Yang H, Wang H, Ren J, Chen Q, Chen ZJ (2017) cGAS is essential for cellular senescence. Proc Natl Acad Sci USA 114(23):E4612–E4620
Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, Jiang Y, Fei Y, Zhu C, Tan R, Jungblut P, Pei G, Dorhoi A, Yan Q, Zhang F, Zheng R, Liu S, Liang H, Liu Z, Yang H, Chen J, Wang P, Tang T, Peng W, Hu Z, Xu Z, Huang X, Wang J, Li H, Zhou Y, Liu F, Yan D, Kaufmann SHE, Chen C, Mao Z, Ge B (2018) Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563(7729):131–136
Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347(6227):aaa2630
Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V (2013) Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503(7477):530–534
Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, Rehwinkel J (2015) Viruses transfer the antiviral second messenger cGAMP between cells. Science 349(6253):1228–1232
Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G, Loew D, Dalod M, Thery C, Manel N (2015) Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science 349(6253):1232–1236
Ishikawa H, Barber GN (2011) The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci 68(7):1157–1165
Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S (2010) The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33(5):765–776
Zhang J, Hu MM, Wang YY, Shu HB (2012) TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 287(34):28646–28655
Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, Yu H, Pan Y, Wang C (2014) The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41(6):919–933
Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, Reinert L, Cai Y, Jensen SB, Skouboe MK, Nyengaard JR, Thompson CB, Lebbink RJ, Sen GC, van Loo G, Nielsen R, Komatsu M, Nejsum LN, Jakobsen MR, Gyrd-Hansen M, Paludan SR (2018) Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J 37(8): e97858
Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S (2009) Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA 106(49):20842–20846
Abe T, Barber GN (2014) Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol 88(10):5328–5341
Stempel M, Chan B, Juranić Lisnić V, Krmpotić A, Hartung J, Paludan SR, Füllbrunn N, Lemmermann NA, Brinkmann MM (2019) The herpesviral antagonist m152 reveals differential activation of STING-dependent IRF and NF-κB signaling and STING’s dual role during MCMV infection. EMBO J. https://doi.org/10.15252/embj.2018100983
Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11(11):997–1004
Dell’Oste V, Gatti D, Gugliesi F, De Andrea M, Bawadekar M, Lo Cigno I, Biolatti M, Vallino M, Marschall M, Gariglio M, Landolfo S (2014) Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage. J Virol 88(12):6970–6982
Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM (2015) cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci USA 112(14):E1773–E1781
Diner BA, Lum KK, Toettcher JE, Cristea IM (2016) Viral DNA sensors IFI16 and Cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. MBio 7(6):e01553–e01516
Jonsson KL, Laustsen A, Krapp C, Skipper KA, Thavachelvam K, Hotter D, Egedal JH, Kjolby M, Mohammadi P, Prabakaran T, Sorensen LK, Sun C, Jensen SB, Holm CK, Lebbink RJ, Johannsen M, Nyegaard M, Mikkelsen JG, Kirchhoff F, Paludan SR, Jakobsen MR (2017) IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat Commun 8:14391
Almine JF, O’Hare CA, Dunphy G, Haga IR, Naik RJ, Atrih A, Connolly DJ, Taylor J, Kelsall IR, Bowie AG, Beard PM, Unterholzner L (2017) IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat Commun 8:14392
Lugrin J, Martinon F (2018) The AIM2 inflammasome: sensor of pathogens and cellular perturbations. Immunol Rev 281(1):99–114
Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12(10):959–965
Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448(7152):501–505
Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang D-W, Lempicki RA, Baseler MW, Veenstra TD, Young HA, Lane HC, Imamichi T (2011) Cutting edge: Ku70 Is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol 186(8):4541–4545
Ishikawa H, Ma Z, Barber GN (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461(7265):788–792
Johnson KE, Bottero V, Flaherty S, Dutta S, Singh VV, Chandran B (2014) IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog 10(11):e1004503
Ansari MA, Dutta S, Veettil MV, Dutta D, Iqbal J, Kumar B, Roy A, Chikoti L, Singh VV, Chandran B (2015) Herpesvirus genome recognition induced acetylation of nuclear IFI16 Is essential for its cytoplasmic translocation, inflammasome and IFN-beta responses. PLoS Pathog 11(7):e1005019
