Abstract
Desmoplastic small round cell tumor (DSRCT) is a rare, biologically aggressive soft tissue neoplasm of uncertain differentiation, most often arising in the abdominal and pelvic cavities of adolescents and young adults with a striking male predominance. Histologically, it is characterized by islands of uniform small round cells in prominent desmoplastic stroma, and it has a polyimmunophenotypic profile, typically expressing WT1 and cytokeratin, desmin, and neural/neuroendocrine differentiation markers to varying degrees. Tumors at other sites and with variant morphology are more rarely described. DSRCT is associated with a recurrent t(11;22)(p13;q12) translocation, leading to the characteristic EWSR1-WT1 gene fusion. Fluorescence in situ hybridization (FISH), to detect EWSR1 rearrangement, and reverse transcription-polymerase chain reaction (RT-PCR) to assess for EWSR1-WT1 fusion transcripts are routine diagnostic ancillary tools. We present a large institutional comparative series of FISH and RT-PCR for DSRCT diagnosis. Twenty-six specimens (from 25 patients) histologically diagnosed as DSRCT were assessed for EWSR1 rearrangement and EWSR1-WT1 fusion transcripts. Of these 26 specimens, 24 yielded positive results with either FISH or RT-PCR or both. FISH was performed in 23 samples, with EWSR1 rearrangement seen in 21 (91.3%). RT-PCR was performed in 18 samples, of which 13 (72.2%) harbored EWSR1-WT1 fusion transcripts. The sensitivity of FISH in detecting DSRCT was 91.3%, and that of RT-PCR was 92.8% following omission of four technical failures. Therefore, both methods are comparable in terms of sensitivity. FISH is more sensitive if technical failures for RT-PCR are taken into account, and RT-PCR is more specific in confirming DSRCT. Both methods complement each other by confirming cases that the other method may not. In isolation, FISH is a relatively non-specific diagnostic adjunct due to the number of different neoplasms that can harbor EWSR1 rearrangement, such as Ewing sarcoma. However, in cases with appropriate morphology and a typical pattern of immunostaining, FISH is confirmatory of the diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Desmoplastic small round cell tumor (DSRCT) is a rare and highly aggressive neoplasm that was first described by Gerald and Rosai in 1989 [1] and subsequently characterized in 1991 in a series of 19 cases [2]. It is most common in adolescents and young adults with a strong male predominance [3, 4]. DSRCT typically arises in the abdominal and pelvic cavities, but has also been described in other organs including lung, pleura, ovary, kidney and parotid gland, as well as paratesticular and intracranial sites [5,6,7,8,9,10,11,12]. Complete excision is often impossible as these tumors tend to spread along mesothelial-lined surfaces. Consequently, clinical presentation is often at an advanced stage with a bulky primary mass, distant metastases, and extensive serosal seeding [13].
Morphologically, DSRCT is characterized by variably sized, well-defined islands of uniform cells separated by desmoplastic stroma [14]. The cells have scant cytoplasm and small to medium-sized, round to oval hyperchromatic nuclei with inconspicuous nucleoli. DSRCT typically shows a polyphenotypic immunohistochemical profile, with co-expression of epithelial (cytokeratins, epithelial membrane antigen), mesenchymal (desmin; typically with perinuclear dot-like positivity), and neural markers. Tumor cells also usually exhibit nuclear expression of the C-terminal part of WT1 [15,16,17,18,19,20,21], while expression of the N-terminal part is typically absent. Both morphologically and immunohistochemically, several variants of DSRCT have been documented, including tumors with tubules and glands [3, 14, 22], marked nuclear atypia [23], rhabdoid features [2], spindle cell or signet ring cell morphology [22], lack of significant stromal desmoplasia [22, 24, 25], and absence of cytokeratin or desmin immunoreactivity [14, 23]. Variation in morphology, immunophenotype, and site of origin can lead to considerable diagnostic difficulty [3], so molecular and molecular cytogenetic investigations are critical ancillary diagnostic tools.
