Abstract
Loss of E-cadherin expression in melanoma correlates with increased tumor thickness and reduced disease-free survival. The molecular mechanisms underpinning its differential expression in melanoma tissue remain elusive. MicroRNAs (miRNAs) have been implicated in tumor progression and regulation of E-cadherin expression. Here, we demonstrate a significant correlation between tumor thickness and loss of expression of miR-200a, miR-200c, and miR-203 in a series of 23 frozen primary melanomas, where it was confirmed in two subsequent validation series (series 1: six nevi, 15 primary melanomas, and 16 metastases; series 2: 11 matched pairs of primary melanomas and metastases). Decreased levels of miR-200a, miR-200c, and miR-203 correlated with increasing thickness in the combined validation series (P = 0.024, 0.033, and 0.031, respectively). In addition, progressive loss of miR-200a expression with disease progression was observed in series 1 (P < 0.001) and in series 2 (P = 0.029). MiR-200 in situ hybridization and E-cadherin immunohistochemistry demonstrated reduced expression of both at the deep invasive margin of the tumor. Furthermore, a functional validation study using an anti-miR200 strategy demonstrated that loss of miR-200 expression in melanoma cell lines reduced E-cadherin expression. Collectively, our data point towards an important role for miR-200 and miR203 expression in regulating E-cadherin during melanoma progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epithelial–mesenchymal transition (EMT) is associated with increased malignancy of carcinomas [1]. EMT increases migratory and invasive capacities of tumor cells and can prevent apoptosis and senescence. Dissolution of E-cadherin-mediated adhesion is a key preliminary step in EMT. Throughout tumor progression, melanoma cells accumulate mesenchymal characteristics [2–4]. In culture, loss of E-cadherin correlates with the metastatic potential of melanoma cells and with the ability of keratinocytes to modulate expression of melanoma progression markers like MelCAM [5]. Accumulation of mesenchymal traits by melanoma cells in tissue is not as evident as in cell lines. This is not only because of the enormous cytological variation between primary cutaneous melanomas, but also because phenotypic differences within a given melanoma in which mesenchymal-like spindle cell areas may alternate with areas in which the melanoma cells assume a more epithelial-like aspect. Furthermore, invasive melanoma cells in tissue commonly lack spindle morphology [6]. Loss of E-cadherin expression has been reported to correlate with increased thickness [7] and reduced survival [8]. However, E-cadherin expression in melanoma may be quite heterogeneous [9].
Canonical and noncanonical Wnt signaling modulate E-cadherin expression [10]. Fibroblast growth factor, Hedgehog, transforming growth factor β (TGFβ), and noncanonical Wnt signals synergistically induce EMT regulators, such as, Snail, Slug, TWIST, and the E-cadherin repressors ZEB1 and 2. High Snail protein expression is observed in melanoma [11] and can induce immune suppression via induction of regulatory T cells and impairment of dendritic cell function [12]. Our previously published gene expression study in primary melanoma demonstrated a correlation between overexpression of Snail and poor 4-year distant metastasis-free survival (DMFS) [13] and supported the hypothesis that mesenchymal traits of melanoma cells is associated with poor prognosis.
In addition to Wnt signaling, E-cadherin expression can be modulated by differential expression of microRNAs (miRNAs). MiR-9 directly targets the E-cadherin encoding messenger, CDH1 [14]. Conversely, miR-200 family members (miR-141; miR-200a, b and c; and miR-429) target transcriptional repressors of E-cadherin, i.e., ZEB1 and ZEB2, in breast, ovarian, and pancreatic cancers [15–17] and various tumor cell lines [18]. Although target prediction algorithms predicted little overlap in the targets of miR-200 family members, experimental approaches suggested that their sets of targets are highly overlapping [18].
Using a large set of fresh frozen nevi, primary melanomas, and melanoma metastases, we demonstrate that expression of miR-200a and miR-200c correlate with melanoma thickness and metastasis. These findings were validated by quantitative PCR, miRNA in situ hybridization, and functional validation studies. Collectively, our data support the hypothesis that loss of miR-200 and miR-203 expression in melanoma increases mesenchymal characteristics and that this can contribute to progression.
Materials and methods
Tissues
All tissue samples were retrieved from the pathology archives at the University Hospital Leuven, Leuven, Belgium. The pertinent histological and clinical features are listed in Supplemental Tables S1–S3. Representative tissue parts were snap frozen in liquid nitrogen-cooled isopentane and stored at −80 °C until used. Sampling, storage, and use of these samples were performed according to requirements of the Local Ethical Committee. For miRNA in situ hybridization, formalin-fixed and paraffin-embedded tissue blocks were retrieved from pathology archives of the Radboud University Nijmegen Medical Centre. Tissues were obtained according to local ethical guidelines and approved by the local regulatory committee.
