Abstract
Pigeons are classic animal models for learning, memory, and cognition. The majority of the current understanding about avian neurobiology outside of the domain of the song system has been established using pigeons. Since MRI represents an increasingly relevant tool for comparative neuroscience, a 3-dimensional MRI-based atlas of the pigeon brain becomes essential. Using multiple imaging protocols, we delineated diverse ascending sensory and descending motor systems as well as the hippocampal formation. This pigeon brain atlas can easily be used to determine the stereotactic location of identified neural structures at any angle of the head. In addition, the atlas is useful to find the optimal angle of sectioning for slice experiments, stereotactic injections and electrophysiological recordings. This pigeon brain atlas is freely available for the scientific community.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Along with rats, pigeons are standard animal models for studies on the mechanisms of learning. Medline lists close to 3,000 papers that used pigeons in learning and/or memory paradigms. Beginning with classic studies on schedules of reinforcement (Ferster and Skinner 1957) up to studies on matching (Kangas et al. 2010), interval timing (Zentall et al. 2004), sequence acquisition (Scarf and Colombo 2010) and equivalence learning (Friedrich et al. 2004), to name just a few, all fields of learning theory have regularly used pigeons. Similarly, studies on cognition and memory and its avian neural substrate have mostly worked with pigeons (e.g. working memory: Browning et al. 2011; Rose et al. 2010; category formation: Yamazaki et al. 2007; spatial cognition: Kahn and Bingman 2009; metacognition: Adams and Santi 2011). In addition, pigeons have traditionally served as a model organism to study perception and its neurobiological and anatomical basis in birds, with a similar wealth of literature as for learning and cognition. Again mentioning just a few areas of research, these studies examined e.g. color perception (Carvalho et al. 2007), optic illusions (Xiao and Güntürkün 2008), visual lateralization (Prior et al. 2004), and magnetoperception (Wilzeck et al. 2010). In addition, the recent reinterpretation of the homology of avian and mammalian forebrains that was followed by a new nomenclature for the bird brain was largely based on studies in pigeons (Reiner et al. 2004; Jarvis et al. 2005).
Within the last decades, the ascending sensory and, to a lesser extent, the descending motor systems of pigeons have been more thoroughly characterized than for any other bird. These structural and hodological data will possibly soon be integrated with new insights from small animal magnetic resonance imaging (MRI). The atlas of the pigeon brain from Karten and Hodos (1967) is still widely used, but obviously has the limitation of all brain atlases that are based on drawings of brain sections: they not only provide only a 2-dimensional perspective of the brain, but are also dependent on the previously specified sectioning angles.
To further neurobiological studies in pigeons, we therefore decided to create the first 3D MRI atlas of the pigeon brain as a freely available tool for the scientific community. The atlas is based on the several scanning protocols, each with their own advantages and disadvantages, to visualize a wide range of neural structures. Reflecting the traditional interest in the sensory and learning-related processes in pigeons, we tried to visualize the ascending visual (thalamofugal, tectofugal, accessory optic pathways, as well as the isthmic nuclei of the brainstem), somatosensory (spinal, trigeminal), auditory, and olfactory systems as complete as possible. In addition, we describe the hippocampal complex and the basal ganglia structures as well as the sources of the descending tractus occipitomesencephalicus, tractus occipitomesencephalicus pars hypothalami, and the tractus septomesencephalis. We hope that this pigeon brain atlas will advance studies in this classic bird model in an era where traditional and MRI-based studies will more and more intermingle.
Materials and methods
Specimen preparation
For this study, one adult female pigeon (Columba livia) of local stock was deeply anesthetized with equithesin and transcardially perfused with a phosphate-buffered saline solution (PBS, 0.12 M), followed by a mixture of paraformaldehyde (PFA, 4%) and Dotarem® (1%), a paramagnetic MR contrast agent. After decapitation, the head was post-fixed in a mixture of PFA (4%) and Dotarem (1%) for at least 5 days at 5°C.
Data acquisition
To obtain a 3-dimensional representation of the skull, the whole birdhead was imaged with a Siemens PET-CT equipped with a rotating 80 kV X-ray source (focal spot size of 50 μm) and a Siemens Inveon PET-CT Camera with a 125 mm X-ray detector. Isotropic voxels were acquired with a resolution of 222 × 223 × 223 μm.
3D MRI datasets of the pigeon brain were acquired with a 9.4 T Biospec® horizontal bore NMR scanner (Bruker BioSpin, Ettlingen, Germany), equipped with a 120 mm BGA12-S actively shielded gradient insert with a maximum gradient strength of 600 mT/m attached to an AVANCE-II Bruker console and a 7 T Pharmascan® horizontal bore NMR scanner (Bruker BioSpin, Ettlingen, Germany) equipped with a 90 mm BGA9-S actively shielded gradient insert with a maximum gradient strength of 400 mT/m attached to an AVANCE-III Bruker console. A Bruker cross-coil setup with a linear transmit volume coil and a parallel receive surface array designed for rat head MRI was used on the 9.4 T. The standard Bruker cross-coil setup with a quadrature volume coil and a quadrature surface coil for rats was used on the 7 T.
T2-weighted 3D images were acquired on the 9.4 T using a RARE sequence with a RARE factor of 2, a spectral bandwidth (BW) of 72 kHz, 8 averages (N av), a repetition time (TR) of 350 ms and an effective echo-time (TE) of 15 ms. With this sequence, a spatial resolution of (78 × 78 × 100) µm3 was obtained with field of view (FOV): (24 × 20 × 17) mm3, and matrix size (M): (308 × 256 × 170) pixels. Acquisition time was 17 h.
T2*-weighted 3D images were acquired on the 7 T using a FISP gradient echo sequence with a 15° flip-angle, a BW of 50 kHz, an N av of 64, a TR of 9.47 ms, a TE of 4.738 ms and a scan repetition time of 5,000 ms. Images had a field of view of (21.504 × 21.504 × 15.36) mm3 with an acquisition matrix of (360 × 360 × 128) zero-filled to (360 × 360 × 256) resulting in an isotropic spatial resolution of 60 μm in all three directions. Acquisition time was 11 h 22 min.