The EWSR1-WT1 fusion is the result of a characteristic reciprocal translocation, t(11;22)(p13;q12), that fuses exon 7 of the Ewing sarcoma gene breakpoint region 1 (EWSR1) on chromosome 22q13 to exon 8 of the Wilms tumor suppressor gene (WT1) on 11p13 [26], with several alternative breakpoints for t(11;22)(p13;q12) described [27,28,29,30,31,32,33,34,35]. The EWSR1 gene encodes the multifunctional EWS protein associated with gene expression, RNA processing, and transport and cell signaling, while WT1 is a tumor suppressor gene encoding a zinc finger protein which regulates several growth factors [36]. The resulting chimeric transcript encodes a protein comprising the N-terminal (transactivation domain) of EWSR1 and the C-terminal (DNA binding domain) of WT1 [37] which acts as an oncogenic transcription factor that alters gene expression and permits tumor growth [26, 38,39,40]. Differentiation of DSRCT from other small round cell neoplasms is crucial to accurately guide the most appropriate management protocol and for prognostication; the outcome for DSRCT is almost universally fatal with an average survival of less than 2 years [2, 22, 41]. As there can be marked clinical, morphologic, and immunohistochemical overlap of DSRCT with other small round cell neoplasms (such as Ewing sarcoma and solid pattern alveolar rhabdomyosarcoma, both of which can occur intra-abdominally), cytogenetic and molecular analyses for EWSR1 rearrangement with fluorescence in situ hybridization (FISH), or EWSR1-WT1 fusion transcripts by reverse transcription-polymerase chain reaction (RT-PCR), are useful routine ancillary diagnostic modalities, which can assist in the diagnosis of DSRCT with appropriate clinical and pathologic correlation. In this study, we compared the utility of FISH for the detection of EWSR1 rearrangement and RT-PCR for EWSR1-WT1 fusion transcripts as ancillary tools in the diagnosis of DSRCT.
Methods
Tissue from all cases was formalin-fixed and paraffin-embedded (FFPE), and comprised consecutive specimens retrieved from the Histopathology Indexing System database which were coded as “desmoplastic small round cell tumor” over a 19-year period from 1996 to 2015. Cases comprised both core biopsy and excision specimens of material biopsied or resected at our center, and external cases, which had been sent for histologic review or second opinion. Only specimens that had FISH or RT-PCR performed for EWSR1 rearrangement and/or EWSR1-WT1 fusion transcripts were included. Clinical information was retrieved for each patient from the electronic patient record (EPR). All diagnoses had been previously made from morphology and immunohistochemistry by one or both of two specialist soft tissue pathologists (KT and CF). FISH and RT-PCR were performed according to standard or previously described methods [42, 46].
Results
The group consisted of 16 males and 9 females, with an age range of 12 to 49 years (median, 22 years) (Table 1). There were 26 specimens in total, from 25 patients (1 patient had a biopsy of an intra-abdominal mass sent as a second opinion case that was shown to have EWSR1 breakpoint on FISH; the EWSR1-WT1 fusion transcript was detected by RT-PCR in the subsequent excision specimen, which together with morphological and immunohistochemical findings confirmed the diagnosis of DSRCT).
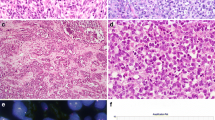
The commonest site was the abdomen (12 cases), followed by the peritoneum (4), omentum (3), pelvis (2), liver (2), retroperitoneum (1), thigh (1), and lymph node and chest wall (1 case; same specimen). Of the 26 specimens, 20 were core biopsies and 3 were excisions, and the nature of the material was not stated in 3 reports. Histologically, 25 tumors were composed of nests and islands of small round cells surrounded by fibrous stroma (Fig. 1a–c) with 1 of these tumors exhibiting focal rhabdoid cytomorphology. One showed predominantly solid morphology (Fig. 1d).
a–d Desmoplastic small round cell tumor (DSRCT). These tumors have characteristic morphology, of sharply demarcated nests of varying sizes, comprising uniform small cells with oval or round nuclei with hyperchromatic or vesicular chromatin, inconspicuous nucleoli, and scant cytoplasm (a–c), dispersed in prominent fibroblastic stroma (a–b). Sometimes, DSRCT can display more uniformly solid features without marked associated stroma, closely resembling other tumors with small round cell morphology such as Ewing sarcoma (d)
Information regarding the immunohistochemical pattern of staining was available for 23 specimens. Of the three specimens with no immunohistochemistry, one was a debulking case where no immunohistochemistry was requested at our center and the EWSR1-WT1 fusion transcript was detected by RT-PCR at another institution. The second specimen was a post-chemotherapy debulking case which was diagnosed at another center, with pathologic features in the debulking specimen consistent with DSRCT. Although RT-PCR failed, FISH was equivocal (described below). The third case was a core biopsy reviewed as a second opinion case; the letter to the referring pathologist stated that immunohistochemistry was “appropriate” for DSRCT although the immunoprofile was not provided. Immunohistochemically, desmin was at least focally positive in 23/23 cases. At least focal cytokeratin expression was present in 16 cases; 6 were negative, and it was not requested in 1 case. WT1 was requested in 17 cases, and at least focal nuclear expression was reported in 10; of these, only 4 reports specified that immunoreactivity was for the C-terminus of WT1. Neuron-specific enolase (NSE) was performed in nine cases, and eight of the nine showed at least focal expression.
