Abstract
The Fgf/Fgfr (Fgf receptor) and Bmp signal pathways are critical for embryonic development and postnatal growth. In order to address their roles in tongue development, preliminary study of expression patterns of some important members in the two families, as well as of apoptosis and proliferation, were carried out in mouse developing tongue. Apoptosis in tongue is a very late event in embryogenesis, restricted to the upper layer of the epithelium whereas proliferation is very vigorous at the early stage of tongue development and remains active throughout embryogenesis. Bmp2, −4 and -5 were localized within the mesenchyme at the early embryonic stage of tongue development (E12 to E13), whereas Bmp3 and Bmp7 were mainly expressed in the epithelium. Most of these molecules were also seen in the tongue muscles at postnatal stages. Among Fgfr isoforms, Fgfr1c, −2b, and -2c were detected in embryogenesis with peak expression at E11 to E13. Fgfr1c and Fgfr2c were localized within the mesenchyme, while Fgfr2b was mainly expressed in the epithelium. High expression of Fgf7 and Fgf10 was also detected in the mesenchyme at the early embryonic stage of tongue development, corresponding to the Fgfr expression, suggesting that they are among the principal ligands functioning at the early embryonic expanding stage. Fgf2 was seen in the tongue muscles at the late embryonic and postnatal stages. These results suggest that Bmp and Fgf signalling regulates tongue development at multiple stages, possibly related to proliferation and differentiation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The tongue arises from the inner lining of the first four branchial arches in the ventral wall of the primitive oropharynx. Paired lateral lingual swellings and one unpaired median eminence (tuberculum impar) fuse in the midline to form the initial tongue bud. Its formation involves diverse embryologic origins that ultimately result in a complex pattern of innervation (Couly et al. 1992; Huang et al. 1999; Noden 1991). This unique process provides the tongue with both motor activity and tactile and gustatory sensation. The early tongue bud undergoes rapid enlargement at the initial developmental stage and differentiates into a muscular organ, covered by a specific epithelium containing several types of papillae including the taste buds (Shuler and Dalrymple 2001).
Rapid withdraw of the tongue in embryogenesis is critical for proper palatogenesis. Delay in this process may disturb proper palatal shelf elevation and hence might lead to cleft palate. The tongue also influences the paths of teeth eruption. Malformation of the tongue, typically macroglossia, is observed in human syndromes and other pathological conditions such as Down’s syndrome, and Beckwith-Wiedemann syndrome (Vogel et al. 1986). Anomalies like microglossia, agglossia and clefted glossia frequently occur in syndromic and non-syndromic mandibular hypoplasia (Singh and Bartlett 2005), but also occur as isolated developmental deformities even though the incidence is very rare. Ankyloglossia, on the other hand, is a fairly common disorder in the tongue.
The tongue is essentially a muscular organ. Its musculature is solely derived from the dermomyotome of occipital somite (Couly et al. 1992; Huang et al. 1999; Noden 1991). Developmental processes of skeletal muscles are not identical. The muscular development of the tongue is characterized by its early maturation and intercalation within tissues of neural crest origin. Molecular events of muscular development in tongue also differ from those of other skeletal muscles (Dalrymple et al. 1999; Yamane et al. 2000). Muscle development typically undergoes a series of processes, including determination, migration, proliferation, differentiation and maturation. These processes are mediated by intrinsic molecular factors, such as Pax3 and myogenic regulatory factors (MRFs); these genes are regulated by a number of other developmental genes, such as members of the Fgf and Bmp families (Amthor et al. 1999; Brunetti and Goldfine 1990).
The Bmp family belongs to the Tgf-β super family and contains more than 20 members that can be subgrouped according to the homology in their sequence. The Bmp signal pathway is crucial for gastrulation and organogenesis. During muscle development, Bmp signalling inhibits myogenic cell proliferation and muscular differentiation (Duprez et al. 1996; Huang et al. 2001; Musgrave et al. 2001; Tzahor et al. 2003). In the head, it has been shown that inhibition of Bmp signalling by its antagonist is required for muscle initiation (Tzahor et al. 2003). During muscle regeneration Bmp signalling was absent from this process, consistent with its negative role in muscle development (Zhao and Hoffman 2004).