Brain area delineation and 3D reconstruction
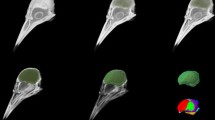
All MRI datasets and the CT dataset were co-registered with the SPM 8 package using normalized mutual information. The MRI images were used as such to co-register to each other, however to co-register the MRI images to the CT image the brain surface was delineated from the MRI image and used as input for co-registration. The position of the ear canal and thus the most likely position of stereotactic earbars was established with the CT data, and all datasets were reoriented to match a 45° angle of the earbars and the most posterior part of the beak opening to the horizontal axis according to Karten and Hodos (1967).
All atlas delineations were performed with Amira 4.0 (Mercury Computer Systems, San Diego, CA, USA). The delineation of the skull was based on the CT dataset and was conducted automatically using a signal intensity high-pass threshold. The brain surface and the neural subdivisions of both hemispheres were manually delineated, based on the MRI signal intensity differences between brain regions. The delineation of each brain structure and fiber tract was performed mostly in frontal plane and subsequently controlled in the two other planes. Regularly Cresyl Violet and Klüver–Barrera sections were consulted in difficult subdelineations.
Histology and validation of MRI data
Following MRI data acquisition, the brain was extracted from the skull and postfixed in 4% paraformaldehyde + 30% sucrose for 2 h at 4°C, cryoprotected in 0.12 M PBS + 30% sucrose at 4°C for 48 h. Then, the brain was sagittally divided along the Fissura longitudinalis cerebri and one half brain was cut at 40 μm on a freezing microtome in frontal and the other in sagittal plane. Sections were collected in five parallel series and stored in 0.12 M PBS containing 0.1% sodium azide at 4°C until they were subjected to histochemistry for Cresyl Violet and Klüver–Barrera staining. After staining was finished, histological sections were coverslipped and brightfield photomicrographs were made with a Zeiss Axio Imager M1 to verify MR-based structural definition. Two kinds of verifications of the MR atlas were then conducted. First, MR-based brain area delineations were controlled for all structures provided by the current atlas. Second, areas identified in histological sections were localized in the atlas of Karten and Hodos (1967), their stereotaxic coordinates were obtained and these were read into the MR atlas to check if the same area identified in histology was found.
Results
Scanning protocols and structural delineations
Different imaging protocols provide different and complementary possibilities to delineate neural structures. X-ray CT imaging is essential to anchor the precise position of the brain within the skull. It thus represents a crucial first step to enable the construction of a stereotaxic atlas since the brain position within the skull can be co-registered with the MR-based structural positions. We first defined a reference plane from three CT imaging based reference points: both ear canals, and the most posterior end of the beak opening. The horizontal plane of the brain atlas is then defined as a plane tilted by 45° to the reference plane about the axis running through both ear canals (Fig. 2). This is the horizontal plane as defined by the atlas of Karten and Hodos (1967). Since a line connecting the commissura posterior and the exit of the third nerve is orthogonal to this horizontal plane, it accords with the Frankfurt, or auriculo-orbital plane. CT imaging is not useful to visualize brain areas. This is best been done with the protocols outlined below (Table 1).
T2-weighted images produce images where in contrast to grey matter structures water-containing areas become brighter and myelinated areas are visible as darker structures. This sequence was used for most of the brain subdelineations since it produced a very good contrast between neuropil and pathways and, to some extent, also between nuclear structures that were partitioned by axonal layers.
T2*-weighted imaging gives rise to a similar contrast as T2, but with a much higher resolution. This advantage comes with the cost of a lower contrast and signal. Thus, T2* was used in the visualization of structures with complex curvatures like the n. isthmo-opticus (ION).
Cresyl Violet and Klüver–Barrera stains were performed to visualize the detailed architecture of the brain and to compare it with delineations obtained from MR procedures. For this, the brain was sagittally divided along the fissura longitudinalis cerebri and one half brain was cut in frontal and the other in sagittal plane. After sectioning and mounting, histological sections were used to verify MR-based structural definition.
Data presentation and validation
The co-registered MRI and CT datasets, including skull, and 42 brain subdivisions are freely available for download from our website: http://www.bio.psy.rub.de/3Dpigeonatlas.
The default data orientation is presented in a similar fashion as previously published atlases, with a head angle of 45° (Fig. 1). The 45° angle has been calculated based on the axis through the earcanal (the most likely position for fixating earbars) and the most posterior end of the beak opening relative to the horizontal plane. When loaded into MRIcro, the stereotactic zero point according to the Karten and Hodos pigeon atlas (1967) is reset to the zero coordinate by default. This reference point can be manually altered however, if another zero coordinate is preferred. In this reference frame, the X-axis represents the brain’s left-to-right axis, the Y-axis corresponds to the posterior–anterior axis, and the Z-axis corresponds to the dorsal–ventral axis of the brain. The stereotactic coordinates of a specific brain area can be easily acquired by moving the cursor onto the desired region.
The accuracy of the coordinate system and the possibility to extrapolate the atlas coordinates to other individuals was substantiated by morphometric comparisons with histological preparations as outlined in the “Methods” section (Fig. 2). The 3D datasets can also be used to determine the optimal head angle for stereotactic operations or the best cutting angle for sectioning. If it is important to have multiple brain areas of interest in one single brain section, rotation and oblique slicing tools can easily be used to estimate the best cutting angle (Fig. 3).
Arbitrary level of sectioning that visualizes the proximity of the trigeminal, visual, and auditory projections towards n. basorostralis palii, entopallium, and Field L2, respectively. The section was placed oblique to a classic frontal level and slightly asymmetric with respect to the left–right orientation. a Plane of sectioning on brain surface, b T2 image of the resulting section, c rotation of b to visualize the section, d corresponding label fields. Note that the level of sectioning is asymmetric such that Field L2 is not visible on the right side
Different datasets and delineations can be superimposed and manipulated synchronously to attain the desired brain image. The delineation sets can further be used to generate a 3-dimensional representation of the brain that gives an accurate description of the relative location, shape and volume of brain areas.