RT-PCR was performed in 18 cases, and EWSR1-WT1 fusion transcripts were detected in 13 (72.2%) (Figs. 2 and 3) with one negative result and four technical failures. Of the 13 cases harboring EWSR1-WT1 fusion transcripts, 10 had FISH performed, all of which showed EWSR1 rearrangement. The case with the negative RT-PCR result did not show EWSR1 rearrangement. FISH was performed in all four cases that were technical failures, with three positive results (75%) and one equivocal result (see below). There was insufficient material in the eight cases that did not undergo RT-PCR analysis, all of which were shown to have EWSR1 rearrangement by FISH.
Real-time quantitative polymerase chain reaction (RT-PCR) amplification plots (left) and melt curve profile (right), showing positive EWSR1-WT1 fusion transcripts of DSRCT. Example of an amplification plot of the SYBR green real-time quantitative polymerase chain reaction assay for EWSR1-WT1 (left). Fluorescence emission delta Rn (DRn) per cycle is shown for the positive control (yellow), a patient sample positive for the EWSR1-WT1 fusion (green), and a cell line known not to harbor the translocation (red). Corresponding melt curve profile (right)
FISH was performed in 23 cases, and EWSR1 rearrangement was seen in 21 (91.3%) with 1 negative and 1 equivocal result. Of the 21 cases that showed EWSR1 rearrangement, 13 had RT-PCR performed with 10 positive results (77%) and 3 cases of technical failure. The case with the negative FISH result was also found by RT-PCR not to harbor EWSR1-WT1 fusion transcripts. The case with the equivocal FISH result included numerous cells with three to five paired signals consistent with derivation from an abnormal clone, but only five nuclei with an identifiable split signal, which was deemed too low to conclude that a translocation involving EWSR1 gene was a primary genetic abnormality in this neoplasm. This sample, which was an excision specimen, was unsuitable for RT-PCR as there was no amplification of the control gene (beta-2 microglobulin (B2M)), indicating poor quality RNA.
FISH was not performed in three cases; RT-PCR was performed first in two of these cases (one internal and one external), both of which yielded positive results, and therefore, subsequent FISH analysis was not required. In the third case, FISH had already been performed on a different sample from the same patient. In all three of these samples, RT-PCR showed the presence of EWSR1-WT1 fusion transcripts.
Of the 26 specimens that had FISH and/or RT-PCR performed, 24 had detectable EWSR1 rearrangement and/or EWSR1-WT1 fusion transcripts. The sensitivity of FISH was 91.3%, and that of RT-PCR was 92.8% following omission of the four technical failures. Of the 2/26 cases that were not confirmed by FISH or RT-PCR, 1 had morphologic appearances and an immunoprofile typical of DSRCT including AE1/AE3 positivity, focal nuclear positivity for WT1 (C-terminus), and focal dot-like positivity for desmin. There was focal immunoreactivity to NB84, which was thought to be of uncertain significance. CK20, TLE1, TTF1, and CD99 were negative. EWSR1-ERG and EWSR1-FLI1 fusion transcripts were not detectable by RT-PCR. Overall, this case was considered to be a rare genetic variant of DSRCT not detectable by RT-PCR primers or FISH probes. The other sample was a post-chemotherapy case of DSRCT. It was not possible to calculate specificity and positive and negative predictive values for either technique, as all the samples were cases of DSRCT initially diagnosed on morphology and immunophenotype by specialist soft tissue pathologists, with no negative controls. In some cases, other FISH probes and RT-PCR primers were used to exclude gene rearrangement and fusion transcripts. FOXO1 rearrangement was undetectable in eight cases, while the SS18 breakpoint was not seen in one case. ASPSCR1-TFE3, SS18-SSX1/2, PAX3/7-FOXO1, EWSR1-ERG, and EWSR1-FLI1 fusion transcripts were undetectable in one, one, three, six, and six cases, respectively.