Preparation of total RNA and cDNA for miRNA profiling
Depending on the size of the sample, 20 to 40 sections (10 μm) were cut from frozen tissue blocks containing more than 75 % melanoma or nevus cells as evaluated from hematoxylin and eosin-stained sections. Sections were lysed immediately in 1 ml TRIzol (Invitrogen) and RNA was extracted subsequently according to the manufacturer’s protocol. Absence of DNA contamination and RNA integrity were analyzed on a Bioanalyzer (Agilent Technologies). RNA integrity number (RIN) higher than 7.5 and, in most cases, higher than 9 were obtained.
Total RNA (8 ng) was reverse transcribed using 50 nM stem–loop RT primer and MultiScribe Reverse Transcriptase (Life Technologies) according to the manufacturer’s protocol. The expression of total 158 mature human miRNAs was analyzed using specific miRNA primers available from Life Technologies (Supplemental Table S4).
Real-time quantitative PCR
Real-time PCR was performed using the standard TaqMan protocol using Applied Biosystems reagents and an Applied Biosystems 7900 High Throughput Sequence Detection System. All reactions were run in duplicate. Mean Treshold Cycle (Ct) value was determined as median of two reads. In order to provide robust data, only values below 32 Ct were considered as specific and values above 32 CT were considered as missing data. This resulted in the exclusion of 29 miRNAs for further analyses, leaving a total of 129 miRNAs in the discovery phase of the study (Supplemental Table S4). Change in expression was determined by calculating the 2−ddCt value [19]. MiRNA expression was normalized to let-7a expression. Normalization to any calibrator sample had very low influence on the overall data (data not shown), demonstrating high quality total RNA purification.
Statistical analysis
The Pearson correlation coefficient between the normalized Ct value of a miRNA and primary tumor thickness was determined. A stratified correlation coefficient was calculated for the analysis of association between expression and tumor thickness after combining all data from the two validation series, thereby controlling for possible batch effects [20]. DMFS was defined as the time interval between the diagnosis of the primary cutaneous melanoma and a distant metastasis and statistical differences assessed by a logrank test. Patients alive without distant metastasis or who died of causes not related to melanoma were censored at the date of last follow-up or at the date of death, respectively. An analysis of variance was applied to test for the difference in expression between nevi, tumors, and metastases. The paired t test was used for paired analysis of miRNA expression in the matched series of primary melanomas and metastases. Because of multiple testing, p values were adjusted for the False Discovery Rate (FDR) criterion [21]. P values from the stratified correlation analyses are based on an empirical permutation distribution and were not further adjusted. The t test for equality of means for independent samples (equal variance not assumed, two-tailed) was used to determine the difference in CDH1 expression in cell lines after miR-200c silencing.
In situ hybridization for miR-200c and E-cadherin immunohistochemistry
Consecutive tissue sections of formalin-fixed and paraffin-embedded primary melanomas (n = 10) were prepared. MiRNA in situ hybridization was performed on 6-μm sections according to our previously described protocol [22] with minor modifications. A 5′- and 3′-end digoxigenin-labeled LNA/DNA oligonucleotide designed to hybridize to miR-200c ([18]; 5′-CCATCATTACCCGGCTGTATTA, underlined nucleotides indicate LNA bases, Exiqon, Vedbaek, Denmark) was hybridized to the tissue section overnight at 55 °C. MiR-200c was visualized using alkaline phosphatase-conjugated anti-digoxigenin Fab fragments (Roche), nitro blue tetrazolium, and 5-bromo-4-chloro-3′-indolyphosphate. The consecutive 4-μm sections were stained for E-cadherin as described previously [23].
