Abstract
Aside from enteroviruses and other viruses, e.g., adenoviruses, which are known to be associated with idiopathic dilated cardiomyopathy (IDC), a cardiac tropism is also attributed to parvovirus B19 (PVB19). The purpose of the present study was to determine the prevalence of enterovirus, adenovirus and PVB19 genomes in the myocardium of adult patients with IDC and to analyze the significance of PVB19 with regard to the course of the disease, as compared to the other cardiotropic viruses. In 52 adult patients with IDC and 10 control patients with normal left ventricular ejection fraction (≥55%) undergoing coronary artery bypass surgery, myocardial tissue samples were investigated for enteroviral RNA using polymerase chain reaction (PCR) and Southern blot hybridization of the PCR product. Specific nested PCR was used to assess the prevalence of adenovirus and PVB19 DNA, in addition to sequencing of the latter. The clinical and echocardiographic course of the disease was followed for a mean (± SD) period of 21.1±9.5 months. Fourteen of the 52 patients (27%) were enterovirus-positive, 2/52 (4%) patients were adenovirus-positive, 14/52 (27%) patients were PVB19-positive, 8/52 (15%) patients were enterovirus plus PVB19-positive, and in 14/52 (27%) patients no viral genomes were found. Six patients died during the follow-up period, without any significant difference between the patient groups: 1/14 (7%) in the enterovirus-positive, 0/2 (0%) in the adenovirus-positive, 2/14 (14%) in the PVB19-positive, 1/8 (12.5%) in the enterovirus plus PVB19-positive, and 2/14 (14%) in the virus-negative group. PVB19 genome was found in 4 of the 10 (40%) control patients, but no enterovirus or adenovirus genomes were detected in these patients. In conclusion, in the myocardium of patients with IDC, PVB19 is detectable as frequently as enteroviral genome. PVB19-positive patients with IDC have a rather favorable prognosis and do not differ significantly from the other virus-positive or virus-negative patient groups with respect to survival. Finally, the pathogenetic and prognostic significance of PVB19 in IDC still remains unclear.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parvovirus B19 (PVB19) is the only known human pathogenic virus with a serological rate of infection of 71% in control groups [13]. It is associated with a broad spectrum of clinical disease manifestations [11, 20]. The most frequent disease caused by PVB19 in immunocompetent individuals is erythema infectiosum.
In patients with hematological disorders PVB19 has been described as a primary cause of aplastic crises [6, 11]. Following organ transplantation, infection with PVB19 can induce hematological disorders in immunocompromised patients [1, 7, 14, 28, 31, 33, 50], or infection may largely be responsible for allograft rejection [53]. In addition, PVB19 DNA was detected in 80–90% of kidney biopsy specimens from immunocompromised patients with focal segmental glomerulonephritis sclerosis [47].
There are several indicators attributing cardiotropic properties to PVB19, particularly in young children, but also in adults. First, PVB19 DNA has been found in the nuclei of fetal myocardial cells and is believed to contribute to the development of a hydrops fetalis due to virus-induced myocarditis [2, 10, 32, 34, 40, 50]. Second, with the use of polymerase chain reaction (PCR), parvoviral DNA has been detected in a small percentage of children with suspected myocarditis as well as in those who had undergone a rejection response following heart transplantation [46]. Third, Heegaard et al. [21] have published a report of a PVB19-associated myocarditis in an adult heart transplant recipient, determined by measuring both the concentration of IgM antibodies in serum and by the more specific method of detecting PVB19 DNA in endomyocardial biopsy tissue using nested-PCR [23]. Recently, PVB19 was visualized in myocardial endothelial cells by in situ hybridization in myocarditis of adult patients [29].
The most frequent viral pathogens found to be associated with idiopathic dilated cardiomyopathy (IDC) in immunocompetent adults are enteroviruses [9, 18, 27, 37, 49] and to a much lesser extent adenoviruses [36, 37]. However, it is as yet not known whether PVB19 plays a pathogenic role in this chronic heart muscle disease. Thus, the aim of the present study was to assess the prevalence of the enterovirus, adenovirus and PVB19 genomes in endomyocardial biopsy tissue samples from patients with IDC, and to analyze the significance of these findings with respect to the clinical and hemodynamic course of the disease and the survival of the patients.
Material and methods
Patients
The study group consisted of 52 consecutive patients, who had been admitted to our clinic due to a chronic heart failure between 1997–1999, showing left ventricular systolic dysfunction with associated cardiomegaly and left ventricular ejection fraction (LVEF) of <55%, assessed angiographically using the area-length method [16].
IDC was diagnosed according to the criteria defined by the World Health Organization/International Society and Federation of Cardiology from 1995 [41], if a hypertensive, coronary and/or a hemodynamically relevant valvular heart disease was excluded. All patients gave their informed written consent regarding cardiac catheterization and endomyocardial biopsy. Patients were prospectively followed at 6–12 month intervals for up to 4 years, thereby documenting the New York Heart Association (NYHA) functional class and assessing the course of LVEF and left ventricular end-diastolic diameter (LVEDD; mm) by echocardiography.