Discussion
This study represents a large comparative series of FISH and RT-PCR in the diagnosis of DSRCT and shows that EWSR1 rearrangement and/or EWSR1-WT1 fusion transcripts were detectable in 24 of 26 specimens (92.3%). In our hands, RT-PCR is of comparable sensitivity to FISH (92.8% compared with 91.3%). RT-PCR was performed in 18 samples, of which 13 (72.2%) were shown to harbor EWSR1-WT1 fusion transcripts, with the sensitivity of RT-PCR rising to 92.8% when the four cases of technical failure were omitted. This is in keeping with RT-PCR detection rates documented in previous studies. Lae et al. [14] demonstrated that 28/30 cases (93%) showed EWSR1-WT1 fusion by RT-PCR with primers for EWSR1 exon 7 and WT1 exon 10, while de Alava et al. [27] detected the chimeric transcript in 11/12 cases (91.6%) using the same primers. In both studies, FISH was not used in the evaluation of these neoplasms.
Of the two cases that lacked EWSR1 rearrangement or EWSR1 fusion transcripts, one had morphological appearances and an immunophenotype typical of DSRCT and was considered to be a genetic variant not detected by commercial RT-PCR primers or FISH probes. The other case was post-chemotherapy, which may have impacted molecular testing. The high percentage of FISH and/or RT-PCR confirmation at our institution likely reflects a degree of tertiary center bias; it is possible that histopathologic diagnosis is more rigorous than those at other centers such that the possibility of DSRCT is not entertained unless the morphologic appearances, immunoprofile, and clinical context truly fit the diagnosis. Therefore, the lack of specificity of FISH may be less of an issue at our center, but may be more so at other institutions, in which case RT-PCR is even more critical for confirming the diagnosis.
There was no correlation between a negative or equivocal FISH/RT-PCR result and the amount of tumor tissue available (i.e., whether the sample was a core biopsy or excision specimen). One potential drawback of RT-PCR is that it may fail to detect the EWSR1-WT1 fusion transcript if variant breakpoints for the t(11;22)(p13;q12) are present, which are not detectable with commercially available primers. Some authors have used specific primer sets in order to detect these variant breakpoints [33, 43,44,45]. Ideally, this would be helpful in confirming RT-PCR negative cases. However, specific RT-PCR primer sets may not be available in standard molecular diagnostic laboratories. Similarly, karyotyping could potentially detect cases missed by RT-PCR. However, on receipt of the samples at the time, we did not have fresh tissue to karyotype, with numerous cases sent to us as referral or second opinion cases from other centers. If variant breakpoints are present, FISH may detect EWSR1 rearrangement in such cases.
Furthermore, FISH has been associated with a higher sensitivity for detecting gene rearrangement and a better success rate than RT-PCR [46], which was less robust a few years ago, due to suboptimal tissue fixation and processing techniques. However, in recent years, there has been a steady improvement in the technical success rates of cases undergoing RT-PCR. This is reflected in our series where two of the four technical fails occurred in 2008 (one second opinion case and one internal case), one in 2010 (second opinion case), and one in 2014 (internal case). No technical fails occurred in 2015. This trend can perhaps be attributed to the increasing experience of laboratory staff at our center and at other institutions, which are now accustomed to optimal tissue handling and processing procedures for FFPE material undergoing molecular analysis. All histopathology departments now recognize the crucial role that molecular pathology plays in assessing genetic aberrations in solid tumors with potential targeted therapies, and consequently, tissue handling and processing techniques have improved accordingly [47].
Of the four technical fails identified, one case was an excision specimen (from 2008) and three were core biopsies. The low cellularity present within the cores presumably did not yield sufficient amounts of extracted RNA [48]. It has been suggested that tumors with prominent fibrous stroma, such as DSRCT, may be more susceptible to technical fails by RT-PCR as the stromal component reduces the overall total density of cells and thus the total amount of RNA to be extracted. Therefore, additional sections are recommended in such cases to ensure sufficient RNA extraction for RT-PCR analysis [48].