Fgfs are also key participants in embryologic development. The Fgf signal is mediated predominately by tyrosine transmembrane receptors. So far, four Fgfrs have been identified (Fgfr1 to Fgfr4) as mediating signals from this large family. For Fgfr1 to Fgfr3, two exons, an invariant IIIa and one of IIIb or IIIc, encode the third Ig-loop producing IIIb and IIIc isoforms for each receptor (Ornitz and Itoh 2001). Those isoforms are differentially expressed and activated during embryogenic development (Ornitz and Itoh 2001). Fgf signalling plays a critical role in skeletal myogenesis (Flanagan-Steet et al. 2000; Patel et al. 1999; Scata et al. 1999). Its general effect seems to be to promote myoblast proliferation and regulate muscular differentiation.
Expression of some members from those two families has been described in murine and avian developing tongue at restricted stages (Huang et al. 2001; Jung et al. 1999). However, very little is known about their roles in tongue development. The patterns of some critical genes, such as Fgfrs, are also unknown. In order to address the roles of the Fgf and Bmp signal pathways in tongue development, a preliminary study of expression patterns of some important members in the two families, as well as of apoptosis and proliferation, was performed in mouse developing tongue. The results show that expression of the studied genes of the Fgf and Bmp families is developmentally regulated in mouse tongue, and the patterns are also remarkably different to that in avian.
Materials and methods
Preparation of tissues
All the procedures involving mouse use were approved by The Animal Welfare Committee of the University of Bergen. Mice (NMRI) embryos aged from embryonic day (ED) 11–18, as well as newborn (P0) and 3-, 5-, 7-days postnatal (P) mice were dissected in PBS and fixed in 4% PFA overnight at 4°C. The mice were killed by cervical dislocation and decapitation. Mice over E15 were decalcified with 12.5% EDTA–2.5% PFA in PBS. They were then embedded in paraffin following standard procedures. Sagital sections were obtained from the midline area of mouse heads. Apoptosis by TUNEL staining was carried out from E11 to E18; BrdU staining and in situ hybridization were performed from E11 to postnatal stages.
Proliferation by BrdU staining
For BrdU incorporation, pregnant and postnatal mice were injected intraperitoneally with 50 mg/kg of body weight. The mice were sacrificed 2 h after injection. Mice heads were fixed, embedded and sectioned following above-mentioned procedures. BrdU positive cells were detected with BrdU labeling and detecting kit according to the manufacture’s instruction. Briefly, tissue sections were deparaffinized by xylene, rehydrated through graded alcohol series. Endogenous peroxidase activity was quenched by incubating the sections in 0.3% H2O2 methanol solution for 30 min. They were then denatured with pre-warmed 2 M HCl for 30 min (37°C), digested by 2 mg/ml pepsin for 15 min and blocked with 10% goat serum and 2% BSA for 30 min. The monoclonal Anti-BrdU (Sigma) diluted in PBS were added to sections and incubated for 2 h at 37°C. After PBS washing, the biotinylated secondary antibody was added and incubated for 1 h at 37°C. Color reaction was detected by ABC kits and AEC or DAB substrate (Vector). The sections were counterstained by hematoxylin.
Apoptosis by TUNEL staining
TUNEL staining was carried out according to previously described method with modification (Gavrieli et al. 1992). The paraffin sections were deparaffinized in xylene, dehydrated in 100% ethanol, then incubated in 0.5% H2O2 in MeOH for 30 min and rehydrated with a series of MeOH in PBS. After washing the sections in PBT (PBS containing 0.1% Triton X-100), they were incubated in proteinase K solution (containing 50 mM Tris–HCl, 5 mM EDTA and 10 mg/ml proteinase K, pH 8.0) for 15 min and then washed three times in PBT for 5 min each.