Ten consecutive patients with normal LVEF (≥55%) undergoing coronary artery bypass surgery served as a control group with respect to the detection of viral genomes in the myocardium.
Endomyocardial biopsy
Six endomyocardial biopsy specimens of the left ventricle were obtained from each patient using a Cordis long sheath bioptome (5.4F, 7F; Cordis). Routine histological investigations as well as immunohistological analyses were performed on the specimens. In addition, these specimens as well as the myocardial tissue samples of the right atrial appendage from the control patients obtained during the surgical procedure were processed for a subsequent analysis of enterovirus RNA and adenovirus and PVB19 DNA.
Histology
Endomyocardial biopsy specimens were fixed in paraffin, sectioned and stained with a standard hematoxylin-eosin stain. A diagnosis of active myocarditis was established according to the Dallas classification [5], when both cellular infiltration and myocytolysis were present.
Histomorphometry
Histomorphometric evaluations were performed on the paraffin sections. The diameter of the monocytes was measured in the hematoxylin-eosin-stained myocardial sections along the longitudinal axis of the nucleus. Cell cross-sections were measured for the shortest diameter value. The mean of 25 diameter measurements per patient was calculated to evaluate the myocyte diameter. In addition, the volume fraction of fibrosis in the myocardium was determined on the basis of the point counting method on paraffin sections stained with Sirius red, a special collagen stain [18].
Immunohistology
T cell infiltration of the myocardium was assessed on acetone-fixed tissue frozen 4-μm cryostat sections employing monoclonal antibodies directed to CD2 (pan T cell marker), CD4 (T helper cells), CD8 (T suppressor cells), CD14 (macrophages) and CD45RO (activated T lymphocytes) (all from DAKO). The reaction was visualized using a biotinylated second antibody and a peroxide-conjugated avidin-biotin complex (Vectastain Elite ABC Kit, Vector Laboratories). As chromogen 3-amino-9-ethylcarbazol (Histoprime, CAMON Laboratory Service) was used. Infiltrating cell counts were performed calculating mean and standard deviation (SD) of a minimum of 15 high power fields. At a 400-fold magnification, one power field of vision was equivalent to 0.125 mm2. In addition, the expression of the major histocompatibility complex antigens of class I (HLA-A, B, C) and the class II (HLA-DR, DP, DQ) was examined on cryostate sections (4 µm) as described above (primary antibodies: anti-HLA-ABC antigen clone W6/32, DAKO, and anti-HLA-DR, Becton Dickinson). The results were evaluated semi-quantitatively under the same microscope at 400-fold magnification.
Detection of enteroviral RNA in endomyocardial biopsy material
Total RNA was isolated from frozen endomyocardial biopsy samples or myocardial tissue of the right atrial appendage from the control patients, using the guanidinium thiocyanate-phenol-chloroform method of Chomczynski and Sacchi [15]. The tissue was homogenized in 4 M guanidine thiocyanate/mercaptoethanol. The RNA was extracted with phenol-chloroform, followed by precipitation and washing with ethanol. The RNA pellet was suspended in 20 µl distilled diethylpyrocarbonate-treated water. cDNA was synthesized in 20 µl of reaction mixture containing a 10-µl sample of total RNA together with oligo(dT) according to the Superscript RT protocol (Life Technologies). A reverse transcription (RT)-PCR of a house keeping gene (GAPDH) was employed to verify the integrity of total RNA extracted from endomyocardial biopsy specimens.
For detection of enteroviral genomic nucleic acid of the RNA virus, one-step RT-PCR (Titan One Tube RT-PCR System, Roche Diagnostics) was carried out. Ten microliters of extracted RNA and modified oligonucleotides of previously published primers [52] that recognize the conserved 5’-region of the enteroviruses (5’-CGGTACCTTTGTGCGCCTGTTTTA-3’ and 5’-CGGACACCCAAAGTAGTCGGTTCC-3’) was used. Thereafter, a Southern blot hybridization was performed using a PCR-generated digoxigenin-labeled DNA probe amplified by means of enterovirus-specific primers (5’-CCCCGGACTGAGTATCAATA-3’ and 5’-CAGTTAGGATTAGCGGCATTC-3’) and DIG-11-dUTP (Roche Diagnostics). To increase the sensitivity and confirm the specificity of the PCR product an antibody conjugate (anti-DIG-AP; Roche Diagnostics) was added, and the reaction visualized with the aid of CSPD (Perkin Elmer-Applied Biosystems) via chemiluminescence. Using this Southern blot assay on PCR products, a sensitivity of 0.023 plaque-performing units (pfu)/ml for coxsackievirus (CV) B3 (total RNA isolated from CVB3-infected HeLa cells) was reached. The primers BG 1 and BG 2 bind completely to sequences of human enteroviruses, including CVs of group B. For diagnosis of human viral heart disease, these primers are suitable because CVB3 is the most common viral agent in myocarditis and IDC. The species human enterovirus (HEV)-A, -C and -D showed single nucleotide differences in sequence analysis with the primers BG 1, enterovirus (EV) 3 and EV 4.