The sensitivity of RT-PCR is high when technical fails are omitted from the analysis. While the failure rate is relatively higher in older cases in our series, its success rate now mirrors that of FISH as fixing and processing techniques have improved. Although numerous neoplasms are associated with EWSR1 gene rearrangement [49,50,51], FISH still plays an important role in confirming the diagnosis. DSRCT is one of the rare sarcomas that can be confidently diagnosed on phenotype in the appropriate clinical and histopathological setting. Desmin, cytokeratin, and WT1 (C-terminus) positivity and the absence of expression of the N-terminal part of WT1 are almost restricted to this entity. Therefore, detection of EWSR1 gene rearrangement by FISH would be confirmatory in this specific setting. Indeed, none of the DSRCT mimics with EWSR1 gene rearrangement, Ewing sarcoma in particular, have this immunophenotype, and none of those entities with similar morphology and an overlapping immunoprofile, such as solid pattern alveolar rhabdomyosarcoma, have EWSR1 gene rearrangement. A probe for WT1, which is now commercially available, could potentially increase the specificity of FISH to that of RT-PCR. However, this probe is expensive with a limited shelf life (up to 2 years) and would need to be purchased in batches. Given the ready availability of RT-PCR, with improved rates of technical success in recent years, the need for a WT1 probe is low, and therefore, the majority of laboratories do not stock it. Hence, RT-PCR has an important role to play in confirming the diagnosis of DSRCT, particularly in cases with limited material, those with atypical morphological features or immunophenotypic findings (e.g., desmin or cytokeratin negative), or those that are clinically unusual.
It should be noted that EWSR1-WT1 is characteristic but no longer specific for DSRCT, as it has been recently described in a behaviorally indolent low-grade small round cell tumor of the cauda equina [52] which comprises nests and cords of small round cells with some rosette-like structures, rare mitotic figures, and a low Ki-67 proliferation index with immunohistochemical features of smooth muscle differentiation, as well as focal CD99 and Neu-N expression. This finding further underlines the importance of clinicopathological correlation when EWSR1-WT1 fusion transcripts are detected by RT-PCR in order to arrive at the correct diagnosis.
Conclusions
The diagnosis of DSRCT can be difficult due to variation in tumor site, morphology, and immunophenotypic spectrum, leading to overlap with other round cell neoplasms, particularly those also harboring EWSR1 rearrangement such as Ewing sarcoma. Hence, ancillary molecular confirmation is a crucial adjunct to histopathology. This study shows that, in comparative analysis, both FISH and RT-PCR are useful in the diagnosis of DSRCT, although technically, RT-PCR was less reliable. If the morphological appearances fit and all immunophenotypic criteria are met, then FISH is confirmatory. RT-PCR, which has a comparable detection rate to FISH for DSRCT (when cases with technical failure are omitted), is able to confirm the diagnosis in the appropriate histologic context if the immunoprofile is incomplete. Furthermore, FISH may detect EWSR1 rearrangement in cases where variant breakpoints for the t(11;22)(p13;q12) are present. Therefore, RT-PCR and FISH complement each other by confirming cases that the other method may miss. In the appropriate morphological setting and if all immunophenotypic criteria are satisfied, it may be contributory to perform FISH, in the first instance with RT-PCR analysis reserved for cases with an atypical immunoprofile that otherwise fit the criteria for DSRCT. A small percentage of cases will not show either EWSR1 rearrangement or EWSR1-WT1 fusion transcripts, and this should not deter a diagnosis of DSRCT if these cases fulfill the morphologic, immunophenotypic, and clinical criteria.
