Abstract
We describe a new methodology for rapid 2D and 3D computer analysis and visualisation of gene expression and gene product pattern in the context of anatomy and tissue architecture. It is based on episcopic imaging of embryos and tissue samples, as they are physically sectioned, thereby producing inherently aligned digital image series and volume data sets, which immediately permit the generation of 3D computer representations. The technique uses resin as embedding medium, eosin for unspecific tissue staining, and colour reactions (β-galactosidase/Xgal or BCIP/NBT) for specific labelling of gene activity and mRNA pattern. We tested the potential of the method for producing high-resolution volume data sets of adult human and porcine tissue samples and of specifically and unspecifically stained mouse, chick, quail, frog, and zebrafish embryos. The quality of the episcopic images resembles the quality of digital images of true histological sections with respect to resolution and contrast. Specifically labelled structures can be extracted using simple thresholding algorithms. Thus, the method is capable of quickly and precisely detecting molecular signals simultaneously with anatomical details and tissue architecture. It has no tissue restrictions and can be applied for analysis of human tissue samples as well as for analysis of all developmental stages of embryos of a wide variety of biomedically relevant species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Modern embryology, biomedical research, and medical diagnosis and treatment rely on a profound understanding of the genetic and molecular pathways underlying normal and abnormal morphogenesis and tissue remodelling. To facilitate the precise analysis of developmental processes and their genetic background, various tools for 3D and 4D visualisations have been developed in the last two decades.
Micro-CT, Micro-MRI, ultrasound, in vivo molecular imaging, and selective plane illumination microscopy (Huisken et al. 2004; Jacobs and Fraser 1994; Louie et al. 2000; Schneider et al. 2003) have all shown considerable potential for analysing embryo development and malformation syndromes. All permit non-destructive imaging of specimens and in vivo examination. But Micro-CT, Micro-MRI, and ultrasound have important limitations. For example, minimal voxel sizes currently do not go below 10×10×10 μm, wherefore even large cells are only represented by a single voxel. In addition, the visualisation of molecular signals from gene expression or gene products is difficult or impossible. In vivo molecular imaging and selective plane illumination microscopy (Blasberg and Tjuvajev 2003; Ewald et al. 2002; Herschman 2003; Huisken et al. 2004; Massoud and Gambhir 2003; Weissleder and Ntziachristos 2003) cannot provide information on overall morphology in histological detail and cannot be used for imaging gene activity in mammalian embryos in utero or avian embryos in ovo.
Analysis of gene activity with respect to morphological detail still relies on post-mortem techniques employing optical sectioning [confocal microscopy, optical projection tomography (OPT)] or physical sectioning [external marker based image congruencing (EMAC), episcopic fluorescence image capturing (EFIC), and serial block-face scanning electron microscopy (SBFSEM)] of histologically prepared embryos and tissue samples (Denk and Horstmann 2004; Kolker et al. 2000; Sharpe et al. 2002; Streicher et al. 2000; Weninger et al. 1998; Weninger and Mohun 2002). However, each of these techniques is limited to a relatively narrow field of potential applications. Optical sectioning techniques and SBFSEM are optimised for cell organelles, small, transparent specimens, and early embryos. Techniques based on physical sections suffer from problems in detecting specifically stained cells and structures, involve complex methods of data generation, and can only be used with relatively thick sections, thereby limiting the resolution of volume data (Streicher et al. 1997, 2000; Weninger et al. 1998). Furthermore data quality is often hampered by distortions and realignment and congruencing procedures. Episcopic imaging techniques such as EFIC (Weninger and Mohun 2002) overcome most of these problems, but rely on the detection of tissue autofluorescence and its extinction by LacZ signals. However, tissue autofluorescence is weak in young embryos and signal detection due to suppression of autofluorescence precludes the simultaneous detection of morphology. Hence, no existing method is presently able to provide high-quality digital image series or volume data sets that can be used for precise 3D analysis of anatomy, tissue architecture, gene expression, and gene product pattern in embryos from early to late developmental stages.
We therefore developed such a method and, in order to demonstrate its robustness and broadness, used it for 3D analysis of tissue architecture in adult human tissue samples and transgene expression and gene product pattern with respect to highly detailed morphology in embryos of various species and developmental stages.