Detection of adenovirus and PVB19 DNA in endomyocardial biopsy specimens: DNA template preparation
Genomic and viral DNA was extracted from frozen myocardial tissue samples of the patients and the control group (1–2 mm3, and weighing ~1.5–2 mg) using the QIAamp Tissue protocol (Qiagen), and purified DNA was eluted in 50 µl AE-buffer. A one-step PCR of a house keeping gene (human β-actin) was carried out to verify the integrity of total DNA extracted from the endomyocardial biopsy samples. Primers corresponding to sequences in the β-actin gene were used as positive controls for the isolation of intact DNA (β-A432–20: 5’-GTGGGGCGCCCCAGGCACCA-3’) and (β-A433–24: 5’-CTCCTTAATGTCACGCACGATTTC-3’, 540 bp).
PVB19 PCR
A two-step amplification method for detection of PVB19 DNA was performed using published previously primers [44]. The first round primers, PVB19/3: 5’-AGCATGTGGAGTGAGGGGGC-3’ and PVB19/4: 5’-AAAGCATCAGGAGCTATACTTCC-3’, produced a 290-bp fragment. This was then amplified using nested primers PVB19/1: 5’-CTAACTCTGTAACTTGTAC-3’ and PVB19/2: 5’-AAATATCTCCATGGGGTTGAG-3’ to produce a 173-bp fragment. Each PCR was performed in a final volume of 50 µl, containing 10 mM TRIS-HCl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, 200 µM deoxyribonucleoside triphosphate (dNTP), 0.1 µM of each primer, 2.0 U Amplitaq DNA polymerase (Perkin Elmer-Applied Biosystems) and either 5 µl of the appropriate DNA sample or sterile distilled water. An initial denaturation step at 94°C for 5 min was followed by 38 cycles at 94°C (50 s), 55°C (50 s) and 72°C (1 min) on a GeneAmp PCR system 9700 (Perkin Elmer-Applied Biosystems). For the secondary amplification, 10 µl of the primary reaction was subjected to a further 38 cycles of PCR amplification as described above. The products from each reaction were analyzed by electrophoresis in a 2% agarose gel containing 0.5 µg/ml ethidium bromide. The DNA product was visualized by UV translumination. The sensitivity of the PVB19 PCR assay permitted reliable detection of 20 genome copies/run of the plasmid Parvo-784 bp we designed for Light Cycler PCR.
Sequence analysis
The parvovirus amplimers of 11 positive virus isolates were re-amplified, the PCR products purified according to the manufacturer’s instructions using a PCR purification kit (Qiagen), and resuspended in 30 µl distilled diethylpyrocarbonate-treated water. Sequence analysis was performed on the purified PCR fragments using either the sense primer PVB19/1 or the antisense primer PVB 19/2, AmpliTaq DNA polymerase, Big Dye terminator sequencing kit (Perkin Elmer-Applied Biosystems) and an ABI PRISM 310 genetic analyzer (Perkin Elmer-Applied Biosystems).
Adenovirus PCR
The adenovirus-specific primers utilized in the experiments were designed by Pauschinger et al. [37] for the amplification of all adenovirus subtypes for which sequence data were available in the National Center for Biotechnology Information GenBank. Adenovirus type 2 DNA, isolated from infected A 549 cells was used as positive virus control for PCR analysis after nucleic acid extraction. The first step-PCR amplification was performed with primers ADH-01: 5’-ACTACAAYATTGGCTACCAGG-3’ and ADH-02: 5’-CAAAACATAAGAAGKGTGGGC-3’, which target a sequence of 440 bp from the hexon gene and ADH-11: 5’-AACTTCCAGCCCATGAGCMG-3’ as well as ADH-12: 5’-CTCAAAAGTCATGTCBAGCGC-3’, which amplify a 330-bp fragment. The PCR reagent mixture is described above, and the concentration of primers was 0.4 µM. For the detection of adenovirus, 5 µl of DNA was subjected to PCR. Both PCR steps were performed with an initial denaturation step at 94°C for 5 min, followed by 38 cycles in the first and 30 cycles in the second round PCR at 94°C (45 s), 64°C (45 s) and 72°C (45 s), respectively. Finally, 10 µl of the first PCR product was amplified using the inner primers. The visualization of the DNA product was performed by UV translumination. The detection limit of the adenovirus-specific PCR was 10-2 pfu/ml for adenovirus type 2 isolated from A549 cells. In the present study, we confirmed successful amplification of the adenovirus serotypes 2, 5 and 8.
Serological investigations
Neither the viral antibody titers nor the viremic status of the patients were determined, since patients with evidence of acute virus infection or histologically proven myocarditis were excluded, and only patients with IDC known to be a chronic cardiac muscle disease were investigated in this study. Thus, acute serological markers such as IgM antibodies against enterovirus, adenovirus or PVB19 could not be expected.
Statistical analysis
Continuous data are given as mean ± SD and categorical data as relative frequencies (percentage). Comparisons between groups were made using unpaired Student’s t-test for continuous data or Mann-Whitney test, if the results were not normally distributed, and chi-square or Fisher exact test, where appropriate, for categorical variables. Baseline and follow-up data within groups were compared with each other by means of the paired Student’s t-test. The survival curves of the patients were tested by Kaplan-Meier analysis, and the log-rank test was used for differences between groups. Statistical significance was defined by a P value of less than 0.05.