The sections were labeled for 1 h at 37°C in a humid box with terminal deoxynucleotidyl transferase (TdT) solution. The reaction was stopped by incubating the slides in a solution containing 300 mM NaCl, 30 mM sodium citrate, 0.1% CHAPS for 15 min. After being blocked by 10% sheep serum and 2% BSA, solution of anti-digoxigenin antibody conjugated with alkaline phosphatase was pipetted onto the sections, which were incubated overnight in a humid box at 4°C. After washing, NBT and BCIP (Roche) solution was pipetted onto the sections and development of the color reaction was monitored under a microscope. The sections were counterstained with hematoxylin or methyl blue and mounted with Aquamount.
Preparation of probes for in situ hybridization
Preparation of the probes has been described previously (Aberg et al. 1997; Kettunen et al. 1998, 2000; Vainio et al. 1993). They were made through in vitro transcriptions from cDNA-containing plasmids, which were linealized and transcribed from T7 or SP6 promoters to generate sense and antisense riboprobes using in vitro transcription kit containing [35S] uridine 5′ triphosphate (Promega). After transcription, cDNA templates were digested with RNase-free DNase I for 15 min at 37°C. The probes were precipitated in ethanol, dried and dissolved in a mix of 0.1 M DTT and hybridization buffer. Bmp2-7, Fgfr1-3 isoforms and Fgf2, −7, −9, −10 probes were synthesized and used in the present study.
In situ hybridization
In situ hybridization on sections was performed as described previously (Wilkinson and Green 1990). Briefly, sections were deparaffinized in xylene, rehydrated through serial ethanol, washed in PBS, treated with proteinase K for 30 min, post-fixed in 4% PFA for 30 min, followed by 1 min in PBS containing glycine at concentration of 2 mg/ml. After washes in PBS for 25 min, sections were acetylated with freshly prepared acetic anhydride in triethanolamine-HCl (pH 8) for 10 min, followed by two times water washing. The sections were dehydrated by dipping into a series of ethanol solution (30, 50, 70 and 95%) for 30 s each, air-dried and used for hybridization. Probes were pipetted onto the sections and covered by parafilm. Hybridization was carried out for about 15 h at 55°C. Following hybridization, the sections were washed under high stringent conditions with 20 mM DTT in 50% formamide and 2× SSC for 1 h at 65°C. The sections were further washed for 1 h at 55°C in 0.1× SSC, dehydrated in 70% ethanol and air-dried. They were then dipped in NTB-2 emulsion (Eastman Kodak, USA) for autoradiography. After three week’s exposure at 4°C, they were developed and fixed, counterstained with hematoxylin and mounted with Depex. No specific signal was detected in sections hybridized with the control sense probes.
Image processing
Images were taken with a SPOT Insight digital camera (Diagnostic Instruments, Sterling Heights, MI, USA) mounted on a Zeiss Axioskop2 microscope (Carl Zeiss Jena GmbH, Jena, Germany). For sections of in situ hybridization, the bright-field and dark-field images of each section were digitized separately and the silver grains from dark fields were selected, colored red, and copied onto the bright-field image using Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA, USA).
Results
Apoptosis and cell proliferation in the developing tongue
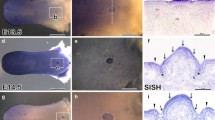
Apoptosis was very rare in the early stage of tongue development (E11 to E15). Sporadic apoptotic cells were detected within the mesenchyme and epithelium (Fig. 1a). At the late embryonic stage (from E16 to E18), more apoptotic cells were detected in the upper layers of the epithelium (Fig. 1b).