MiR-200c silencing
E-cadherin-expressing human melanoma cell lines 888Mel, 1 F6, and SK-Mel28 were cultured as described previously [23, 24]. Exponentially growing cells were transfected with 20 nM thioacetylated LNA–DNA oligo towards miR-200c (5′-CCATCATTACCCGGCTGT ATTA) or a thioacetylated LNA–DNA scrambled oligo (Exiqon) using X-tremeGENE siRNA transfection reagent (Roche) according to the supplier’s protocol. Two days after transfection, cells were harvested and plated in a 6-well plate at 60 % confluence and the transfection protocol repeated. The remaining cells were used for total RNA extraction (NucleoSpin RNA II, Macherey-Nagel, Düren, Germany). A total of three transfections were performed. cDNA was prepared using 0.5 μg total RNA, oligo-dT primers, and Moloney Murine Leukemia Virus Reverse Transcriptase (Promega) according to the supplier’s protocol. cDNA was diluted five times and 5 μl was used for qPCR using Power SYBR Green Universal Master Mix (Applied Biosystems), and a CDH1-specific primer pair (NM_004360.3, 5′-ATGAGTGTCCCC CGGTATCTT and 5′-CAGCCGCTTTCAGATTTTCATC) in 25 μl. All samples were normalized for GAPDH expression (NM_002046.3, 5′-GAACCTGACCTGCCGTCTAG, 5′-CCTGCTTCACCACCTTCTTGA). A preliminary primer validation study revealed that the slope of the standard curves for both CDH1 and GAPDH was between 2.9 and 3.2. Normalized Ct values for CDH1 expression for scrambled and anti-miR-200 oligo-transfected cells at t = 0 was set at 100 % and used to calculate change in CDH1 expression for all other time points. The experiment was performed three times in triplicate.
Results
Discovery phase for miRNAs associated with progression
For the discovery phase of the study, miRNAs were isolated from fresh frozen primary cutaneous melanomas from 23 different patients (median Breslow thickness 3.68 mm; range 0.44–15.64; Table S1). Of these patients, 12 patients did not develop distant metastases during a follow-up of at least 4 years. Expression of 159 different miRNAs was measured in triplicate by qPCR. Thirty miRNAs were subsequently excluded from further analyses because reproducible quantification failed. Statistical analyses of the remaining 129 miRNAs (Supplemental Table S4) identified 11 miRNAs which expression strongly correlated with tumor thickness (p < 0.01 and FDR < 0.08, Table 1). The most significant miRNA was miR-203: its expression strongly decreased with increasing tumor thickness (p < 0.001, FDR < 0.002).
Two miRNAs demonstrated a significant correlation with DMFS or death due to the disease. Loss of miR-203 expression was associated with reduced DMFS (logrank p = 0.006), whereas increased expression of miR-130b was associated with better DMFS (logrank p = 0.008). However, for both miRNAs, the FDR was high (FDR = 0.25).
Validation phase: unmatched and matched series
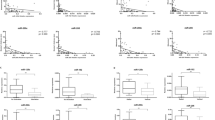
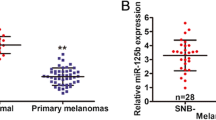
To replicate the possible associations with progression, we subsequently analyzed expression of nine miRNAs (miR-17.5p, 106a, 130b, 150, 200a, 200c, 203, 205, and 335) in a second unmatched series of melanocytic lesions (six acquired nevi, 15 primary melanomas (median thickness 4.20 mm, range 0.94–9.28), and 16 melanoma metastases). qPCR analyses revealed significantly different expressions of miR-200a, miR-200c, and miR-203 (Fig. 1; ANOVA, p < 0.001, FDR < 0.001), with miR-203 being the most significant. To confirm the progressive decrease in expression with progression to metastasis, their expression was determined in a third matched set of 11 primary cutaneous melanomas (median thickness 4.27 mm, range 0.59–5.57) and their corresponding locoregional (n = 10) or distant (n = 3) metastases (Supplemental Table S3). A paired t test confirmed a significant difference for miR-200a (p = 0.029, FDR < 0.08), but not for miR-203 (p = 0.08) and miR-200c (p = 0.07). However, the expressions of the three miRNAs were very highly correlated between each other (all three pairwise correlation coefficients > 0.84, p < 0.002).
We next analyzed whether expression of the three miRNAs correlated with tumor thickness by combining the two validation studies (26 primary melanomas, median thickness 4.1 mm (range 0.59–9.28)). For each miRNA, a correlation coefficient stratified by series was calculated in order to control possible batch effects [20] and found a significant correlation with tumor thickness for each miRNA (p = 0.024, 0.033, and 0.031 for miR-200a, miR-200c, and miR-203, respectively).
In situ detection of miR-200c and E-cadherin expression in primary melanoma
It has been established previously that downregulation of miR-200a and miR-200c increases ZEB1/2 expression and that it downregulates E-cadherin expression in vitro [25]. To demonstrate this correlation in melanoma tissue, a chromogenic in situ hybridization assay for miR-200c was performed using a LNA–DNA oligo [18]. A total of ten superficial spreading primary melanomas with clear vertical growth phase were analyzed. Consecutive tissue slides were stained for E-cadherin. Strong miR-200c and E-cadherin expressions were observed in epidermis and hair follicles of the skin and served as an internal positive control. This strong and positive correlation between miR-200c and E-cadherin expressions in nontumor tissue strongly suggests specific hybridization signals. These analyses demonstrated that loss of miR-200c expression coincides with loss of E-cadherin expression in all tumors analyzed. The weakest hybridization signal at the deep invasive margin of the melanoma (Fig. 2, representative staining) confirmed the progressive loss of miR-200c with increasing thickness. Loss of miR-200c and E-cadherin expression did not correlate with clear mesenchymal cell morphology: cells lacking E-cadherin did not display an obvious spindled shape. However, a case of spindle cell melanoma was entirely negative (Supplemental Fig. S1).