Results
Detection of viral genomes in the myocardium
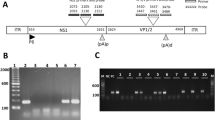
A total of 52 patients (44 men, 8 women; mean age ± SD 50.1±10.7 years; range 23–69 years) diagnosed with IDC after excluding myocarditis, were enrolled into the study. On the basis of the viral genomes detected in the endomyocardial biopsy samples by PCR analysis, the patients were divided into five groups. Enterovirus genomes were detected in 14 of the 52 patients (27%; mean age ± SD 55.2±9.4 years) and adenovirus in 2 (4%; 60.5±4.9 years). Fourteen of the 52 patients (27%; 50.9±11.6 years) were positive for PVB19 only (Fig. 1), and in 8 patients (15%; 47.0±13.5 years) both enteroviral RNA and PVB19 DNA were detected in the endomyocardial biopsy specimens. In the sequence analysis of 11 of the 14 PVB19-positive isolates, changes in the third nucleotide were seen in 7 cases (Fig. 2), which do not result in differences in the amino acid sequence.
PCR detection of PVB19 DNA sequence in endomyocardial biopsy samples. Products were detected by ethidium bromide staining in 2% agarose gel. Presence of bands at 173 bp represents PCR amplification of PVB19 DNA in the patient sample number 805/99 and in a PVB19-positive control. A Phi X 174 HaeIII fragment as size marker is shown in the first lane and a negative patient sample in the last lane. (PVB19 parvovirus B19, bp base pairs, LVEMB left ventricular endomyocardial biopsy)
The PVB19 isolates obtained from 11 endomyocardial biopsy samples were sequenced for 173 bp, corresponding to the PCR product with PVB19/1 and PVB19/2 (primer sequences at both ends excluded). The sequence of human PVB19 DNA (gb/M13178.1/PVBAUA) is indicated at the top, and the differences are highlighted
In another 14 of the 52 patients (27%; 44.4±10.6 years) neither enterovirus or adenovirus genomes nor PVB19 genomic DNA were found. This group was thus classified as virus negative.
In 4 of the 10 (40%) control patients with normal LVEF, who underwent coronary artery bypass surgery, we found PVB19 gene copies showing less then 50 copies/µg total DNA (data not shown). However, neither enterovirus nor adenovirus genomes were detected in the myocardial tissue samples from these control subjects.
Clinical data
With respect to the age- and sex-matched data, no significant difference was observed between the groups (Table 1). Further, there was no significant difference between patient groups regarding the clinical parameters of NYHA functional class and medication for chronic heart failure, including angiotensin-converting inhibitors, diuretics, digitalis and β-blocker. Interestingly, the group positive for the presence of enterovirus and PVB19 DNA and the virus-negative group consisted of younger patients. The duration of symptoms varied widely within the groups, but the differences of the data between the groups were not statistically significant.
Echocardiograpic and hemodynamic findings
No significant difference was observed on comparing LVEF and LVEDD, as measured by echocardiography, among the groups. Moreover, comparison of the other hemodynamic parameters, which were assessed invasively, between the groups was not statistically significant (Table 1).
Histomorphometry and Immunohistology
There was no significant difference between the virus-positive groups and the group negative for virus genomes with respect to the myocyte diameter or the volume fraction of interstitial fibrosis. In addition, there was no significant difference regarding the numbers of T cells, macrophages or activated lymphocyte per high power field between the patient groups. The semi-quantitative evaluation of the expression of the MHC class I and II antigens showed no significant difference.
Follow-up: mortality and NYHA functional class
The mean follow-up period (± SD) in all patients was 21.1±9.5 months (range 6–46 months) and was not significantly different between the individual groups. Six patients died during the follow-up period: one patient from the enterovirus-positive group (7%) after 1 month; one from the enterovirus plus PVB19-positive group (12.5%) after 10 months; two from the PVB19-positive group (14%) after 1 and 28 months, respectively; two from the virus-negative group (14%) after 4 and 5 months, respectively. The differences between the groups, as calculated by Kaplan-Meier analysis, were not statistically significant (log-rank test, P=0.80). Patients positive for enterovirus and enterovirus plus PVB19 experienced a significant improvement in NYHA functional class during the follow-up period (NYHA class 2.6±0.5 vs 1.9±0.6, P<0.01, and 2.6±0.8 vs 1.3±0.8, P<0.05, respectively). No significant functional improvement was observed in the adenovirus- and PVB 19-positive groups (NYHA class 2.5±0.7 vs 2.0±0.0, ns, and 2.6±0.9 vs 2.1±0.4, ns, respectively) or the virus-negative group (NYHA class 2.5±0.7 vs 2.2±0.9, ns).