Materials and methods
We used human tissue samples obtained from anatomy specimens, adult porcine tissues (Sus scrofa domesticus), wild-type and transgenic (XMLC2a.rtTA:teTO.lacZ) mouse embryos (Mus musculus) of 8.5–14.5 dpc, chick embryos (Gallus domesticus) of stages 5 to 34 according to Hamburger and Hamilton (HH) (Hamburger and Hamilton 1992), quail embryos (Coturnix japonica) comparable to HH 20, frog tadpoles (Xenopus laevis) of approximately stage 36, and wild-type and mutant dominant dishevelled mRNA-injected zebrafish embryos (Matsui et al. 2005) (Danio rerio) of 28 hpf or prim stage 5 (Kimmel et al. 1995). All specimens were fixed in 4% PBS-buffered formaldehyde. Some specimens were whole mount in situ hybridised using BCIP/NBT labelling techniques (Ng et al. 2002). Transgenic mouse embryos were whole mount stained for transgene activity using standard β-galactosidase/Xgal staining techniques.
For sectioning and data generation, the specimens were dehydrated in ethanol and embedded in standard resin (JB4, Polyscience) that contained 0.40 mg/100 ml eosin (Chroma, Stuttgart). After hardening, the resin blocks were mounted on a rotary microtome (Microtec “CUT 4060E”). Due to a refinement of the mechanics of the microtome the stopping point of the block holder after each rotation step was kept constant with an accuracy of approximately 1 μm. The optical axis of a modified microscope (Leica “DM LM”) equipped with fluorescence filter sets and a digital video camera (Leica DC 480) was aligned with the upper stopping point of the block holder excursion (Fig. 1). During sectioning, episcopic images of every freshly cut block surface were captured, resulting in a series of precisely aligned images. Section thicknesses ranged from 0.5 to 10 μm, but were kept constant for each specimen. With section thicknesses greater than 2 μm the sections could be mounted on glass slides for later histological inspection, although this step is not required for the image capture and reconstruction technique HREM itself.
Some specimens were devoted to the analysis of tissue architecture and morphology alone. From these specimens, only one block surface image using the GFP filter set (excitation filter 470/40; emission filter 525/50) of the microscope was captured after each section until the entire block was sectioned. The resulting digital image series was converted to a volume data set and further processed and analysed using state-of-the-art image processing and 3D reconstruction software. Some specimens were devoted to the analysis of specifically labelled mRNA and transgenes in the context of anatomical detail and tissue architecture. From such specimens two images of each freshly cut block surface were captured until the entire block was sectioned. One image was captured with the Leica GFP filter set, again for depicting tissue architecture, but the second image was captured using the Leica TX2 filter set (excitation filter 560/40; emission filter 645/75). The TX2 filter enhances the molecular signal allowing an almost selective detection of the marked areas of gene activity and subsequent analysis with simple thresholding algorithms.
Images were processed using Photoshop version 8.0 (Adobe) and EmVis (developed by M. Knapp, Technical University of Vienna; http://www.cg.tuwien.ac.at/staff/MichaelKnapp.html). Surface- and volume-rendered 3D models were obtained using EmVis and Volocity (Improvision).
Results
Our new imaging and 3D analysing technique is based on volume data sets composed of subsequent digital images obtained from the block surface of resin-embedded embryos and tissue samples, as they are physically sectioned. Since the images are obtained from the block face rather than from glass-mounted sections, they are inherently aligned and free of sectioning and mounting artefact. Although the technique is based on episcopic images, these are of high resolution and almost true histological quality. Therefore we named this technique high-resolution episcopic microscopy (HREM). HREM uses the intrinsic fluorescence of resin-embedding media, such as JB4 (Polyscience), and the histological stain “eosin” for providing morphology. With the exception of adding eosin to the embedding medium, unstained or whole mount stained specimens are prepared according to standard protocols for resin embedding. The addition of eosin prior to polymerisation of the resin results in a reddish coloration, but the solution remains translucent and proper specimen positioning is possible. Camera images with pixel sizes from 0.25 μm×0.25 μm upwards can be generated. However, for most queries, pixel resolutions of approximately 0.5 μm×0.5 μm or larger proved to be sufficient. A section thickness of 0.5 μm can be achieved, but for routine purposes 2 or 3 μm thick sections are sufficient. Such sections can be retained and mounted on glass slides for subsequent light microscopy viewing.
To demonstrate the potential of HREM and the breadth of its application, we produced volume data sets of zebrafish embryos (D. rerio), frog tadpoles (X. laevis), quail (C. japonica), chick (G. domesticus), and mouse embryos (M. musculus) of different developmental stages, and tissue samples from pigs (Sus scrofa domesticus), and humans. The digital 2D images, the virtual 3D data sets, and the resulting 3D computer models proved to be of high resolution, providing details of overall morphology, topology, tissue architecture, and cellular structure (Fig. 2 and Table 1). Histological sectioning, HREM image capture, and section mounting were no more arduous or time consuming than the standard procedure for generating a resin block section series. 3D information and volume-rendered 3D models were available within a few hours after mounting the blocks on the microtome.