References
Gerald WL, Rosai J (1989) Desmoplastic small cell tumor with divergent differentiation. Pediatr Pathol 9:177–183
Gerald WL, Miller HK, Battifora H, Miettinen M, Silva EG, Rosai J (1991) Intra-abdominal desmoplastic small round-cell tumor. Report of 19 cases of a distinctive type of high-grade polyphenotypic malignancy affecting young individuals. Am J Surg Pathol 15:499–513
Gerald WL, Ladanyi M, de Alava E, Cuatrecasas M, Kushner BH, LaQuaglia MP, Rosai J (1998) Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): desmoplastic small round-cell tumor and its variants. J Clin Oncol 16:3028–3036
Gerald WL, Rosai J (1993) Desmoplastic small cell tumor with multi-phenotypic differentiation. Zentralbl Pathol 139:141–151
Young RH, Eichhorn JH, Dickersin GR, Scully RE (1992) Ovarian involvement by the intra-abdominal desmoplastic small round cell tumor with divergent differentiation: a report of three cases. Hum Pathol 23:454–464
Bian Y, Jordan A, Rupp M, Cohn H, McLaughlin CJ, Miettinen M (1993) Effusion cytology of desmoplastic small round cell tumor of the pleura. A case report. Acta Cytol 37:77–82
Parkash V, Gerald WL, Parma A, Miettinen M, Rosai J (1995) Desmoplastic small round cell tumor of the pleura. Am J Surg Pathol 19:659–665
Zaloudek C, Miller TR, Stern JL (1995) Desmoplastic small cell tumor of the ovary: a unique polyphenotypic tumor with an unfavorable prognosis. Int J Gynecol Pathol 14:260–265
Tison V, Cerasoli S, Morigi F, Ladanyi M, Gerald WL, Rosai J (1996) Intracranial desmoplastic small-cell tumor. Report of a case. Am J Surg Pathol 20:112–117
Cummings OW, Ulbright TM, Young RH, Dei Tos AP, Fletcher CD, Hull MT (1997) Desmoplastic small round cell tumors of the paratesticular region. A report of six cases. Am J Surg Pathol 21:219–225
Wolf AN, Ladanyi M, Paull G, Blaugrund JE, Westra WH (1999) The expanding clinical spectrum of desmoplastic small round-cell tumor: a report of two cases with molecular confirmation. Hum Pathol 30:430–435
Cho KJ, Roy J, Choi J, Choi SH, Nam SY, Kim SY (2008) Mesenchymal neoplasms of the major salivary glands: clinicopathological features of 18 cases. Eur Arch Otorhinolaryngol 265(Suppl 1):S47–S56
Hassan I, Shyyan R, Donohue JH, Edmonson JH, Gunderson LL, Moir CR, Arndt CA, Nascimento AG, Que FG (2005) Intraabdominal desmoplastic small round cell tumors. A diagnostic and therapeutic challenge. Cancer 104:1264–1270
Lae ME, Roche PC, Jin L, Lloyd RV, Nascimento AG (2002) Desmoplastic small round cell tumor: a clinicopathologic, immunohistochemical, and molecular study of 32 tumors. Am J Surg Pathol 26:823–835
Ordonez NG (1998) Desmoplastic small round cell tumor: II: an ultrastructural and immunohistochemical study with emphasis on new immunohistochemical markers. Am J Surg Pathol 22:1314–1327
Zhang PJ, Goldblum JR, Pawel BR, Fisher C, Pasha TL, Barr FG (2003) Immunophenotype of desmoplastic small round cell tumors as detected in cases with EWS-WT1 gene fusion product. Mod Pathol 16:229–235
Hill DA, Pfeifer J, Marley EF, Dehner LP, Humphrey PA, Zhu X, Swanson PE (2000) WT1 staining reliably differentiates desmoplastic small round cell tumor from Ewing sarcoma/primitive neuroectodermal tumor. An immunohistochemical and molecular diagnostic study. Am J Clin Pathol 114:345–353
Arnold MA, Schoenfield L, Limketkai BN, Arnold CA (2014) Diagnostic pitfalls of differentiating desmoplastic small round cell tumor (DSRCT) from Wilms tumor (WT): overlapping morphologic and immunohistochemical features. Am J Surg Pathol 38:1220–1226
Charles AK, Moore IE, Berry PJ (1997) Immunohistochemical detection of the Wilms’ tumour gene WT1 in desmoplastic small round cell tumour. Histopathology 30:312–314
Antonescu CR, Ladanyi M (2013) Desmoplastic small round cell tumour. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F (eds) WHO Classification of Tumours of Soft Tissue and Bone, 4th edn. Lyon, IARC, pp 225–227