Echocardiographic course
All patients showed an improvement in the echocardiographic LVEF value during the follow-up period. The difference between the LVEF 1 at baseline and the final LVEF 2 value for the enterovirus- and the enterovirus plus PVB19-positive group (26.8±9.4 vs 40.2±9.0%, P<0.001, and 35.9±10.7 vs 53.0±8.9%, P<0.01, respectively) compared to the changes in the adenovirus-positive (24.5±9.2% vs 44.5±6.4%, ns) and the PVB19-positive group (29.3±8.1 vs 39.8±15.3%, P=0.05) was highly significant. The difference between LVEF 1 (39.6±12.3%) and LVEF 2 (34.5±11.4%) in the virus-negative group was not significant. The mean increase in the LVEF value (LVEF 2−LVEF 1) in the enterovirus- and the enterovirus plus PVB19-positive patients was significantly greater than in the virus-negative group (13.4±10.8% vs 4.9±9.3%, P<0.05, and 17.1±10.1% vs 4.9±9.3%, P<0.05, respectively). In contrast, there was no difference between the PV B19-positive patients and the virus-negative patients regarding the changes in the LVEF during the follow-up period (10.6±17.0% vs 4.9±9.3%, ns).
Concerning the LVEDD, only the enterovirus plus PVB19-positive patients showed a significant improvement in this value during follow-up (63.4±4.5 mm vs 55.6±5.2 mm, P<0.01). In the other groups, the mean decrease of LVEDD (LVEDD 2−LVEDD 1) during follow-up was not significant. The mean difference between LVEDD 2 and LVEDD 1 in the enterovirus plus PVB19-positive group was significantly greater in comparison to the corresponding values of the PVB19-positive group (−7.8±5.8 vs −0.4±4.8, P<0.01). The mean decrease in LVEDD in the remaining patient groups was not significant.
Discussion
In the present study, we demonstrated the presence of PVB19 DNA in endomyocardial biopsy tissue obtained from adult patients with IDC. The prevalence of parvoviral genome in myocardial tissue was 42%, and thus was equal to that observed for enteroviral genomes, which are known to be the most common viral pathogens associated with IDC with a prevalence rate of 10–67% [4, 18, 19, 25, 26, 37, 39, 48, 49].
The prevalence of 42% obtained for the detection of enteroviral RNA in IDC patients in our study is somewhat high, and may be due to factors such as the heterogenic nature of the patient groups, the different primers used and the varying conditions, under which RT-PCR was performed [19, 25, 26, 37, 39, 48].
In 15% of the cases both enteroviral and parvoviral genomes were detected in myocardial tissue. The prevalence of adenoviral genomes in only 4% of the cases in our study, including the adenovirus serotypes 2, 5 and 8, was markedly lower than that obtained by Pauschinger et al. [37], who found adenovirus DNA in endomyocardial biopsy samples in 12 of 94 (13%) patients with IDC using the nested PCR technique. In another even larger study consisting of 860 patients with IDC, adenovirus DNA was detected in 6% of the cases [36]. In contrast, in a smaller study including 16 patients with IDC no adenoviral genome was detected in the endomyocardial specimens by Grumbach et al. also using PCR [19]. This discrepancy may be due to the different patient selection criteria, the size of the study group, the different detection techniques, and, possibly, the various adenovirus serotypes found in our and the other studies [19, 36, 37]. Interestingly, no viral genome was detected in only about a quarter of the patients (27%) with IDC.
The detection of PVB19 genome in 40% of the control patients with normal LVEF undergoing coronary artery bypass surgery possibly indicates a comparable prevalence rate of PVB19 genome in patients with IDC and control subjects with significant coronary artery disease, but normal LVEF. However, due to the small control group, further investigation including a greater number of patients with other heart disease or healthy individuals is needed to confirm this observation.
The investigation presented here demonstrates the detection of PVB19 DNA in myocardial tissue obtained from IDC patients using PCR. This technique is both highly specific and sensitive, but is liable to false-positive results due to contamination problems [23]. One way of demonstrating the lack of contamination here was detection of nucleotide variability in the VP1 region of the genome by sequence analysis of 11 myocardial probes positive for PVB19. Another procedure undertaken involved performing repeated PCR on all positive PVB19 DNA results. Thus, we could prove as far as possible that, despite the relatively high percentage of positive PCR results for PVB19, our endomyocardial specimens were free of any contamination.
Our findings demonstrate that PVB19 is not only associated with fetal myocarditis [2, 10, 32, 34, 40], myocarditis [8, 17, 30, 43, 46] and cardiac allograft rejection in children [46], or with myocarditis following adult heart transplantation [21] and myocarditis of immunocompetent adults [29], but can also be frequently detected in the myocardium of adult patients with IDC.
Of central importance is the clinical significance of the detection of parvoviral genomes in endomyocardial biopsies from patients with IDC. The three main questions to be asked are: (1) does the virus hibernate in the myocardium after infection, as in other organs without any pathogenetic impact, (2) does it undergo constant replication and, if so, does this continuous virus replication in cardiac tissue affect myocardial contractility, and (3) is any myocardial disease such as IDC prone to PVB19, hibernating or as an epiphenomenon?