Discussion
The present pigeon brain atlas provides the possibility to visualize all outlined structures from a 3-dimensional and thus whole-brain point of view. By combining several scanning protocols, we could capture a large number of different brain regions along the neuraxis, making this atlas suitable for diverse lines of research on the pigeon brain. In combination with the atlases for zebra finches (Poirier et al. 2008) and canaries (Vellema et al. 2011), the scientific community now also has an MR-based atlas for a non-song bird with an emphasis on sensory and motor systems. In the following, we first dwell on the relevance of an MR-based atlas to then shortly outline the intra-systems connectivity of the recovered areas and comment on the structural delineations of the present work.
General considerations
For generations of bird neuroscientists, the Karten and Hodos (1967) atlas of the pigeon brain has been an excellent and meanwhile classic resource. But like all 2D atlases, it is restricted to predefined orientations, making it difficult to extrapolate to other angles of sectioning. The present 3-dimensional atlas can be used to study the anatomy of certain structures from any possible angle. This is highly relevant when planning injections of tracer substances or the insertion of recording electrodes from unorthodox angles. In this case, the coordinate system of the 3D brain can be reset to any preferred orientation, making highly idiosyncratic surgical approaches easy. Similarly, the planning of the best angle for brain sections that contain multiple regions of interest is now easy to perform. This is an essential advance for slice recordings where not only two or more structures, but also the pathways interconnecting them have to be in one plane of view. But also diverse histological staining procedures or in situ hybridization protocols can benefit from this possibility.
Possibly, the most important application of the present atlas is for MRI research itself. Functional MRI is firmly established for human subjects. Using monkeys, awake animal MRI experiments are also common meanwhile (Joly et al. 2011). Small animals, however, are usually scanned under sedated conditions. The results of these stimulation paradigms (Poirier et al. 2009; Van Meir et al. 2005) and diffusion tensor imaging studies (De Groof et al. 2006) are of high relevance to analyze, e.g. the avian song system. But certainly, more complex experiments will be possible when new procedures for awake small animals become established (Desai et al. 2011). Along with the 3D brain atlas of the zebra finch (Poirier et al. 2008), and of the canary (Vellema et al. 2011), this atlas of a non-song bird and of a classic animal model for learning and cognition will then possibly become an important tool.
Visual pathways
Tectofugal system
The tectofugal pathway is composed of optic nerve axons which decussate in the chiasma opticum and synapse in a strictly topographical way in layers 2–7 of the optic tectum (Remy and Güntürkün 1991). The tectum projects bilaterally to the thalamic n. rotundus (Rt) and n. triangularis (T) (Güntürkün et al. 1993, 1998), which itself sends efferent fibers to the ipsilateral entopallium (E) (Fredes et al. 2010). The projection from tectum to Rt/T is constituted by functionally and morphologically distinct tectal subpopulations that terminate in different thalamic territories, thereby creating a transfer from a retinotopic to a functionotopic organization (Hellmann and Güntürkün 2001; Marín et al. 2003). Since the rotundal and, to a lesser extent, the triangular projection onto the entopallium constitutes a point-to-point transfer; the entopallium seems to be constituted by a similar functionotopy as its thalamic afferents (Fredes et al. 2010). Some of these functionally and anatomically defined subareas of the E project to the perientopallium (Ep), a surrounding shelf area, while other subfields project to the mesopallium ventrolaterale (MVL) (Krützfeldt and Wild 2005) (Fig. 4a).
Sensory systems within a transparent 3D pigeon brain: a ascending visual thalamofugal (dark red colors) and tectofugal structures (light red colors). Note that the transition between the caudal visual and the somatosensory rostral Wulst at A 13.00 represents an estimate, b accessory optic (yellow and orange colors ) and visual isthmic structures (red and magenta colors), c somatosensory spinal (blue colors) and trigeminal systems (turquoise), d auditory (green colors) and olfactory system structures (lilac colors). An n. angularis, Bas n. basorostralis palii, BO bulbus olfactorius, CPi cortex prepiriformis, CPP cortex piriformis, DIVA n. dorsalis intermedius ventralis anterior, DLP n. dorsolateralis posterior thalami, Entop entopallium, Ex n. externus, GC n. gracilis et cuneatus, GLd n. geniculatus lateralis, pars dorsale, HA hyperpallium apicale, HD hyperpallium dorsal, HI hyperpallium intercalatum, IHA interstitial nucleus of the HA, Imc n. isthmi pars magnocellularis, IO n. isthmo-opticus, Ipc n. isthmi pars parvocellularis, La n. laminaris, LL n. of the lateral lemniscus, LM n. lentiformis mesencephali, MC n. magnocellularis, MLD n. mesencephalicus lateralis pars dorsalis, nBOR n. of the basal optic root, OS oliva superior, Ov n. ovoidalis, PM n. pontis medialis, PrV n. sensorius principalis nervi trigemini, Rt n. rotundus, Slu n. semilunaris, T n. triangularis, TnA n. taeniae amygdalae
The tectum was easily visible in all scanning protocols, while R and T could best be visualized using T2*. The entopallium with its jagged dorsal rim becomes nicely visible with T2 and T2*. It is important to emphasize that we could not differentiate the internal and the external subdivision of the entopallium. Thus, we herein present the entopallial complex without the Ep and the MVL, which were not visible with MR.
Thalamofugal system
The thalamofugal pathway consists of the retinal projection onto the n. geniculatis lateralis pars dorsalis (GLd), a group of nuclei in the contralateral dorsal thalamus, and the bilateral projection of the GLd onto the Wulst (“bulge”) in the anterodorsal forebrain (Miceli et al. 1990; Güntürkün 2000). Due to its anatomical, physiological, and functional properties, the avian thalamofugal pathway corresponds to the mammalian geniculostriate system (Shimizu and Karten 1993). The GLd is composed of four retinorecipient components (Güntürkün and Karten 1991). We were unable to delineate these subdivisions with our protocols and collectively labeled them as GLd. The posterior border of the GLd is difficult to draw with cytoarchitectonic means. Güntürkün and Karten (1991) defined it at A 6.00 since this is the posterior border of the retinal projections within the dorsal thalamus. Using T2* a fiber bundle was visible that clearly took a position similar to the retinal projections. Using this landmark, we defined the posterior end of the GLd and located it also at A 6.00 (Fig. 4a).