Apoptosis and proliferation in mouse tongue development. Sporadic apoptotic cells were detected in the mesenchyme and epithelium of the tongue at E16 (A). At E18, active apoptosis was detected on the upper layer of the epithelium (B). Framed areas in (A) and (B) were magnified 4 times and superimposed onto the images. At E13, cell proliferation was very vigorous (C). At E16, following muscular differentiation, proliferation was decreased within the tongue; however, in the epithelium, proliferation was still vigorous in the basal layers (D). At P7, active cell proliferation was maintained in the basal layers of epithelium, but very few proliferative cells were detected within the body of the tongue (E). Arrows indicates apoptotic and proliferative cells respectively. Scale bar in (A) represents 400 μm and applies to (B); scale bar in (C) represents 200 μm and applies to (D) and (E)
Cell proliferation, on the other hand, was very vigorous from the early initiation stage of the tongue. BrdU labeled cells were widespread within the tongue. The peak of proliferation was observed during E12 to E14 (Fig. 1c). It appeared that most of the cells were proliferating during this period. Vigorous cell proliferation persisted to the end of embryogenesis (Fig. 1d). At this stage, even though active cell proliferation was still observed, the overall amount of proliferative cells was decreased. At postnatal stages proliferative activity was further decreased from the mesenchyme, and BrdU labeled cells were gradually restricted to the basal epithelium at P7 (Fig. 1e).
Expression of Bmps in the developing tongue
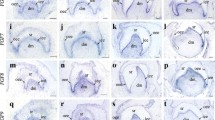
At E11, Bmp2 was moderately expressed within the mesenchyme of the tongue bud. One day later, Bmp2 transcripts were detected in the posterior mesenchyme and restricted domains of the epithelium (Fig. 2a). At E13, transcripts were seen in an anterior–posterior myogenic condensation within the tongue, adjacent to the dorsum (Fig. 2d). At E14, the expression was downregulated and later no longer detectable in embryonic development of the tongue (Fig. 2g). At postnatal stages, Bmp2 expression was reinitiated in the tongue muscles (Fig. 2j).
Bmp2, -4 and -5 expression in mouse tongue development. Bmp2 was highly expressed in restricted domains in the mesenchyme and epithelium at E12 (A). At E13 it was seen in the myogenic mesenchyme condensation at a high level (B). Bmp4 were moderately expressed in connective tissues within the tongue at E12 and E13 (B and E). Bmp 5 expression was widespread in the mesenchyme at E12 and intensified at E13 (C and F). At E14, their expression was downregulated (G, H and I). At P0, weak expression of Bmp2 and Bmp5 was reinitiated in the tongue muscles (J and l). Arrows indicate mRNA expression. Scale bar represents 400 μm
Bmp4 was widespread within the mesenchyme at E11 in the tongue bud. At E12, its expression was restricted in the mesenchyme and epithelium (Fig. 2b). One day later, Bmp4 expression was seen in the connective tissues among the myogenic condensations within the tongue body. It was also clearly seen in the ventral mesenchyme of the tongue (Fig. 2e). This expression was maintained to E14, and thereafter downregulated. After E16, it was on longer detectable (Fig. 2h, K).
Bmp5 was moderately expressed in the mesenchyme at E11 and E12 (Fig. 3c). At E13, this expression was intensified and widespread within the body of the tongue (Fig. 2f). Later, the expression was sharply downregulated. After E16, Bmp5 was no longer detectable in embryogenesis. At postnatal stages, its expression was weakly detected in the tongue muscles (Fig. 2l).
Bmp3, Bmp7 and Bmp6 expression in mouse tongue development. Bmp3 were expressed in both the epithelium and mesenchyme during E12 to E13 (A and B). Bmp7 was mainly expressed in the epithelium, and also weakly detected in the mesenchyme at E12 (C). At P5, it showed moderate expression in the muscles (D). Bmp6 was weakly expressed in the mesenchyme at E13 (E); at P5, it showed weak expression in the tongue muscles also. Arrows indicate expression. Scale bar represents 400 μm. Scale bar in (A) applies to (B), (C) and (E)
Bmp3 was expressed within the dorsum of the tongue bud at E11. At E12 and E13, it was moderately expressed in the epithelium and sub-epithelial mesenchyme; it was also weakly expressed in the central mesenchyme during this period (Fig. 3a, b). Thereafter it was no longer detectable. At postnatal stages, its expression was weakly reinitiated in the tongue muscles. Bmp7 was also moderately expressed in the epithelium at E11. One day later, transcripts were also weakly localized in the mesenchyme (Fig. 3c). This expression was maintained at E13 and was no longer detectable during the rest of embryonic stage. At postnatal stages, it was moderately expressed in the tongue muscles (Fig. 3d). Bmp6 was mainly detected between E13 and E16 within the mesenchyme (Fig. 3e). It also showed moderate expression in the tongue muscles at postnatal stages (Fig. 3f).