It has been established that the presence of the erythrocyte-P antigen, serving as a virus receptor, facilitates infection of tissue cells with PVB19 [12, 13]. Since myocardial cells also contain this antigen receptor protein [42] and PVB19 has been found to infect intracardial endothelial cells in myocarditis [29], it is conceivable that infection of the myocardium with PVB19 may cause direct damage to the heart [29]. Although the initial acute myocarditis may be potentially fatal in both children and adults [17, 30, 43], according to published reports, a PVB19-associated myocarditis tends to have a more favorable outcome in the majority of pediatric cases [8, 21, 24, 35, 45, 46]. In our study, the percentage mortality in the group positive for PVB19 did not significantly differ from that in the other groups during long-term follow-up.
Following the initial infection of the myocardium, it might be that PVB19 is still detectable, but undergoes little or no replication, and thus possibly causes minimal or no further damage to the myocardial tissue during the stage of the chronic cardiac muscle disease associated with dilated cardiomyopathy. In some patients, a spontaneous elimination of the virus may occur. However, to prove this hypothesis serial endomyocardial biopsy samples would be required.
The significant improvement in the clinical and hemodynamic parameters in the two groups of patients positive for enterovirus only or enterovirus plus PVB19, as compared to the virus-negative group during follow-up, suggests that enterovirus-associated IDC may have a more favorable prognosis [18]. This may be an indicator of a possible elimination of the enterovirus, at least in some patients, which was, however, not investigated by a second endomyocardial biopsy during the follow-up period. Valuable information would be obtained by studying the difference between latent persistence of the enterovirus and active virus replication to explain the lack of improvement or even a deterioration in clinical symptoms of some patients with enterovirus-associated dilated cardiomyopathy [22, 38].
In conclusion, in myocardial tissue of patients with IDC, the prevalence rate of PVB19 genome of 42% is comparable to that of enterovirus RNA in IDC. PVB19-associated dilated cardiomyopathy tends to have a rather favorable prognosis and did not differ in survival from other virus-positive or virus-negative patients. Due to the low patient numbers, it is impossible to assess definitely the prognostic value of myocardial PVB19 in IDC, particularly since PVB19 genome was also found quite frequently in a small control group with normal LVEF. Finally, the significance of PVB19 in the pathogenesis of IDC still remains unclear and will be the issue of further investigation.
References
Amiot L, Langnay T, Drenou B, Lelong B, Le Prise PY, Logeais Y, Fauchet R (1998) Spontaneous recovery from severe parvovirus B19 pure red cell aplasia, in a heart transplant recipient, as demonstrated my marrow culture. Hematol Cell Ther 40:71–73
Anderson MJ, Khousam MN, Maxwell DJ, Gould SJ, Happerfield LC, Smith WJ (1988) Human parvovirus B19 and hydrops fetalis. Lancet I:535
Andreoletti L, Hober D, Becquart P, Belaich S, Copin MC, Lambert V, Wattre P (1997) Experimental CVB3-induced chronic myocarditis in two murine strains: evidence of interrelationships between virus replication and myocardial damage in persistent cardiac infection. J Med Virol 52:206–214
Archard LC, Bowles NE, Cunningham L, Freeke CA, Olsen EGJ, Rose ML, Meany B, Why HJF, Richardson PJ (1991) Molecular probes for detection of persisting enterovirus infection of human heart and their prognostic value. Eur Heart J 12 (Suppl 12):56–59
Aretz HT, Billingham ME, Edwards WD, Factor SM, Fallon JT, Fenoglio JF, Olsen EGJ, Schoen FJ (1987) Myocarditis. A histopathologic definition and classification. Am J Cardiovasc Pathol 1:3–14
Beland SS, Danile GK, Menard LC, Miller NM (1997) Aplastic crisis with parvovirus B19 in an adult heriditary spherocytosis. J Ark Med Soc 94:163–164
Bertoni E, Rosati A, Zanazzi M, Azzi A, Zakrzewska K, Guidi S, Fanci R, Salvadori M (1997) Aplastic anemia due to B19 parvovirus infection in cadaveric renal recipients: an underestimated infectious disease in the immunocompromised host. J Nephrol 10:152–156
Borreda D, Palomera S, Gilbert B, Lienhardt A, Lumley L de (1992) A propos de vingt-quatre observations d’infections a parvovirus humain B19 chez l’enfant (24 cases of human parvovirus B19 infection in children). Ann Pediatr (Paris) 3:543–549
Bowles NE, Richardson PJ, Olson EGJ, Archard LC (1986) Detection of coxsackie-B-virus-specific RNA sequences in myocardial biopsy samples from patients with myocarditis and dilated cardiomyopathy. Lancet 17:1120–1123