The visual Wulst is organized from dorsal to ventral in four pseudolaminae: hyperpallium apicale (HA), interstitial nucleus of the HA (IHA), hyperpallium intercalatum (HI), and hyperpallium dorsale (HD). These subdivisions are based on the cytoarchitectonics of the Wulst and do not reflect the full complexity of the structure, since Shimizu and Karten (1990) were able to distinguish at least eight subdivisions using immunocytochemical techniques. The granular IHA and probably also lateral HD are the major recipients of the GLd input (Watanabe et al. 1983). The extratelencephalic Wulst efferents arise from the HA (Miceli et al. 1987). We were not able to distinguish HA and IHA and combined them into an HA complex. Similarly, the distinction between HI and HD could not be drawn with certainty, especially along their lateral border. We therefore decided to combine them into an HI-HD complex.
The Wulst can be subdivided into a rostral somatosensory and a caudal visual division with the transition zone being located at about A 13.00 (Wild 1987). Possibly, the border between visual and somatosensory territories is not strict, since bimodal visual and somatosensory neurons can be recorded in a broader area around this transitional region (Deng and Wang 1992). The rostral somatosensory HA also harbors in its medial and deep aspects descending motor projections to the nucleus ruber from where fibers descend to the spinal cord (Wild 1992). Cytoarchitectonically, the visual and the somatosensory components of the Wulst are indistinguishable. The same is true for our MR images. Although we are fully aware that the border between the visual and the somatosensory Wulst is in no way sharp, we nevertheless decided to divide this structure at A 13.00 to visualize the presence of a posterior visual and an anterior somatosensory component.
It should be noted that the visual thalamofugal pathway is possibly also the system that mediates at least a part of the magnetic compass information. In migrating bird species, a subcomponent of the HA plays a critical role in compass mediated orientation (Zapka et al. 2009). Magnetic compass signals are possibly transferred via the GLd towards the Wulst (Heyers et al. 2007). Since pigeons also sense the magnetic compass (Wilzeck et al. 2010), it is likely that also in this species the thalamofugal system could mediate magnetic information.
The accessory optic system
The accessory optic system (AOS) is constituted by a group of brainstem structures that control gaze stabilization by analyzing self motion. In pigeons, this system consists of the n. of the basal optic root (nBOR) and the pretectal n. lentiformis mesencephali (LM). The NBOR receives direct retinal input from displaced ganglion cells and can be subdivided in at least three components (Brecha et al. 1980). The mammalian pretectal nucleus of the optic tract (NOT) is also retinorecipient and consists of two subdivisions. The nBOR complex projects to diverse structures like the LM, the vestibulocerebellum, and the n. pontis medialis (MP) (Wylie et al. 1997). Diekamp et al. (2001) also reported a small projection to the n. rotundus. The nBOR and the LM could easily be recognized by the T2 protocol, but since their respective subdivisions were not visible, we delineated them as a whole. In addition we delineated the n. pontis medialis (MP), which was best visible in T2* (Fig. 4b).
The isthmic nuclei
The isthmic nuclei of birds consist of the four midbrain structures: n. isthmo-opticus (IO), n. isthmi pars parvocellularis (Ipc), n. isthmi pars magnocellularis (Imc) and n. semilunaris (SLu). While the IO is part of the centrifugal visual system, Ipc, Imc, and SLu constitute a network of midbrain nuclei that enables fast selection of relevant visual objects and subsequent switches of attention. The ION consists of a highly convoluted lamina in which two perikaryal layers are separated by a neuropil in which the dendrites from opposing layers ramify toward the middle of the two layers (Güntürkün 1987). Afferent tectal axons synapse topographically on these dendrites while axons from ION cells emerge at opposing ends of the two lamina and proceed to the contralateral retina. These centrifugal axons terminate in the retina and could possibly modulate attentional switches between retinal subfields and could “highlight” relevant visual objects (Catsicas et al. 1987; Uchiyama 1989). Due to its complex internal structure, ION was best visualized by T2*. Neither T2 nor T2* enabled us to see the scatter of ectopic ION neurons that surround the nucleus ventrally and laterally (Fig. 4b).
Ipc and SLu are cholinergic structures that have precise homotopic reciprocal connections with the ipsilateral optic tectum (Güntürkün and Remy 1990; Gruberg et al. 2006). In contrast, the GABAergic Imc neurons receive only a coarse topographical tectal projection and projects back in a heteotopical manner. Some Imc neurons also innervate Ipc and SLu (Wang et al. 2004, 2006). This isthmo-tectal network enables a fast switch of visual attention to the most salient visual targets and their features (Knudsen 2011). These three structures were easily visible in T2 and T2*. However, the T2* protocol provided a higher contrast of nuclear areas against the dense brachium colliculi superioris.
The somatosensory pathways
Spinal system and body representation
Primary sensory neurons of the somatosensory pathway transmit the signal of mechanoreceptors of the body below the head to the nuclei of the dorsal column (n. gracilis et cuneatus, GC; n. cuneatus externus, CE) of the medulla oblongata (Wild 1985). The dorsal column nuclei project to the thalamus via the medial lemniscus. The main somatosensory thalamic nucleus is the n. dorsalis intermedius ventralis anterior (DIVA) (Wild 1987). A smaller contingent of dorsal column nuclei afferents reaches the n. dorsolateralis posterior thalami (DLP) (Korzeniewska and Güntürkün 1990). DIVA projects to the rostrally located somatosensory Wulst (see the discussion on the thalamofugal visual pathway). Efferents of the DLP ascend to a somatosensory area in the intermediate and caudomedial nidopallium (Wild 1987; Güntürkün and Kröner 1999). We could not separate GC and CE with T2 or T2* with certainty at all levels. These two structures were therefore combined. While the delineation of DLP was straightforward using T2, the recognition of the borders of DIVA was more difficult with either scanning protocol. The dorsal border was partly reconstructed by referring to Cresyl Violet sections. The nidopallial projection zone of the DLP was not visible with our scanning protocols (Fig. 4c).
Trigeminal system and beak representation
The trigeminal nerve, which innervates the beak of all birds, is topographically represented in n. sensorius principalis nervi trigemini PrV (Wild and Zeigler 1996). The PrV projects via the quintofrontal tract to the n. basorostralis palii (Bas), bypassing the thalamus (Wild 1985). As described below, Bas also receives auditory input from the brainstem (Arends and Zeigler 1986; Schall et al. 1986). Both the PrV and the Bas were best visualized with the T2 protocol (Fig. 4c).