Expression of Fgfr isoforms in the developing tongue
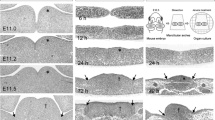
Fgfr1c was weakly expressed in the tongue bud at E11. The expression was increased during E12 and E13, widespread within the mesenchyme (Fig. 4a, b). Thereafter the expression was downregulated, and undistinguishable from background level at the late embryonic stage. At postnatal stages, its expression was barely detectable in the tongue muscles.
Fgfr1c, −2b and -2c expression in mouse tongue development. Fgfr1c and Fgfr2c were both detected in the mesenchyme at E12 and E13 (A, B, C and D). While expression of Fgfr1c was upregulated during this period, expression of Fgfr2c was downregulated. Fgfr2b was mainly expressed in the epithelium during this period (E and F). Arrows indicate expression. Scale bar represents 400 μm
High expression of Fgfr2c was detected at E11 within the mesenchyme of the tongue bud. One day later, the expression decreased sharply (Fig. 4c). At E13 and E14, its expression could only be weakly detected within the mesenchyme (Fig. 4d) and thereafter was no longer detectable in embryogenesis. At postnatal stages, Fgfr2c were also weakly detected in the tongue muscles.
Fgfr2b was moderately expressed in both the epithelium and mesenchyme at E11. One day later, its expression was seen in the epithelium and sub-epithelial mesenchyme (Fig. 4e). At E13, its expression was only detected within the epithelium (Fig. 4f). Later, Fgfr2b expression was no longer detectable.
The developing tongue was devoid of Fgfr1b, Fgfr3b and Fgfr3c expression in embryologic and early postnatal stages.
Expression of Fgfs in the developing tongue
Fgf7 was highly expressed in the central mesenchyme at E12, and the expression was downregulated from E13 (Fig. 5a, b). High expression of Fgf10 was also seen within the body of the tongue during E12 to E13 (Fig. 5c, d). Thereafter, they were both undetectable. Fgf9 were also detected in the mesenchyme in this period at a moderate level (data not shown). Fgf2 was first seen at E14 in individual myogenic cells in the body of the tongue (Fig. 6a). The transcripts were widely clustered in the geniohyoid muscle at this time. Later Fgf2 transcripts were also widely distributed within the body of the tongue following the orientation of myogenesis at a very high level (Fig. 6b, c). This expression was maintained to the postnatal stages (Fig. 6d).
Fgf2 expression in mouse tongue development. Fgf2 transcripts were clustered in the mesenchyme and oral epithelium at E14 (A). At E15 and E16, its expression was more widespread within the body of the tongue and intensified in the geniohyoid muscle (B and C). At postnatal stage, Fgf2 was highly expressed in the tongue muscles (D). Arrows indicate expression. Scale bar represents 400 μm
Discussion
In developing tongue, a rapid cell turnover in the epithelium from late embryonic stages was indicated by constant cell death in the upper layer and persistent proliferation in the basal layers. However, in the mesenchyme and later the myogenic cells, apoptosis was very rarely detected. Proliferation, on the other hand, was extremely vigorous in embryogenesis, concurring with another study (Nagata and Yamane 2004). The mesenchyme cells, after populating into the tongue primordia, first underwent a process of extensive proliferation that caused the rapid enlargement of the tongue during this period. High expression of both Fgfs/Fgfrs and Bmps was concentrated in this period, suggesting their roles in this rapid growth process. Thereafter the mesenchyme begins to differentiate at around E15 (Shuler and Dalrymple 2001). MRFs play a critical role in this process. Expression of MRFs occurs earlier in tongue than in other muscles (Yamane et al. 2000); correspondingly, maturation of myofibres in tongue also occurs earlier, reflecting the demands of early functions like sucking.