Brandenburg H, Los FJ, Cohen-Overbeck TE (1996) A case of early intrauterine parvovirus B19 infection. Prenat Diagn 16:75–77
Brown KE, Young NS (1997) Parvovirus B19 in human disease. Annu Rev Med 48:59–67
Brown KE, Anderson SM, Young NS (1993) Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114–117
Brown KE, Hibbs JR, Gallinella G, Anderson SM, Lehmann ED, Mc Carthy P, Young NS (1994) Resistance to parvovirus B19 infection due to lack of virus receptor (erythrocyte P antigen). N Engl J Med 330:1192–1196
Calvet A, Pujol MO, Bertocchi M, Bastien O, Boissonat P, Mornex JF (1999) Parvovirus B19 infection in thoracic organ transplant recipients. J Clin Virol 13:37–42
Chomczynski P, Sacchi N (1987) Single step method of RNA isolation by guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–162
Dogde HT, Sheehan FH (1983) Quantitative contrast angiography for assessment of ventricular performance in heart disease. J Am Coll Cardiol 1:73–81
Enders G, Dötsch J, Bauer J, Nützenadel W, Hengel H, Haffner D, Schalasta G, Searle K, Brown KE (1998) Life-threatening parvovirus B19-associated myocarditis and cardiac transplantation as possible therapy: two case reports. Clin Infect Dis 26:355–358
Figulla HR, Stille-Siegener M, Mall G, Heim A, Kreuzer H (1995) Myocardial enterovirus infection with left ventricular dysfunction: a benign disease compared with idiopathic dilated cardiomyopathy. J Am Coll Cardiol 74:1170–1175
Grumbach IM, Heim A, Pring-Akerblom P, Vonhof S, Hein WJ, Müller G, Figulla HR (1999) Adenoviruses and enteroviruses as pathogens in myocarditis and dilated cardiomyopathy. Acta Cardiol 54:83–88
Heegaard ED, Hornsleth A (1995) Parvovirus: the expanding spectrum of disease. Acta Paediatr 84:109–117
Heegaard ED, Eiskjaer H, Baandrup U, Hornsleth A (1998) Parvovirus B19 infection associated with myocarditis following adult cardiac transplantation. Scand J Infect Dis 30:607–610
Hohenadl C, Klingel K, Mertsching J, Hofschneider PH, Kandolf R (1991) Strand-specific detection of enteroviral RNA in myocardial tissue by in situ hybridization. Mol Cell Probes 5:11–20
Hornsleth A, Carlsen KM, Christensen LS, Gundestrup M, Heegaard ED, Myhre J (1994) Estimation of serum concentration of parvovirus B19 DNA by PCR in patients with chronic anaemia. Res Virol 145:379–386
Janner D, Bork J, Baum M, Chinnock R (1994) Severe pneumonia after heart transplantation as a result of human parvovirus B19. J Heart Lung Transplant 13:336–338
Jin O, Sole MJ, Butany JW, Chia WK, McLaughlin PR, Liu P, Liew CC (1990) Detection of enterovirus RNA in myocardial biopsies from patients with myocarditis and cardiomyopathy using gene amplification by polymerase chain reaction. Circulation 82:8–16
Kämmerer U, Kunkel B, Korn K (1994) Nested PCR for specific detection and rapid identification of human picornaviruses. J Clin Microbiol 32:285–291
Kandolf R, Ameis D, Kirschner P, Canu A, Hofschneider PH (1987) In situ detection of enteroviral genomes in myocardial cells by nucleic acid hybridization: an approach to the diagnosis of viral heart disease. Proc Natl Acad Sci USA 84:6272–6276
Kariyawasam HH, Gyi KM, Hodson ME, Cohen BJ (2000) Anaemia in lung transplant patient caused by parvovirus B19. Thorax 55:619–620
Klingel K, Selinka HC, Sauter M, Bock CT, Szalay G, Kandolf R (2002) Molecular mechanisms in enterovirus and parvovirus B19 associated myocarditis and inflammatory cardiomyopathy. Eur Heart J Supplements 4 (Suppl I):I8-I12
Knisely AS, O’Shea PA, Anderson LJ, Gary GW (1988) Parvovirus B19 infection, myocarditis and death in a 3-year old boy. Pediatr Pathol 8:665
Marchand S, Tchernia G, Hiesse C, Tertian G, Cartron J, Kriaa F, Boubenider S, Goupy C, Lecointe D, Charpentier B (1999) Human parvovirus B19 infection in organ transplant recipients. Clin Transplant 13:17–24
Morey AL, Keeling JW, Porter HJ, Fleming KA (1992) Clinical and histopathological features of parvovirus B19 infection in the human fetus. Br J Obstet Gynaecol 99:566–574
Moudgil A, Shidban H, Nast CC, Bagga A, Aswad S, Grahma SL, Mendez R, Jordan SC (1997) Parvovirus B19 infection-related complications in renal transplant recipients: treatment with intravenous immunoglobulin. Transplantation 64:1847–1850
Naides SJ, Weiner CP (1989) Antenatal diagnosis and palliative treatment of non-immune hydrops fetalis secondary to fetal parvovirus B19 infection. Prenat Diagn 9:105–114
Nour B, Green M, Michaelis M, Reyes J, Tzakis A, Gartner JC, McLoughlin L, Starzl TE (1993) Parvovirus B19 infection in pediatric transplant patients. Transplantation 56:835–838
Pankuweit S, Portig I, Eckhardt H, Crombach M, Hufnagel G, Maisch B (2000) Prevalence of viral genome in endomyocardial biopsies from patients with inflammatory heart disease. Herz 25:221–226