Auditory pathway
The fibers of the nervus octavus project to both the n. angularis (An) and the n. magnocellularis (MC). The MC projects bilaterally to the n. laminaris (La), which is the first nucleus to receive an input from both ears (Necker 2000). The An projects mainly contralaterally to the midbrain n. mesencephalicus lateralis pars dorsalis (MLD) to give off collaterals to the oliva superior (OS) and to the n. of the lateral lemniscus (Ll; Arends and Zeigler 1986). The La has a similar projection as the An and both of these structures have segregated terminal fields within the MLd of pigeons (Wild 1995). The superior olive (OS) also projects to the MLD (Wild 1995). The three nuclei of the lateral lemniscus have differential projections. While the central one projects to Bas (see section on the “Trigeminal system”), the dorsal and ventral nuclei project to the MLD and to the thalamic n. semilunaris parvovoidalis (Wild 1987). The midbrain MLD projects ipsilaterally to the thalamic n. ovoidalis (Ov) and SPO. Ov and SPO project to the ventral and dorsal parts of Field L2 in the forebrain, respectively (Wild et al. 1993). Field L has been divided into three laminae (Ll, L2, L3) with L1 and L3 receiving input from L2 (Bonke et al. 1979) (Fig. 4d).
An, MC, and La are very difficult to differentiate. Our delineation was done in T2* with the aid of histological sections. The delineation of OS and LL were also performed in T2*, but did not follow the locations given in the atlas of Karten and Hodos (1967). Instead, we followed the anatomical redefinition of Arends and Zeigler (1986). These authors subdivided the LL in three components. Since we were unable to see them with certainty in MR, we herewith provide the whole LL as a single structure. Similarly, the MLd is shown as a single entity, without the various subdivisions described by Logerot et al. (2011). While it was rather easy to depict Ov in T2 and T2*, we could not delineate SPO with certainty and therefore left it out. Field L as shown here very likely is confined to Field L2a plus L2b (Wild et al. 1993).
Olfactory pathway
The olfactory nerve projects to olfactory bulb (BO) from where efferent mitral cells have efferents to various telencephalic structures. The main areas of termination are the cortex prepiriformis (CPP), cortex piriformis (Cpi), n. taeniae amygdalae (TnA) (Reiner and Karten 1985; Bingman et al. 1994; Patzke et al. 2011). All of these structures were visible with T2 (Fig. 4d).
Hippocampal complex
The pigeons’ hippocampal complex occupies the medial and dorsomedial wall of the medial and caudal telencephalon. Hippocampal lesions produce impairments on a variety of spatial tasks, such as navigation, maze learning, and the retention of spatial information (Gagliardo et al. 1999; Vargas et al. 2004). Since both birds and mammals with damage to the hippocampus are not impaired on a variety of other diagnostic tasks, Colombo and Broadbent (2000) suggested that the avian hippocampus is a functional homolog of the mammalian hippocampus. Also neurogenetic (Reiner et al. 2004), developmental (Redies et al. 2001), immunohistochemical (Székely 1999) and hodological (Casini et al. 1986; Atoji et al. 2002, 2006; Kahn et al. 2003) evidence makes a homology to the mammalian hippocampal formation likely (Fig. 5).
Atoji and Wild (2004) have presented the most sophisticated analysis of the hippocampal subdivisions in pigeons and we used their results for the present study. These authors have classified seven subdivisions of the pigeon hippocampal formation. The most prominent one in nissl sections is the V-shaped area close to the medial wall of the telencephalon that extents over a long anterioposterior distance. We barely could see this structure with our scanning protocols and were unable to see the small longitudinal nuclei at the most dorsomedial edge of the forebrain. The parahippocamapal sulcus, however, that runs rostrocaudally on the ventricular face and is used as a boundary between DL and DM was a useful landmark for the present study. Also the distinction from the hyperpallium apicale was possible. Therefore, we delineated the hippocampal formation as a single entity that is constituted by the triangular area, its two adjacent lamina, the dorsomedial hippocampal region, and the three subnuclei that constitute the dorsomedial wall of the hippocampal formation.
Descending systems
Basal ganglia
The basal ganglia play a key role in the selection and control of movement patterns as well as the learning of new behavioral routines (Reiner et al. 1998). The striatal component of the basal ganglia receives input from diverse pallial fields (Veenman et al. 1995) and projects to the globus pallidus, the substantia nigra, the area ventralis tegmentalis and the n. lateralis spiriformis (Reiner and Karten 1985). We have incorporated the telencephalic components of the basal ganglia into the atlas. The striatum as delineated here incorporates both the lateral as well as the medial striatum. Although several cellular features make these two components distinct from each other, they were included into the terminus “striatum” in the new avian nomenclature and so were depicted as a unity here (Reiner et al. 2004). The globus pallidus is mostly encapsulated by the striatum with the exception of its ventromedial component where the fasciculus prosencephali lateralis and other fiber bundles enter of leave the basal ganglia system. Neither the n. accumbens nor the bed nucleus of the stria terminalis or the intrapeduncular nucleus could be discerned properly with our scanning protocols and were therefore left out (Fig. 5).
Arcopallium and amygdala
The arcopallium consists of the arcopallium anterius, dorsale, intermedium, and mediale and constitutes a collection of somatomotor structures in the caudoventral forebrain. It receives afferents from a large number of sensory and associative structures. Possibly the largest input arrives via the tractus dorsoarcopallialis from the nidopallium caudolaterale, an associative forebrain structure that is supposed to be functionally equivalent to the mammalian prefrontal cortex (Herold et al. 2011). The tractus occipitomesencephalicus (OM), a compact bundle of thick descending fibers, arises in the AI and sweeps down into the diencephalon and towards the brainstem. Extending laterally, OM fibers enter the optic tectum, the mesencephalic and medullary formatio reticularis, the substantia nigra, and a large number of sensory and brainstem structures up to the level of the spinal cord (Zeier and Karten 1971; Dubbeldam et al. 1997) (Fig. 5).