Gariano GR, Dell’Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S (2012) The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog 8(1):e1002498
Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, Stetson DB (2016) The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity 45(2):255–266
Biolatti M, Dell’Oste V, Pautasso S, Gugliesi F, von Einem J, Krapp C, Jakobsen MR, Borgogna C, Gariglio M, De Andrea M, Landolfo S (2018) Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J Virol 92:(6)
Paijo J, Doring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, Witte G, Messerle M, Hornung V, Kaever V, Kalinke U (2016) cGAS senses human cytomegalovirus and induces Type I interferon responses in human monocyte-derived cells. PLoS Pathog 12(4):e1005546
Lio CW, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, Pham E, Benedict CA, Sharma S (2016) cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. J Virol 90(17):7789–7797
Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM (2014) Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505(7485):691–695
Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B (2015) Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci USA 112(31):E4306–E4315
Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF (2016) Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci USA 113(8):E1034–E1043
Huang J, You H, Su C, Li Y, Chen S, Zheng C (2018) Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J Virol 92:(15)
Zhang J, Zhao J, Xu S, Li J, He S, Zeng Y, Xie L, Xie N, Liu T, Lee K, Seo GJ, Chen L, Stabell AC, Xia Z, Sawyer SL, Jung J, Huang C, Feng P (2018) Species-specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host Microbe 24(2):234–248 e235
Su C, Zheng C (2017) Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff protein, UL41. J Virol 91(6):JVI-02414
Huang ZF, Zou HM, Liao BW, Zhang HY, Yang Y, Fu YZ, Wang SY, Luo MH, Wang YY (2018) Human Cytomegalovirus protein UL31 Inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microbe 24(1):69–80 e64
Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F (2015) Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe 18(3):333–344
Orzalli MH, DeLuca NA, Knipe DM (2012) Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA 109(44):E3008–E3017
Cuchet-Lourenco D, Anderson G, Sloan E, Orr A, Everett RD (2013) The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J Virol 87(24):13422–13432
Li T, Chen J, Cristea IM (2013) Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14(5):591–599
Deschamps T, Kalamvoki M (2017) Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J Virol 91(16):JVI-00535
Kim JE, Kim YE, Stinski MF, Ahn JH, Song YJ (2017) Human cytomegalovirus IE2 86 kda protein induces STING degradation and inhibits cGAMP-Mediated IFN-beta Induction. Front Microbiol 8:1854
Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, Luo WW, Li S, Luo MH, Wang YY, Shu HB (2017) Human Cytomegalovirus tegument protein UL82 inhibits STING-Mediated signaling to evade antiviral immunity. Cell Host Microbe 21(2):231–243
Choi HJ, Park A, Kang S, Lee E, Lee TA, Ra EA, Lee J, Lee S, Park B (2018) Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses. Nat Commun 9(1):125
Kumari P, Saha I, Narayanan A, Narayanan S, Takaoka A, Kumar NS, Tailor P, Kumar H (2017) Essential role of HCMV deubiquitinase in promoting oncogenesis by targeting anti-viral innate immune signaling pathways. Cell Death Dis 8(10):e3078
Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, O’Hare P, Adler H, Sun R, Ploegh HL, Damania B, Upton JW, Fitzgerald KA, Paludan SR (2015) Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol 194(4):1819–1831
Ma Y, Jin H, Valyi-Nagy T, Cao Y, Yan Z, He B (2012) Inhibition of TANK binding kinase 1 by herpes simplex virus 1 facilitates productive infection. J Virol 86(4):2188–2196
Manivanh R, Mehrbach J, Knipe DM, Leib DA (2017) Role of herpes simplex virus 1 γ34.5 in the regulation of IRF3 signaling. J Virol 91(23):e01156–e01117
Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, Mettenleiter T, Chen ZJ, Knipe DM, Sandri-Goldin RM, Enquist LW, Hartmann R, Mogensen TH, Rice SA, Nyman TA, Matikainen S, Paludan SR (2016) HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 35(13):1385–1399
Liu X, Main D, Ma Y, He B (2018) Herpes Simplex Virus 1 Inhibits TANK-Binding Kinase 1 through Formation of the Us11-Hsp90 Complex. J Virol 92(14):e00402–e00418