As mentioned above, high expression of Fgfs and Fgfrs was concentrated at the early stage of tongue development in embryogenesis; when cell proliferation was also most vigorous. Fgf/Fgfr signalling is mitogenic to most cell lineages including myogenic cells (Patel et al. 1999). The coincidence of Fgf/Fgfr expression and vigorous proliferation implies that this signalling is mitogenic for tongue growth at this stage. Fgfr1 has been shown to be critical for skeletal muscle development, and deletion or overexpression of this gene causes abnormal muscle development (Flanagan-Steet et al. 2000; Patel et al. 1999; Scata et al. 1999). During the development of tongue, Fgfr1c is likely the most important signal for mesenchyme proliferation and differentiation among different isoforms. Peak expression of Fgfr2c was earlier than Fgfr1c and sharply downregulated during the expanding period of the tongue, suggesting that this signal mainly acts at an early stage. Fgfr2c seems to be functionally redundant with Fgfr1c, as deletion of this gene results in no obvious defect in the tongue except the general small size seen in all head structures (Eswarakumar et al. 2002).
Fgf10 is a preferred ligand for Fgfr2b. The Fgf10/Fgfr2b signalling pathway plays crucial roles in the developmental processes of many organs (Alappat et al. 2005; Mailleux et al. 2002; Ohuchi et al. 2000). Deletion of these two genes shows a similar phenotype in mouse (De Moerlooze et al. 2000; Sekine et al. 1999). In developing tongue, expression of Fgf10 overlaps with Fgfr in the rapid expanding period. Spatiotemporal pattern of Fgf10 also implies that, besides being a ligand of Fgfr2b, it is a principal ligand for other receptors in the mesenchyme. In addition, Fgf7 appears to be another principal ligand within the mesenchyme.
Following differentiation, expression of Fgfrs and Fgf10 was downregulated; while Fgf2 was upregulated in the muscles. This expression pattern is different to that of other skeletal muscle, where Fgf2 was mainly expressed at proliferating myoblasts (Hannon et al. 1996). Fgf2 expressed from late embryonic stage is likely to regulate myogenic differentiation or maintain a certain proliferation rate. Hence, it appears that Fgf signalling has dual roles in regulating tongue development, promoting cell proliferation and regulating myogenic differentiation, consistent with its general roles in muscle development.
Expression of Bmps was also concentrated at an early stage of tongue development in embryogenesis, coincident with that of Fgfs and Fgfrs. The Bmp signal inhibits myogenic cell proliferation and differentiation (Huang et al. 2001; Tzahor et al. 2003). Therefore, it is likely Bmp and Fgf signal pathways have both antagonistic and synergistic role in early tongue development. The Fgf/Fgfr signal pathway predominates in cell proliferation, whereas the Bmp signal inhibits early muscular differentiation and maturation to maintain a high proliferation rate at the early developing stage. Bmp signalling also regulates connective tissue formation among tongue muscles, as demonstrated by specific expression of Bmp4 in these tissues. Following muscular differentiation, Bmp and Fgf/Fgfr expression was greatly decreased or disappeared from the tongue. At the postnatal stage, reinitiation of the Bmps in the tongue muscles implies that they are needed for regulation of the rate of muscular differentiation in postnatal development. Thus, functions of Fgf/Fgfr and Bmp signalling are important in regulating the pace of proliferation and differentiation in order to meet the specific growth need at different developmental stages.