The avian amygdala consists of the posterior pallial amygdala, the subpallial amygdala, and the n. taeniae amygdalae (TnA). Descending projections of the amygdalar complex are bundled in the tractus occipitomesencephalicus, pars hypothalami (HOM). The HOM connects the amygdala with the bed nucleus of the stria terminalis, preoptic and hypothalamic nuclei, as well as noradrenergic and dopaminergic structures of the brainstem (Zeier and Karten 1971; Dubbeldam et al. 1997; Cheng et al. 1999).
The TnA had been delineated as part of the olfactory system. With none of our scanning protocols could we further delineate the remaining arcopallial or amygdalar substructures with certainty. We therefore decided to incorporate them into a single group.
Wulst
The hyperpallium apicale (HA) gives rise to the tractus septomesencephalicus (TSM) which sweeps down along the medial wall of the telencephalon to innervate thalamic and brainstem nuclei (Karten et al. 1973). The TSM also innervates the tectum to modulate ascending and descending components of the tectal efferent systems (Miceli et al. 1987). The HA complex has been delineated as part of the thalamofugal visual and the ascending somatosensory system (Fig. 5).
References
Adams A, Santi A (2011) Pigeons exhibit higher accuracy for chosen memory tests than for forced memory tests in duration matching-to-sample. Learn Behav 39:1–11
Arends JJ, Zeigler HP (1986) Anatomical identification of an auditory pathway from a nucleus of the lateral lemniscal system to the frontal telencephalon (nucleus basalis) of the pigeon. Brain Res 398:375–381
Atoji Y, Wild JM (2004) Fiber connections of the hippocampal formation and septum and subdivisions of the hippocampal formation in the pigeon as revealed by tract tracing and kainic acid lesions. J Comp Neurol 475:426–461
Atoji Y, Wild JM, Yamamoto Y, Suzuki Y (2002) Intratelencephalic connections of the hippocampus in pigeons. J Comp Neurol 447:177–199
Atoji Y, Saito S, Wild JM (2006) Fiber connections of the compact division of the posterior pallial amygdala and lateral part of the bed nucleus of the stria terminalis in the pigeon (Columba livia). J Comp Neurol 499:161–182
Bingman VP, Casini G, Nocjar C, Jones TJ (1994) Connections of the piriform cortex in homing pigeons (Columba livia) studied with fast blue and WGA-HRP. Brain Behav Evol 43:206–218
Bonke BA, Bonke D, Scheich H (1979) Connectivity of the auditory forebrain nuclei in the guinea fowl (Numida meleagris). Cell Tissue Res 200:101–121
Brecha N, Karten HJ, Hunt SP (1980) Projections of the nucleus of basal optic root in the pigeon: an autoradiographic and horseradish peroxidase study. J Comp Neurol 189:615–670
Browning R, Overmier JB, Colombo M (2011) Delay activity in avian prefrontal cortex—sample code or reward code? Eur J Neurosci 33:726–735
Carvalho LS, Cowing JA, Wilkie SE, Bowmaker JK, Hunt DM (2007) The molecular evolution of avian ultraviolet- and violet-sensitive visual pigments. Mol Biol Evol 24:1843–1852
Casini G, Bingman VP, Bagnoli P (1986) Connections of pigeon dorsomedial forebrain studied with HRP and 3H-proline. J Comp Neurol 245:454–470
Catsicas S, Catsicas M, Clarke PGH (1987) Long-distance intraretinal connections in birds. Nature 326:186–187
Cheng M-F, Chaiken M, Zuo M, Miller H (1999) Nucleus taeniae of the amygdala of birds: anatomical and suctional studies in ring doves (Streptopelia risoria) and European starlings (Sturnus vulgaris). Brain Behav Evol 53:243–270
Colombo M, Broadbent NJ (2000) Is the avian hippocampus a functional homologue of the mammalian hippocampus? Neurosci Biobehav Rev 24:465–484
De Groof G, Verhoye M, Van Meir V, Tindemans I, Leemans A, Van der Linden A (2006) In vivo diffusion tensor imaging (DTI) of brain subdivisions and vocal pathways in songbirds. Neuroimage 29:754–763
Deng C, Wang B (1992) Overlap of somatic and visual response areas in the Wulst of pigeon. Brain Res 582:320–322
Desai M, Kahn I, Knoblich U, Bernstein J, Atallah H, Yang A, Kopell N, Buckner RL, Graybiel AM, Moore CI, Boyden ES (2011) Mapping brain networks in awake mice using combined optical neural control and fMRI. J Neurophysiol 105:1393–1405
Diekamp B, Hellmann B, Troje NF, Wang SR, Güntürkün O (2001) Electrophysiological and anatomical evidence for a direct projection from the nucleus of the basal optic root to the nucleus rotundus in pigeons. Neurosci Lett 305:103–106
Dubbeldam JL, Den Boer-Visser AM, Bout RG (1997) Organization and efferent connections of the archistriatum of the Mallard, Anas platyrhynchos L.: an anterograde and retrograde tracing study. J Comp Neurol 388:632–657
Ferster CB, Skinner BF (1957) Schedules of reinforcement. Appleton-Century-Crofts, New York
Fredes F, Tapia S, Letelier JC, Marín G, Mpodozis J (2010) Topographic arrangement of the rotundo-entopallial projection in the pigeon (Columba livia). J Comp Neurol 518(21):4342–4361
Friedrich AM, Clement TS, Zentall TR (2004) Functional equivalence in pigeons involving a four-member class. Behav Process 67(3):395–403
Gagliardo A, Ioale P, Bingman VP (1999) Homing in pigeons: the role of the hippocampal formation in the representation of landmarks used for navigation. J Neurosci 19(1):311–315
Gruberg E, Dudkin E, Wang Y, Marín G, Salas C, Sentis E, Letelier J, Mpodozis J, Malpeli J, Cui H, Ma R, Northmore D, Udin S (2006) Influencing and interpreting visual input: the role of a visual feedback system. J Neurosci 26(41):10368–10371