Barnoud R, Sabourin JC, Pasquier D, Ranchere D, Bailly C, Terrier-Lacombe MJ, Pasquier B (2000) Immunohistochemical expression of WT1 by desmoplastic small round cell tumor: a comparative study with other small round cell tumors. Am J Surg Pathol 24:830–836
Ordonez NG (1998) Desmoplastic small round cell tumor: I: a histopathologic study of 39 cases with emphasis on unusual histological patterns. Am J Surg Pathol 22:1303–1313
Murphy AJ, Bishop K, Pereira C, Chilton-MacNeill S, Ho M, Zielenska M, Thorner PS (2008) A new molecular variant of desmoplastic small round cell tumor: significance of WT1 immunostaining in this entity. Hum Pathol 39:1763–1770
Dorsey BV, Benjamin LE, Rauscher F 3rd, Klencke B, Venook AP, Warren RS, Weidner N (1996) Intra-abdominal desmoplastic small round-cell tumor: expansion of the pathologic profile. Mod Pathol 9:703–709
Argatoff LH, O’Connell JX, Mathers JA, Gilks CB, Sorensen PH (1996) Detection of the EWS/WT1 gene fusion by reverse transcriptase-polymerase chain reaction in the diagnosis of intra-abdominal desmoplastic small round cell tumor. Am J Surg Pathol 20:406–412
Ladanyi M, Gerald W (1994) Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res 54:2837–2840
de Alava E, Ladanyi M, Rosai J, Gerald WL (1995) Detection of chimeric transcripts in desmoplastic small round cell tumor and related developmental tumors by reverse transcriptase polymerase chain reaction. A specific diagnostic assay. Am J Pathol 147:1584–1591
Liu J, Nau MM, Yeh JC, Allegra CJ, Chu E, Wright JJ (2000) Molecular heterogeneity and function of EWS-WT1 fusion transcripts in desmoplastic small round cell tumors. Clin Cancer Res 6:3522–3529
Antonescu CR, Gerald WL, Magid MS, Ladanyi M (1998) Molecular variants of the EWS-WT1 gene fusion in desmoplastic small round cell tumor. Diagn Mol Pathol 7:24–28
Shimizu Y, Mitsui T, Kawakami T, Ikegami T, Kanazawa C, Katsuura M, Obata K, Yamagiwa I, Hayasaka K (1998) Novel breakpoints of the EWS gene and the WT1 gene in a desmoplastic small round cell tumor. Cancer Genet Cytogenet 106:156–158
Werner H, Idelman G, Rubinstein M, Pattee P, Nagalla SR, Roberts CT Jr (2007) A novel EWS-WT1 gene fusion product in desmoplastic small round cell tumor is a potent transactivator of the insulin-like growth factor-I receptor (IGF-IR) gene. Cancer Lett 247:84–90
Nakanishi Y, Oinuma T, Sano M, Fuchinoue F, Komatsu K, Seki T, Obana Y, Tabata M, Kikuchi K, Shimamura M, Ohmori K, Nemoto N (2006) Coexpression of an unusual form of the EWS-WT1 fusion transcript and interleukin 2/15 receptor betamRNA in a desmoplastic small round cell tumour. J Clin Pathol 59:1108–1110
Adsay V, Cheng J, Athanasian E, Gerald W, Rosai J (1999) Primary desmoplastic small cell tumor of soft tissues and bone of the hand. Am J Surg Pathol 23:1408–1413
Su MC, Jeng YM, Chu YC (2004) Desmoplastic small round cell tumor of the kidney. Am J Surg Pathol 28:1379–1383
Hamazaki M, Okita H, Hata J, Shimizu S, Kobayashi H, Aoki K, Nara T (2006) Desmoplastic small cell tumor of soft tissue: molecular variant of EWS-WT1 chimeric fusion. Pathol Int 56:543–548
Lee SB, Kolquist KA, Nichols K, Englert C, Maheswaran S, Ladanyi M, Gerald WL, Haber DA (1997) The EWS-WT1 translocation product induces PDGFA in desmoplastic small round-cell tumour. Nat Genet 17:309–313
Brodie SG, Stocker SJ, Wardlaw JC, Duncan MH, McConnell TS, Feddersen RM, Williams TM (1995) EWS and WT-1 gene fusion in desmoplastic small round cell tumor of the abdomen. Hum Pathol 26:1370–1374
Chan AS, MacNeill S, Thorner P, Squire J, Zielenska M (1999) Variant EWS-WT1 chimeric product in the desmoplastic small round cell tumor. Pediatr Dev Pathol 2:188–192
Rodriguez E, Sreekantaiah C, Gerald W, Reuter VE, Motzer RJ, Chaganti RS (1993) A recurring translocation, t(11;22)(p13;q11.2), characterizes intra-abdominal desmoplastic small round-cell tumors. Cancer Genet Cytogenet 69:17–21
Sawyer JR, Tryka AF, Lewis JM (1992) A novel reciprocal chromosome translocation t(11;22)(p13;q12) in an intraabdominal desmoplastic small round-cell tumor. Am J Surg Pathol 16:411–416