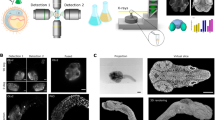
Images in different resolution, obtained with HREM using GFP filter sets. For high-resolution versions we refer to Table 1. Note that these are not images of glass-mounted sections, but episcopic images captured from a resin block surface. These images, in their original resolution are available at http://www.meduniwien.ac.at/3D-Rekonstr/HREM. a Human skin. Note the erythrocytes (approximate diameter 7 μm) inside the blood vessels (BV). b Porcine liver. Note the cell nuclei (N) are clearly visible. The inlay at the top left shows a digital zoom in of the outlined area of this very image in order to demonstrate its resolution. c Thorax of a mouse embryo (14 dpc). Sagittal section, ventral to the right. d Thorax of a chick embryo (HH 34). Sagittal section, ventral to the left. e Thorax of a quail embryo (HH 20). f Tadpole (approximately stage 36). Shrinkage artefacts are due to fixation and dehydration. E epidermis, D dermis, H hair follicle, AT adipose tissue, HL hepatic lobule, CT connective tissue, RV right ventricle, RA right atrium, RIC right common carotid artery, P lung, C rib, ICM intercostal muscles, Cl clavicle, T thyroid gland, TP transversal process of vertebra, SN spinal nerve, SG spinal ganglion, HE liver, r3 rd and r4 th right third and fourth pharyngeal arch arteries, NTL neural tube lumen, A atrium, AP atrial appendix, V ventricle, OT outflow tract, PPA pharyngeal arch arteries, PE pericardium, YS yolk sac, NT neural tube, M muscle, NC notochord, COR heart, CL pigment layer of the eye, L lens. Scalebars 200 μm
In addition to exploring the capacities of HREM to visualise anatomy and tissue architecture, we tested its potential to provide high-resolution images of specifically labelled gene expression signals and gene product patterns in developing embryos. For this purpose, we used zebrafish embryos stained for mlc2a and nkx2.5 products (Matsui et al. 2005) (nkx2.5 data not shown), chick embryos of various developmental stages in situ hybridised for qik, nkx2.5, and tbx5 gene products (nkx2.5 and tbx5 data not shown), and transgenic mouse embryos (XMLC2a.rtTA:teTO.lacZ) stained for LacZ activity. From each specimen two images of each freshly cut block surface were captured. The first image was captured using GFP-fluorescence excitation and emission filters. The second image was captured using an emission filter of 560/40 nm wavelength and an excitation filter of 645/75 nm (Leica TX2 filter set). Specifically stained cells and tissues could be detected in both images. However, the GFP images showed excellent morphology but the signal was hard to discern, while the images captured with the Leica TX2 filter set showed the signal in a specificity high enough to permit automatic thresholding (Fig. 3 and Table 1). Virtual reslices and 3D models obtained from the combined GFP and TX2 volume data sets revealed the 3D transgene and RNA distribution patterns superimposed upon remarkable anatomical detail and tissue architecture (Figs. 4, 5, 6).
Block surface images of whole mount stained specimens obtained with HREM. Images to the left were taken with a GFP filter set; images to the right were captured from the identical block surface with a Leica TX2 filter set. a, b Head region of a wild-type zebrafish embryo (28 hpf). Sagittal section. The GFP image (a) shows details of anatomical structures such as the lens (L), neural tube and its lumen (NTL), otic vesicle (OV), and developing heart tube (HT). The Leica TX2 image (b) enhances mlc2a products (mlc2a) in the developing heart tube. c, d Heart of a transgenic (XMLC2a.rtTA:tetO.lacZ) mouse embryo (12 dpc). Sagittal section. The GFP image (c) shows morphological details, such as the trabeculation of the forming ventricles (V), architecture of cushions forming at the atrioventricular junction (AVC) and of the cushions starting to divide the outflow tract (OTC). The Leica TX2 image (d) shows LacZ expression in circumscript areas of the myocardium. Note that sites of weakly stained myocardium surround sites of strongly stained myocardium. YS yolk sac, LA left atrial appendix. Scalebars 200 μm
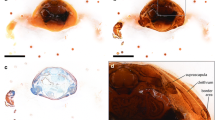
Surface-rendered 3D models of a mutant zebrafish embryo (double heart mutant) of approximately 28 hpf. Left panels show the morphology of the surface (grey) of the embryo sitting on the yolk sac (YS). Right panels show identical views with translucent surface (translucent grey), the otic vesicles (green), and mlc2a products in two forming heart tubes, the right (blue) smaller than the left (red). a, b Oblique view from dorsal left. c, d View of the head region from dorsal. e, f View from left. T tail. Scalebar 100 μm