Güntürkün O (1987) A Golgi study of the isthmic nuclei in the pigeon (Columba livia). Cell Tissue Res 248:439–448
Güntürkün O (2000) Sensory physiology: vision. In: Whittow GC (ed) Sturkie’s avian physiology. Academic Press, Orlando, pp 1–19
Güntürkün O, Karten HJ (1991) An immunocytochemical analysis of the lateral geniculate complex in the pigeon (Columba livia). J Comp Neurol 314:721–749
Güntürkün O, Kröner S (1999) A polysensory pathway to the forebrain of the pigeon: the ascending projections of the n. dorsolateralis possterior thalami (DLP). Eur J Morphol 37:124–128
Güntürkün O, Remy M (1990) The topographical projection of the n. isthmi pars parvocellularis (Ipc) onto the tectum opticum in the pigeon. Neurosci Lett 111:18–22
Güntürkün O, Melsbach G, Hörster W, Daniel S (1993) Different sets of afferents are demonstrated by the two fluorescent tracers fast blue and rhodamine. J Neurosci Methods 49:103–111
Güntürkün O, Hellmann B, Melsbach G, Prior H (1998) Asymmetries of representation in the visual system of pigeons. NeuroReport 9:4127–4130
Hellmann B, Güntürkün O (2001) The structural organization of parallel information processing within the tectofugal visual system of the pigeon. J Comp Neurol 429:94–112
Herold C, Palomero-Gallagher N, Hellmann B, Kröner S, Theiss C, Güntürkün O, Zilles K (2011) The receptorarchitecture of the pigeons’ nidopallium caudolaterale—an avian analogue to the prefrontal cortex. Brain Struct Funct 216:239–254
Heyers D, Manns M, Luksch H, Güntürkün O, Mouritsen H (2007) A visual pathway links brain structures active during magnetic compass orientation. PLoS ONE 2(9):e937
Jarvis ED, Güntürkün O, Bruce L, Csillag A, Karten HJ, Kuenzel W, Medina L, Paxinos G, Perkel DJ, Shimizu T, Striedter G, Wild M, Ball GF, Dugas-Ford J, Durand S, Hough G, Husband S, Kubikova L, Lee D, Mello CV, Powers A, Siang C, Smulders TV, Wada K, White SA, Yamamoto K, Yu J, Reiner A, Butler AB (2005) Avian brains and a new understanding of vertebrate brain evolution. Nat Rev Neurosci 6:151–159
Joly O, Ramus F, Pressnitzer D, Vanduffel W, Orban GA (2011) Interhemispheric differences in auditory processing revealed by fMRI in awake rhesus monkeys. Cereb Cortex [Epub ahead of print]
Kahn MC, Bingman VP (2009) Avian hippocampal role in space and content memory. Eur J Neurosci 30:1900–1908
Kahn MC, Hough GE II, Eyck GRT, Bingman VP (2003) Internal connectivity of the homing pigeon (Columba livia) hippocampal formation: an anterograde and retrograde tracer study. J Comp Neurol 459:127–141
Kangas BD, Vaidya M, Branch MN (2010) Titrating-delay matching-to-sample in the pigeon. J Exp Anal Behav 94:69–81
Karten HJ, Hodos W (1967) A stereotaxic atlas of the brain of the pigeon. Johns Hopkins Press, Baltimore
Karten HJ, Hodos W, Nauta WJ, Revzin AM (1973) Neural connections of the “visual wulst” of the avian telencephalon. Experimental studies in the pigeon (Columba livia) and owl (Speotyto cunicularia). J Comp Neurol 150:253–278
Knudsen EI (2011) Control from below: the role of a midbrain network in spatial attention. Eur J Neurosci 33:1961–1972
Korzeniewska E, Güntürkün O (1990) Sensory properties and afferents of the n. dorsolateralis posterior thalami (DLP) of the pigeon. J Comp Neurol 292:457–479
Krützfeldt NO, Wild JM (2005) Definition and novel connections of the entopallium in the pigeon (Columba livia). J Comp Neurol 490:40–56
Logerot P, Krützfeldt NO, Wild JM, Kubke MF (2011) Subdivisions of the auditory midbrain (n. mesencephalicus lateralis, pars dorsalis) in zebra finches using calcium-binding protein immunocytochemistry. PLoS ONE 6:e20686
Marín G, Letelier JC, Henny P, Sentis E, Farfán G, Fredes F, Pohl N, Karten H, Mpodozis J (2003) Spatial organization of the pigeon tectorotundal pathway: an interdigitating topographic arrangement. J Comp Neurol 45:361–380
Miceli D, Repérant J, Villalobos J, Dionne L (1987) Extratelencephalic projections of the avian visual Wulst. A quantitative autoradiographic study in the pigeon Columba livia. J Hirnforsch 28:45–57
Miceli D, Marchand L, Repérant J, Rio J-P (1990) Projections of the dorsolateral anterior complex and adjacent thalamic nuclei upon the visual Wulst in the pigeon. Brain Res 518:317–323
Necker R (2000) The avian ear and hearing. In: Whittow GC (ed) Sturkie’s avian physiology. Academic Press, Orlando, pp 21–38
Patzke N, Manns M, Güntürkün O (2011) Telencephalic organisation of the olfactory system in homing pigeons (Columba livia). Neuroscience 194:53–61
Poirier C, Vellema M, Verhoye M, Van Meir V, Wild JM, Balthazart J, Van Der Linden A (2008) A three-dimensional MRI atlas of the zebra finch brain in stereotaxic coordinates. Neuroimage 41:1–6
Poirier C, Boumans T, Verhoye M, Balthazart J, Van der Linden A (2009) Own-song recognition in the songbird auditory pathway: selectivity and lateralization. J Neurosci 29:2252–2258
Prior H, Wiltschko R, Stapput K, Güntürkün O, Wiltschko W (2004) Visual lateralization and homing in pigeons. Behav Brain Res 154:301–310
Redies C, Medina L, Puelles L (2001) Cadherin expression by embryonic divisions and derived gray matter structures in the telencephalon of the chicken. J Comp Neurol 438:253–285
Reiner A, Karten HJ (1985) Comparison of olfactory bulb projections in pigeons and turtles. Brain Behav Evol 27:11–27
Reiner A, Medina L, Veenman CL (1998) Structural and functional evolution of the basal ganglia in vertebrates. Brain Res Brain Res Rev 28:235–285