Bmp signalling was also detected in the epithelium of the tongue and might play a role in epithelial differentiation. Bmp2 and -4 were localized in restricted areas of the epithelium, whereas Bmp3 and -7 were mostly seen in epithelium in embryogenesis. Bmp2 and Bmp4 have been suggested to play a role in papilla initiation in the tongue (Jung et al. 1999). Expression of Bmp3, a unique member in the Bmp family because of its negative role in osteogenesis by antagonizing osteogenic Bmps (Daluiski et al. 2001), implies that modulation of the Bmp signal through interaction of Bmp members might be an important mechanism in tongue epithelial development.
Some members of the Fgf and Bmp families have also been previously examined during tongue development (Jung et al. 1999; Huang et al. 2001). The results of this study show remarkable difference in the expression of some important genes between avian and mouse tongue. In avian developing tongue expression of Fgf8, Fgf10 and Bmp2 were not detected (Huang et al. 2001), whereas the latter two molecules were clearly detected in developing mouse tongue in this study. Bmp2 and Fgf8 expression was also reported in early mouse tongue in another study (Jung et al. 1999). This difference might be a species-specific feature in genetic events underlying tongue development. The mammalian tongue is far more advanced and complex than avian due to new evolutionary functions, such as tasting and aiding chewing. Therefore, there might be genetic differences governing their development. If so, the tongue is a useful model in unraveling genetic changes in evolution.
In conclusion, the results of this study show that Fgf/Fgfr and Bmp signal pathways play important roles in tongue development, involved in both the formation of muscle and papillae. Their expression patterns suggest that they co-operate at the early developing stage to maintain a rapid enlargement by promoting proliferation and inhibiting differentiation. They are also present in postnatal growth, possibly regulating the balance of myogenic proliferation and differentiation. This study also provides data for comparative and evolutionary studies. However, the molecular mechanism of the Fgf and Bmp signal pathways in tongue development, especially their regulatory roles on MRFs, is unknown. Further study is needed to elucidate their specific roles within the genetic cascades of tongue development.
References
Aberg T, Wozney J, Thesleff I (1997) Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn 210(4):383–396
Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen Y (2005) The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev Biol 277(1):102–113
Amthor H, Christ B, Patel K (1999) A molecular mechanism enabling continuous embryonic muscle growth—a balance between proliferation and differentiation. Development 126(5):1041–1053
Brunetti A, Goldfine ID (1990) Role of myogenin in myoblast differentiation and its regulation by fibroblast growth factor. J Biol Chem 265(11):5960–5963
Couly GF, Coltey PM, Le Douarin NM (1992) The developmental fate of the cephalic mesoderm in quail-chick chimeras. Development 114(1):1–15
Dalrymple KR, Prigozy TI, Mayo M, Kedes L, Shuler CF (1999) Murine tongue muscle displays a distinct developmental profile of MRF and contractile gene expression. Int J Dev Biol 43(1):27–37
Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM (2001) Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet 27(1):84–88
De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C (2000) An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127(3):483–492
Duprez DM, Coltey M, Amthor H, Brickell PM, Tickle C (1996) Bone morphogenetic protein-2 (BMP-2) inhibits muscle development and promotes cartilage formation in chick limb bud cultures. Dev Biol 174(2):448–452
Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P (2002) The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development 129(16):3783–3793
Flanagan-Steet H, Hannon K, McAvoy MJ, Hullinger R, Olwin BB (2000) Loss of FGF receptor 1 signaling reduces skeletal muscle mass and disrupts myofiber organization in the developing limb. Dev Biol 218(1):21–37
Gavrieli Y, Sherman Y, Ben-Sasson SA (1992) Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol 119(3):493–501