Surface-rendered 3D models of the head region of a wild-type zebrafish embryo of approximately 28 hpf. Note that 2D images of the data sets underlying these models are displayed in Fig. 2. The models demonstrate details of morphology along with topology of the embryo’s surface (translucent grey), lumen of the neural tube (yellow), lens (orange), otic vesicle (green), and mlc2a products in the developing heart tube (translucent red). The yolk sac is not displayed. a, b View from left. c View from dorsal. Note the leftward shift of the anterior end of the heart tube, its shape and lumen. Scalebar 100 μm
Surface-rendered 3D models of the heart of a transgenic mouse embryo (XMLC2a.rtTA:tetO.lacZ). Note that 2D images of the data sets underlying these models are displayed in Fig. 2. a–c View from ventral. d–f Oblique view from right ventral. g–i Oblique view from left ventral. j–l View from left. Left-row panels (a, d, g, j) show the morphology of the cardiac wall and myocardial trabeculation (translucent red). Middle-row panels (b, e, h, k) show sites of weak myocardial LacZ expression (blue) along with morphology. Right-row panels (c, f, j, l) show sites of weak myocardial LacZ expression along with sites of strong myocardial LacZ expression (green) and morphology. Note the inhomogeneous expression pattern with enhanced transgene expression in the forming anterior interventricular groove (asterisk in b) and left atrial appendix (LA). RA right atrial appendix, OT outflow tract, RV forming right ventricle, LV forming left ventricle. Scalebar 200 μm
Discussion
Our data show that HREM, beside its simplicity and cost-efficiency due to the use of standard laboratory equipment, enables precise high-resolution analysis and 3D representation of positively contrasted gene expression and RNA pattern simultaneously with detailed anatomical structure. It uses the advantages of episcopic imaging for data generation. But in contrast to existing episcopic methods such as EFIC (Weninger and Mohun 2002), HREM is successful with all developmental stages of all the principal biomedically relevant model organisms, including specimens in which the autofluorescence is too low to allow proper analysis with EFIC (e.g. quail embryos or early embryonic stages). In addition, HREM overcomes the limited ability of EFIC for visualisation of specifically contrasted tissues. [EFIC is reduced to analyses of transgene expression pattern inside solid organs because it uses the extinction of tissue autofluorescence by LacZ (i.e. a “negative” contrast method).] HREM directly detects the product of colour reactions (i.e. a “positive” contrast method) and preserves the morphological information of stained structures since the staining reaction does not obscure tissue morphology. Furthermore, HREM enables distinction of strong and weak signals (Fig. 5) and thus, in principle, permits visualisation of gene product quantities.
HREM analysis requires histologically prepared specimens or tissue samples and cannot be applied to living specimens. However, compared to in vivo imaging methods such as CT, MRI, US, and in vivo molecular imaging (Huisken et al. 2004; Jacobs and Fraser 1994; Louie et al. 2000; Massoud and Gambhir 2003; Schneider et al. 2003), HREM data sets allow a precise and comprehensive detection of gene expression and gene product patterns including a high-resolution cellular detail. In contrast to confocal microscopy, OPT, and SBFSEM (Denk and Horstmann 2004; Sharpe et al. 2002)—techniques that are optimised for the subcellular level or for small, transparent embryos—HREM has no such restrictions. It can be used to obtain 3D data sets of both early and late embryos, obtained either from small or large organisms.
The digital images have histological quality and accurately show tissue architecture and cell morphology. 2D and 3D information of the sectioned specimens is available in nearly the same time frame as required for generating a series of traditional glass-mounted resin sections. The digital images of one volume data set are inherently aligned and do not show distortions caused by the sectioning or glass slide preparation process. Thus HREM has advantages over traditional microscopy and 3D reconstruction techniques based on glass-mounted sections such as EMAC (Streicher et al. 1997), which requires complex specimen and image processing procedures.
Besides its potential applications in embryo and biomedical research, HREM can be used to rapidly obtain 3D information of neonatal (data not shown) and adult tissue samples of humans, whilst at the same time preserving the sectioned material. The technique therefore can also be used as an analytical tool in pathology and forensic medicine.