Pauschinger M, Bowles NE, Fuentes-Garcia FJ, Pham V, Kühl U, Schwimmbeck PL, Schultheiss H-P, Towbin JA (1999) Detection of adenoviral genome in the myocardium of adults with idiopathic left ventricular dysfunction. Circulation 99:1348–1354
Pauschinger M, Dörner A, Kühl U, Schwimmbeck PL, Poller W, Kandolf R, Schultheiss H-P (1999) Enteroviral RNA replication in the myocardium of patients with left ventricular dysfunction and clinically suspected myocarditis. Circulation 99:889–895
Petitjean J, Kopecka H, Freymuth F, Langlard JM, Scanu P, Galateau F, Bouhour JB, Ferriere M, Charbonneau P, Komajada M (1992) Detection of enteroviruses in endomyocardial biopsy by molecular approach. J Med Virol 37:76–82
Porter HJ, Quantrill AM, Fleming KA (1988) B19 parvovirus infection of myocardial cells. Lancet I:535–536
Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O’Connell J, Olsen E, Thiene G, Goodwin J, Gyarfas I, Martin I, Nordet P (1996) Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definiton and Classification of Cardiomyopathies. Circulation 93:841–842
Rouger P, Gane P, Salmon C (1987) Tissue distribution of H, Lewis and P antigen as shown by a panel of 18 monoclonal antibodies. Rev Fr Transfus Immunohematol 30:699–708
Saint-Martin J, Choulot JJ, Bonnaud E (1990) Myocarditis caused by parvovirus. J Pediatr 116:1007–1008
Salimans MM, Holsappel S, Rijke FM van de, Jiwa NM, Raap AK, Weiland HT (1989) Rapid detection of human parvovirus B19 DNA by dot-hybridization and the polymerase chain reaction. J Virol methods 23:19–28
Schowengerdt KO, Ni J, Denfield SW, Gajarski RJ, Radovancevic B, Frazier OH, Demmler GJ, Kearney D, Bricker JT, Towbin JA (1996) Diagnosis, surveillance, and epidemiologic evaluation of viral infections in pediatric cardiac transplant recipients with the use of the polymerase chain reaction. J Heart Lung Transplant 15:111–123
Schowengerdt KO, Ni J, Denfield SW, Gajarski RJ, Bowles NE, Rosenthal G, Kearney DL, Price JK, Rogers BB, Schauer GM, Chinnock RE, Towbin JA (1997) Association of parvovirus B19 genome in children with myocarditis and cardiac allograft rejection: diagnosis using the polymerase chain reaction. Circulation 96:3549–3554
Tanawattanacharoen S, Falk RJ, Jenette JC, Kopp JB (2000) Parvovirus B19 DNA in kidney tissue of patients with focal segmental glomerulosclerosis. Am J Kidney Dis 35:1166–1174
Weiss LW, Liu XF, Chang KL, Billingham ME (1992) Detection of enteroviral RNA in idiopathic dilated cardiomyopathy and other human cardiac tissue. J Clin Invest 90:156–159
Why HJF, Meany BT, Richardson PJ, Olsen EGJ, Bowles NE, Cunningham L, Freeke CA, Archard LC (1994) Clinical and prognostic significance of detection of enteroviral RNA in the myocardium of patients with myocarditis and dilated cardiomyopthy. Circulation 89:2582–2589
Wicki J, Samii K, Cassinotti P, Voegeli J, Rochat T, Beris P (1997) Parvovirus B19-induced red cell aplasia in solid organ transplant recipients. Two case reports and review of the literature. Hematol Cell Ther 39:199–204
Yaegashi N (2000) Pathogenesis of nonimmune hydrops fetalis caused by intrauterine B19 infection. Tokuyu J Exp Med 190:65–82
Zell R, Klingel K, Bültmann B, Kandolf R (1995) Detection of enteroviral RNA from formalin-fixed paraffin-embedded myocardial specimens by nested PT-PCR. Pathol Res Pract 191:196
Zolnourian ZR, Curran MD, Rima BK, Coyle PV, O’Neill HJ, Middleton D (2000) Parvovirus B19 in kidney transplant patients. Transplantation 69:2198–2202
Acknowledgements
We are grateful to Roland Zell, PhD (Institute of Virology and Antiviral Therapy, Friedrich-Schiller-University, Jena, Germany) for performing the sequence analysis of 11 positive PVB19 isolates. We are indebted to Prof. Gerhard Mall, MD (Department of Pathology, Städtische Kliniken Darmstadt, Germany) for histological and immunohistological investigations of the cardiac tissue samples and for assessing the myocyte diameter and the volume fraction of interstitial fibrosis. Furthermore, we are grateful to Prof. Thorsten Wahlers, MD and colleagues (Department of Cardiothoracic and Vascular Surgery, Friedrich-Schiller-University, Jena, Germany) for providing myocardial tissue samples from patients undergoing coronary artery bypass surgery. Finally, we thank Mrs. Birgit Lotze for her help in the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lotze, U., Egerer, R., Tresselt, C. et al. Frequent detection of parvovirus B19 genome in the myocardium of adult patients with idiopathic dilated cardiomyopathy. Med Microbiol Immunol 193, 75–82 (2004). https://doi.org/10.1007/s00430-003-0211-0
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-003-0211-0