Kang HR, Cheong WC, Park JE, Ryu S, Cho HJ, Youn H, Ahn JH, Song MJ (2014) Murine gammaherpesvirus 68 encoding open reading frame 11 targets TANK binding kinase 1 to negatively regulate the host type I interferon response. J Virol 88(12):6832–6846
Zhang D, Su C, Zheng C (2016) Herpes Simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J Virol 90(12):5824–5829
Sen N, Sommer M, Che X, White K, Ruyechan WT, Arvin AM (2010) Varicella-zoster virus immediate-early protein 62 blocks interferon regulatory factor 3 (IRF3) phosphorylation at key serine residues: a novel mechanism of IRF3 inhibition among herpesviruses. J Virol 84(18):9240–9253
Vandevenne P, Lebrun M, El Mjiyad N, Ote I, Di Valentin E, Habraken Y, Dortu E, Piette J, Sadzot-Delvaux C (2011) The varicella-zoster virus ORF47 kinase interferes with host innate immune response by inhibiting the activation of IRF3. PLoS One 6(2):e16870
Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL (2011) Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J Virol 85(21):11079–11089
Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ Jr, Barber GN, Hiscott J (2001) HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20(7):800–811
Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L (2007) Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J Virol 81(20):10950–10960
Wang JT, Doong SL, Teng SC, Lee CP, Tsai CH, Chen MR (2009) Epstein-Barr virus BGLF4 kinase suppresses the interferon regulatory factor 3 signaling pathway. J Virol 83(4):1856–1869
Hwang S, Kim KS, Flano E, Wu TT, Tong LM, Park AN, Song MJ, Sanchez DJ, O’Connell RM, Cheng G, Sun R (2009) Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5(2):166–178
Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L (2007) Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J Virol 81(11):5737–5748
Ye R, Su C, Xu H, Zheng C (2017) Herpes simplex Virus 1 ubiquitin-specific protease UL36 abrogates NF-kappaB activation in DNA sensing signal pathway. J Virol 91(5):e02417–e02416
Whitmer T, Malouli D, Uebelhoer LS, DeFilippis VR, Fruh K, Verweij MC (2015) The ORF61 protein encoded by Simian varicella virus and Varicella-Zoster virus inhibits NF-kappaB signaling by interfering with IkappaBalpha degradation. J Virol 89(17):8687–8700
Xu H, Su C, Pearson A, Mody CH, Zheng C (2017) Herpes Simplex Virus 1 UL24 Abrogates the DNA Sensing Signal Pathway by Inhibiting NF-kappaB Activation. J Virol 91(7):e00025–e00017
Zhang J, Wang S, Wang K, Zheng C (2013) Herpes simplex virus 1 DNA polymerase processivity factor UL42 inhibits TNF-alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. Med Microbiol Immunol 202(4):313–325
Mathers C, Schafer X, Martinez-Sobrido L, Munger J (2014) The human cytomegalovirus UL26 protein antagonizes NF-kappaB activation. J Virol 88(24):14289–14300
Wang K, Ni L, Wang S, Zheng C (2014) Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-kappaB activation. J Virol 88(14):7941–7951
Wang S, Wang K, Lin R, Zheng C (2013) Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol 87(23):12814–12827
Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C (2013) Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol 87(17):9788–9801
Chan B, Goncalves Magalhaes V, Lemmermann NAW, Juranic Lisnic V, Stempel M, Bussey KA, Reimer E, Podlech J, Lienenklaus S, Reddehase MJ, Jonjic S, Brinkmann MM (2017) The murine cytomegalovirus M35 protein antagonizes type I IFN induction downstream of pattern recognition receptors by targeting NF-kappaB mediated transcription. PLoS Pathog 13(5):e1006382
Steain M, Slobedman B, Abendroth A (2010) Experimental models to study varicella-zoster virus infection of neurons. Curr Top Microbiol Immunol 342:211–228
Markus A, Lebenthal-Loinger I, Yang IH, Kinchington PR, Goldstein RS (2015) An in vitro model of latency and reactivation of varicella zoster virus in human stem cell-derived neurons. PLoS Pathog 11(6):e1004885
Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455(7213):674–678
Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC (2008) MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol 28(16):5014–5026
Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA 106(21):8653–8658
Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29(4):538–550
Tanaka M, Kato A, Satoh Y, Ide T, Sagou K, Kimura K, Hasegawa H, Kawaguchi Y (2012) Herpes simplex virus 1 VP22 regulates translocation of multiple viral and cellular proteins and promotes neurovirulence. J Virol 86(9):5264–5277