In conclusion, with HREM, we provide a technique for rapidly generating high-resolution digital volume data sets of histologically sectioned material. Since HREM uses “positive” imaging for detecting both anatomical structure and molecular signal, the spectrum of potential applications includes analysis of embryos of all developmental stages and biomedially relevant model organisms and the examination of neonatal and adult human tissue samples. HREM is able to accurately detect cells, tissues and structures specifically contrasted with molecular staining techniques, which are in routine laboratory use. The high contrast and precise alignment of the data sets enable the reconstruction and analysis of comprehensive 3D and 4D models using state-of-the-art visualisation and morphometry software. Furthermore, HREM data quality is suitable for the mathematical characterisation of 3D gene expression patterns (Costa et al. 2004, 2005), which will be essential for developing quantitative models for analysing the dynamics of gene activity in development.
References
Blasberg RG, Tjuvajev JG (2003) Molecular-genetic imaging: current and future perspectives. J Clin Invest 111(11):1620–1629
Costa LdaF, Barbosa MS, Manoel ET, Streicher J, Müller GB (2004) Mathematical characterization of three-dimensional gene expression patterns. Bioinformatics 20(11):1653–1662
Costa LdaF, Beletti ME, Müller GB, Rasskin-Gutman D, Sternik G, Ibanes M, Belmonte JCI (2005) Field approach to three-dimensional gene expression pattern characterization. Appl Phys Lett 86(14):143901
Denk W, Horstmann H (2004) Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol 2(11):e329
Ewald AJ, McBride H, Reddington M, Fraser SE, Kerschmann R (2002) Surface imaging microscopy, an automated method for visualizing whole embryo samples in three dimensions at high resolution. Dev Dyn 225(3):369–375
Hamburger V, Hamilton HL (1992) A series of normal stages in the development of the chick embryo. 1951. Dev Dyn 195(4):231–272
Herschman HR (2003) Molecular imaging: looking at problems, seeing solutions. Science 302(5645):605–608
Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH (2004) Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305(5686):1007–1009
Jacobs RE, Fraser SE (1994) Magnetic resonance microscopy of embryonic cell lineages and movements. Science 263(5147):681–684
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203(3):253–310
Kolker SJ, Tajchman U, Weeks DL (2000) Confocal imaging of early heart development in Xenopus laevis. Dev Biol 218(1):64–73
Louie AY, Huber MM, Ahrens ET, Rothbacher U, Moats R, Jacobs RE, Fraser SE, Meade TJ (2000) In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol 18(3):321–325
Massoud TF, Gambhir SS (2003) Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev 17(5):545–580
Matsui T, Raya A, Kawakami Y, Callol-Massot C, Capdevila J, Rodriguez-Esteban C, Izpisua Belmonte JC (2005) Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev 19(1):164–175
Ng JK, Kawakami Y, Buscher D, Raya A, Itoh T, Koth CM, Rodriguez Esteban C, Rodriguez-Leon J, Garrity DM, Fishman MC, Izpisua Belmonte JC (2002) The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development 129(22):5161–5170
Schneider JE, Bamforth SD, Farthing CR, Clarke K, Neubauer S, Bhattacharya S (2003) High-resolution imaging of normal anatomy, and neural and adrenal malformations in mouse embryos using magnetic resonance microscopy. J Anat 202(2):239–247
Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sørensen J, Baldock R, Davidson D (2002) Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296(5567):541–545
Streicher J, Weninger WJ, Müller GB (1997) External marker-based automatic congruencing: a new method of 3D reconstruction from serial sections. Anat Rec 248(4):583–602
Streicher J, Donat MA, Strauss B, Sporle R, Schughart K, Müller GB (2000) Computer-based three-dimensional visualization of developmental gene expression. Nat Genet 25(2):147–152
Weissleder R, Ntziachristos V (2003) Shedding light onto live molecular targets. Nat Med 9(1):123–128
Weninger WJ, Mohun T (2002) Phenotyping transgenic embryos: a rapid 3-D screening method based on episcopic fluorescence image capturing. Nat Genet 30(1):59–65
Weninger WJ, Meng S, Streicher J, Müller GB (1998) A new episcopic method for rapid 3-D reconstruction: applications in anatomy and embryology. Anat Embryol (Berl) 197(5):341–348
Acknowledgements
The authors are grateful to M. Knapp, E. Gröller, K. Dorfmeister, G.M. Gruber, B. Maurer, and M.T. Nödl for technical support. This work was supported by the HFSP grant, RGP 0039/2002-C203.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Weninger, W.J., Geyer, S.H., Mohun, T.J. et al. High-resolution episcopic microscopy: a rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat Embryol 211, 213–221 (2006). https://doi.org/10.1007/s00429-005-0073-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-005-0073-x