Hannon K, Kudla AJ, McAvoy MJ, Clase KL, Olwin BB (1996) Differentially expressed fibroblast growth factors regulate skeletal muscle development through autocrine and paracrine mechanisms. J Cell Biol 132(6):1151–1159
Huang R, Zhi Q, Izpisua-Belmonte JC, Christ B, Patel K (1999) Origin and development of the avian tongue muscles. Anat Embryol (Berl) 200(2):137–152
Huang R, Lang ER, Otto WR, Christ B, Patel K (2001) Molecular and cellular analysis of embryonic avian tongue development. Anat Embryol (Berl) 204(3):179–187
Jung HS, Oropeza V, Thesleff I (1999) Shh, Bmp-2, Bmp-4 and Fgf-8 are associated with initiation and patterning of mouse tongue papillae. Mech Dev 81(1–2):179–182
Kettunen P, Karavanova I, Thesleff I (1998) Responsiveness of developing dental tissues to fibroblast growth factors: expression of splicing alternatives of FGFR1, −2, −3, and of FGFR4; and stimulation of cell proliferation by FGF-2, −4, −8, and −9. Dev Genet 22(4):374–385
Kettunen P, Laurikkala J, Itaranta P, Vainio S, Itoh N, Thesleff I (2000) Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn 219(3):322–332
Mailleux AA, Spencer-Dene B, Dillon C, Ndiaye D, Savona-Baron C, Itoh N, Kato S, Dickson C, Thiery JP, Bellusci S (2002) Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development 129(1):53–60
Musgrave DS, Pruchnic R, Wright V, Bosch P, Ghivizzani SC, Robbins PD, Huard J (2001) The effect of bone morphogenetic protein-2 expression on the early fate of skeletal muscle-derived cells. Bone 28(5):499–506
Nagata J, Yamane A (2004) Progress of cell proliferation in striated muscle tissues during development of the mouse tongue. J Dent Res 83(12):926–929
Noden DM (1991) Cell movements and control of patterned tissue assembly during craniofacial development. J Craniofac Genet Dev Biol 11(4):192–213
Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N (2000) FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun 277(3):643–649
Ornitz DM, Itoh N (2001) Fibroblast growth factors. Genome Biol 2(3), REVIEWS3005
Patel SG, Funk PE, DiMario JX (1999) Regulation of avian fibroblast growth factor receptor 1 (FGFR-1) gene expression during skeletal muscle differentiation. Gene 237(1):265–276
Scata KA, Bernard DW, Fox J, Swain JL (1999) FGF receptor availability regulates skeletal myogenesis. Exp Cell Res 250(1):10–21
Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S (1999) Fgf10 is essential for limb and lung formation. Nat Genet 21(1):138–141
Shuler CF, Dalrymple KR (2001) Molecular regulation of tongue and craniofacial muscle differentiation. Crit Rev Oral Biol Med 12(1):3–17
Singh DJ, Bartlett SP (2005) Congenital mandibular hypoplasia: analysis and classification. J Craniofac Surg 16(2):291–300
Tzahor E, Kempf H, Mootoosamy RC, Poon AC, Abzhanov A, Tabin CJ, Dietrich S, Lassar AB (2003) Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev 17(24):3087–3099
Vainio S, Karavanova I, Jowett A, Thesleff I (1993) Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell 75(1):45–58
Vogel JE, Mulliken JB, Kaban LB (1986) Macroglossia: a review of the condition and a new classification. Plast Reconstr Surg 78(6):715–723
Wilkinson DG, Green J (1990) In situ hybridization and the three-dimensional reconstruction of serial sections. In: Copp AJ, Cockroft DL (eds) Postimplantation mammalian embryos: a practical approach. IRL Press, Oxford, pp 155–171
Yamane A, Mayo M, Shuler C, Crowe D, Ohnuki Y, Dalrymple K, Saeki Y (2000) Expression of myogenic regulatory factors during the development of mouse tongue striated muscle. Arch Oral Biol 45(1):71–78
Zhao P, Hoffman EP (2004) Embryonic myogenesis pathways in muscle regeneration. Dev Dyn 229(2):380–392
Acknowledgements
This work was carried out in the department of Biomedicine and supported by Faculty of Dentistry, University of Bergen. I would like to express my thanks to Kjellfrid Haukanes and Anne Nyhaug for their skilful technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nie, X. Apoptosis, proliferation and gene expression patterns in mouse developing tongue. Anat Embryol 210, 125–132 (2005). https://doi.org/10.1007/s00429-005-0009-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-005-0009-5