Zhao J, Zeng Y, Xu S, Chen J, Shen G, Yu C, Knipe D, Yuan W, Peng J, Xu W, Zhang C, Xia Z, Feng P (2016) A Viral Deamidase targets the helicase domain of RIG-I to block RNA-induced activation. Cell Host Microbe 20(6):770–784
Esclatine A, Taddeo B, Evans L, Roizman B (2004) The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc Natl Acad Sci USA 101(10):3603–3608
Johnson KE, Chikoti L, Chandran B (2013) Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol 87(9):5005–5018
Kalamvoki M, Roizman B (2014) HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci USA 111(5):E611–E617
Browne EP, Shenk T (2003) Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci USA 100(20):11439–11444
Abate DA, Watanabe S, Mocarski ES (2004) Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol 78(20):10995–11006
Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O’Keefe ES, Rout MP, Chait BT, Shenk T (2010) Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol 84(15):7803–7814
Biolatti M, Dell’Oste V, Pautasso S, von Einem J, Marschall M, Plachter B, Gariglio M, De Andrea M, Landolfo S (2016) Regulatory interaction between the cellular restriction factor IFI16 and viral pp65 (pUL83) modulates viral gene expression and IFI16 protein stability. J Virol 90(18):8238–8250
Weidner-Glunde M, Mariggio G, Schulz TF (2017) Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen: replicating and shielding viral DNA during viral persistence. J Virol 2017 91(14):e01083–e01016
Taylor RT, Bresnahan WA (2005) Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J Virol 79(6):3873–3877
Taylor RT, Bresnahan WA (2006) Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression. J Virol 80(21):10763–10771
Shen W, Westgard E, Huang L, Ward MD, Osborn JL, Chau NH, Collins L, Marcum B, Koach MA, Bibbs J, Semmes OJ, Kerry JA (2008) Nuclear trafficking of the human cytomegalovirus pp71 (ppUL82) tegument protein. Virology 376(1):42–52
Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL (2003) NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med 197(10):1245–1253
Krmpotic A, Messerle M, Crnkovic-Mertens I, Polic B, Jonjic S, Koszinowski UH (1999) The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J Exp Med 190(9):1285–1296
Ziegler H, Thale R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski UH (1997) A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6(1):57–66
Fink A, Renzaho A, Reddehase MJ, Lemmermann NA (2013) The p36 isoform of murine cytomegalovirus m152 protein suffices for mediating innate and adaptive immune evasion. Viruses 5(12):3171–3191
Krmpotic A, Busch DH, Bubic I, Gebhardt F, Hengel H, Hasan M, Scalzo AA, Koszinowski UH, Jonjic S (2002) MCMV glycoprotein gp40 confers virus resistance to CD8 + T cells and NK cells in vivo. Nat Immunol 3(6):529–535
Tanaka Y, Chen ZJ (2012) STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5(214):ra20
Chou J, Chen JJ, Gross M, Roizman B (1995) Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 134.5- mutants of herpes simplex virus 1. Proc Natl Acad Sci USA 92(23):10516–10520
Chou J, Roizman B (1992) The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc Natl Acad Sci USA 89(8):3266–3270
Boname JM, May JS, Stevenson PG (2005) Murine gammaherpesvirus 68 open reading frame 11 encodes a nonessential virion component. J Virol 79(5):3163–3168
Wang S, Wang K, Li J, Zheng C (2013) Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J Virol 87(21):11851–11860
Lin SF, Robinson DR, Miller G, Kung HJ (1999) Kaposi’s sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol 73(3):1909–1917
Acknowledgements
This work was supported by the SMART BIOTECS alliance between the Technische Universität Braunschweig and the Leibniz Universität Hannover, funded by the Ministry of Science and Culture (MWK) of Lower Saxony, Germany, and the Deutsche Forschungsgemeinschaft (DFG), BR3432/3-1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by Matthias J. Reddehase.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Special Issue on Immunological Imprinting during Chronic Viral Infection.
Rights and permissions
About this article
Cite this article
Stempel, M., Chan, B. & Brinkmann, M.M. Coevolution pays off: Herpesviruses have the license to escape the DNA sensing pathway. Med Microbiol Immunol 208, 495–512 (2019). https://doi.org/10.1007/s00430-019-00582-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-019-00582-0