Kempson RL, Fletcher CDM, Evans HL, Henrickson MR, Sibley RS (2001) Desmoplastic small round cell tumor. In: Tumors of the Soft Tissue. Atlas of Tumor Pathology, Fascicle 30, 3rd series. Armed Forces Institute of Pathology, Washington, DC, pp 452–458
Thway K, Gonzalez D, Wren D, Dainton M, Swansbury J, Fisher C (2015) Angiomatoid fibrous histiocytoma: comparison of fluorescence in situ hybridization and reverse transcription polymerase chain reaction as adjunct diagnostic modalities. Ann Diagn Pathol 19:137–142
Cliteur VPM, Szuhai K, Baelde HJ, van Dam J, Gelderblom H, Hogendoorn PCW (2012) Paratesticular desmoplastic small round cell tumour: an unusual tumour with an unusual fusion; cytogenetic and molecular genetic analysis combining RT-PCR and COBRA-FISH. Clin Sarcoma Res 2:3
Yamaguchi U, Hasegawa T, Morimoto Y, Tateishi U, Endo M, Nakatani F, Kawai A, Chuman H, Beppu Y, Endo M, Kurotaki H, Furuta K (2005) A practical approach to the clinical diagnosis of Ewing’s sarcoma/primitive neuroectodermal tumour and other small round cell tumours sharing EWS rearrangement using new fluorescence in situ hybridisation probes for EWSR1 on formalin fixed, paraffin wax embedded tissue. J Clin Pathol 58:1051–1056
Wang LL, Perlman EJ, Vujanic GM, Zuppan C, Brundler MA, Cheung CRLH, Calicchio ML, Dubois S, Cendron M, Murata-Collins JL, Wenger GD, Strzelecki D, Barr FG, Collins T, Perez-Atayde AR, Kozakewich H (2007) Desmoplastic small round cell tumor of the kidney in childhood. Am J Surg Pathol 31:576–584
Thway K, Rockcliffe S, Gonzalez D, Swansbury J, Min T, Thompson L, Fisher C (2010) Utility of sarcoma-specific fusion gene analysis in paraffin-embedded material for routine diagnosis at a specialist centre. J Clin Pathol 63:508–512
Vroobel K, Gonzalez D, Wren D, Thompson L, Swansbury J, Fisher C, Thway K (2016) Ancillary molecular analysis in the diagnosis of soft tissue tumours: reassessment of its utility at a specialist centre. J Clin Pathol 69:505–510
Thway K, Wren D, Lee J, Thompson L, Fisher C, Gonzalez D (2017) Evaluation of the optimal provision of formalin-fixed, paraffin-embedded material for reverse transcription-PCR in soft-tissue tumour diagnosis. J Clin Pathol 70:20–24
Ordonez JL, Osuna D, Herrero D, de Alava E, Madoz-Gurpide J (2009) Advances in Ewing’s sarcoma research: where are we now and what lies ahead? Cancer Res 69:7140–7150
Fisher C (2014) The diversity of soft tissue tumours with EWSR1 gene rearrangements: a review. Histopathology 64:134–150
Thway K, Fisher C (2012) Tumors with EWSR1-CREB1 and EWSR1-ATF1 fusions: the current status. Am J Surg Pathol 36:e1–e11
Ud Din N, Pekmezci M, Javed G, Horvai AE, Ahmad Z, Faheem M, Navarro AL, López-Terrada D, Perry A (2015) Low-grade small round cell tumor of the cauda equina with EWSR1-WT1 fusion and indolent clinical course. Hum Pathol 46:153–158
Acknowledgments
We acknowledge support from the NIHR Royal Marsden/ICR Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Research involving human participants and/or animals
No research involving human participants or animals. All materials were formalin-fixed, paraffin-embedded archival surplus biopsy or excision material from the pathology archive.
Informed consent
All specimens were formalin-fixed, paraffin-embedded archival surplus biopsy or excision material from the pathology archive. No patient informed consent was required for this study, and there is no identifiable patient information.
Funding
No funding to disclose.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Mohamed, M., Gonzalez, D., Fritchie, K.J. et al. Desmoplastic small round cell tumor: evaluation of reverse transcription-polymerase chain reaction and fluorescence in situ hybridization as ancillary molecular diagnostic techniques. Virchows Arch 471, 631–640 (2017). https://doi.org/10.1007/s00428-017-2207-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-017-2207-y