Reiner A, Bruce L, Butler A, Csillag A, Kuenzel W, Medina L, Paxinos G, Perkel D, Powers A, Shimizu T, Striedter G, Wild M, Ball G, Durand S, Güntürkün O, Lee D, Mello C, White S, Hough G, Kubikova L, Smulders T, Wada K, Dugas-Ford J, Husband S, Yamamoto K, Yu J, Siang C, Jarvis ED (2004) Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol 473:377–414
Remy M, Güntürkün O (1991) Retinal afferents of the tectum opticum and the nucleus opticus principalis thalami in the pigeon. J Comp Neurol 305:57–70
Rose J, Schiffer A-M, Dittrich L, Güntürkün O (2010) The role of dopamine in maintenance and distractability of attention in the ‘prefrontal cortex’ of pigeons. Neuroscience 167:232–237
Scarf D, Colombo M (2010) Representation of serial order in pigeons (Columba livia). J Exp Psychol Anim Behav Process 36:423–429
Schall U, Güntürkün O, Delius JD (1986) Sensory projections to the nucleus basalis prosencephali of the pigeon. Cell Tissue Res 245:539–546
Shimizu T, Karten HJ (1990) Immunohistochemical analysis of the visual Wulst of the pigeon (Columba livia). J Comp Neurol 300:346–369
Shimizu T, Karten HJ (1993) The avian visual system and the evolution of the neocortex. In: Zeigler HP, Bischof H-J (eds) Vision, brain and behavior in birds. MIT Press, Cambridge
Székely AD (1999) The avian hippocampal formation: subdivisions and connectivity. Behav Brain Res 98:219–225
Uchiyama H (1989) Centrifugal pathways to the retina: influence of the optic tectum. Visual Neurosci 3:183–206
Van Meir V, Boumans T, De Groof G, Van Audekerke J, Smolders A, Scheunders P, Sijbers J, Verhoye M, Balthazart J, Van der Linden A (2005) Spatiotemporal properties of the BOLD response in the songbirds’ auditory circuit during a variety of listening tasks. Neuroimage 25:1242–1255
Vargas JP, Petruso EJ, Bingman VP (2004) Hippocampal formation is required for geometric navigation in pigeons. Eur J Neurosci 20:1937–1944
Veenman CL, Wild JM, Reiner A (1995) Organization of the avian ‘corticostriatal’ projection system: a retrograde and anterograde pathway tracing study in pigeons. J Comp Neurol 354:87–126
Vellema M, Verschuerena J, Van Meira V, Van der Linden A (2011) A customizable 3-dimensional digital atlas of the canary brain in multiple modalities. Neuroimage 57:352–361
Wang Y, Major DE, Karten HJ (2004) Morphology and connections of nucleus isthmi pars magnocellularis in chicks (Gallus gallus). J Comp Neurol 469:275–297
Wang Y, Luksch H, Brecha NC, Karten HJ (2006) Columnar projections from the cholinergic nucleus isthmi to the optic tectum in chicks (Gallus gallus): a possible substrate for synchronizing tectal channels. J Comp Neurol 494:7–35
Watanabe M, Ito H, Masai H (1983) Cytoarchitecture and visual receptive neurons in the Wulst of the Japanese quail (Coturnix coturnix japonica). J Comp Neurol 213:188–198
Wild JM (1985) The avian somatosensory system. I. Primary spinal afferent input to the spinal cord and brainstem in the pigeon (Columba livia). J Comp Neurol 240:377–395
Wild JM (1987) The avian somatosensory system: connections of regions of body representation in the forebrain of the body. Brain Res 412:205–223
Wild JM (1992) Direct and indirect “cortico”-rubral and rubro-cerebellar cortical projections in the pigeon. J Comp Neurol 326:623–636
Wild JM (1995) Convergence of somatosensory and auditory projections in the avian torus semicircularis, including the central auditory nucleus. J Comp Neurol 358:465–486
Wild JM, Zeigler HP (1996) Central projections and somatotopic organisation of trigeminal primary afferents in pigeon (Columba livia). J Comp Neurol 368:136–152
Wild JM, Karten HJ, Frost BJ (1993) Connections of the auditory forebrain in the pigeon (Columba livia). J Comp Neurol 337:32–62
Wilzeck C, Wiltschko W, Güntürkün O, Wiltschko R, Prior H (2010) Lateralization of magnetic compass orientation in pigeons. J R Soc Interface 7:235–240
Wylie DRW, Linkenhoker B, Lau KL (1997) Projections of the nucleus of the basal optic root in pigeons (Columba livia) revealed with biotinylated dextrane amine. J Comp Neurol 384:517–536
Xiao Q, Güntürkün O (2008) Do pigeons perceive the motion aftereffect? A behavioral study. Behav Brain Res 187:327–333
Yamazaki Y, Aust U, Huber L, Güntürkün O (2007) Lateralized cognition: asymmetrical and complementary strategies of pigeons during discrimination of the “human” concept. Cognition 104:315–344
Zapka M, Heyers D, Hein CM, Engels S, Schneider NL, Hans J, Weiler S, Dreyer D, Kishkinev D, Wild JM, Mouritsen H (2009) Visual but not trigeminal mediation of magnetic compass information in a migratory bird. Nature 461:1274–1277
Zeier H, Karten HJ (1971) The archistriatum of the pigeon: organization of afferent and efferent connections. Brain Res 31:313–326
Zentall TR, Weaver JE, Clement TS (2004) Pigeons group time intervals according to their relative duration. Psychon Bull Rev 11(1):113–117
Acknowledgments
Supported by the Deutsche Forschungsgemeinschaft through its SFB 874 (O.G.). We are grateful to Steven Staelens, Steven Deleye and Philippe Joye from MICA (Universiteit Antwerpen) for support during CT scans and Ariane Schwarz as well as Felix Ströckens for help with histology and microscopy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Güntürkün, O., Verhoye, M., De Groof, G. et al. A 3-dimensional digital atlas of the ascending sensory and the descending motor systems in the pigeon brain. Brain Struct Funct 218, 269–281 (2013). https://doi.org/10.1007/s00429-012-0400-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-012-0400-y