In situ detection of miR-200 and E-cadherin. Consecutive tissue sections were subjected to E-cadherin immunohistochemistry (a–d) and miR-200c in situ hybridization (e–h). A subcutaneous metastasis (a, e) displays no E-cadherin (a) and miR-200c (e), whereas high E-cadherin (red AEC staining) and miR-200 expression (blue NBT-BCIP staining) was observed in the adjacent hair shaft (asterisk). In primary melanoma (b–f), a progressive loss of both E-cadherin (b) and miR-200c (f) can be seen with increasing thickness. At higher power view of the subepidermal component (c, g), clear membranous E-cadherin staining of the overlying epidermis and strong membranous staining of melanoma cells can be seen (c). Strong miR-200c expression was observed in epidermis and melanoma cells (g). In the deep dermal component, low and predominantly membranous E-cadherin expression was detected (d, arrow) but miR-200c expression was not observed (h)
Downregulation of miR-200 in melanoma cell lines
To demonstrate a possible causal relation between loss of miR-200c and E-cadherin expressions, E-cadherin expressing melanoma cell lines 888Mel, 1 F6, and SKMel28 were transfected with a LNA–DNA oligo that was previously shown to antagonize miR-200c [18]. Three rounds of transfection with anti-miR-200c resulted in decreased CDH1 expression in all three cell lines, whereas transfection with the scrambled control LNA–DNA oligo did not (Fig. 3). At 6 days post-transfection, mean CDH1 expression was reduced to 43 % (SEM = 5.8, n = 3, P = 0.008) in 1F6 cells compared to scrambled control (Fig. 3a), 44 % for 888Mel cells (SEM = 6.9, n = 3, P = 0.014, Fig. 3b), and 60 % in SKMel28 cells (SEM = 6.9, n = 3, P = 0.031, Fig. 3c). These data suggest a causal relationship between miR-200c and E-cadherin expressions in melanoma. Additional transfections did not further decrease CDH1 expression and expression returned to normal levels within 1 week after the last transfection (data not shown).
Anti-miR-200c reduced E-cadherin expression in melanoma. Consecutive rounds of transfection of three different E-cadherin-expressing melanoma cell lines 1F6 (a), 888Mel (b), and SKMel28 (c) with the LNA–DNA oligo-targeting miR-200c resulted in a progressive decrease in E-cadherin (CDH1) expression as measured by qPCR in all cell lines (solid line) compared to scrambled control transfected cells (dashed line).Six days after transfection, mean CDH1 expression levels in 1F6, 888Mel, and SKMel28 cells were reduced to 43 % (P = 0.008), 44 % (P = 0.014), and 60 % (P = 0.031) of that of control cells, respectively. Arrows indicate time points of transfection. Data is expressed as the percentage of expression of scrambled control transfected cells. Error bars indicate the standard deviation of three different experiments, each in triplicate
Discussion
In this study, we demonstrate that loss of miR-200a, miR-200c, and miR-203 expression in melanoma correlates with tumor thickness, and their progressive loss in a series of matched primary tumors and associated metastases strongly suggest a possible association with disease progression. Loss of miR-200c and E-cadherin expression was exclusively observed that the invasive front and in in vitro experiments support a causal relation between loss of miR-200c and E-cadherin expression.
Loss of miR-200 family members in melanoma has been described previously [26], but not in the context of tumor thickness, localization of expression with the tumor, and progression towards a metastatic melanoma phenotype. Recently, reduced expression of miR-200 family members was observed in a series of melanoma metastases compared to a series of unrelated primary melanoma [27]. Our work significantly refines these observations by visualizing the gradual loss of miR-200c and E-cadherin expression in primary melanoma, with maximal loss of expression at the most invasive component of the tumor. Furthermore, using pairs of melanoma metastases and the primary melanomas from which they originated, we could demonstrate loss of miR-200a, c, and miR-203 expression with advanced stages of disease for individual. Therefore, our data support the hypothesis that loss miR-200a, c, and miR-203 correlates with melanoma progression towards metastases.
The mechanism by which loss of miR-200a, c, and miR-203 contributes to melanoma metastasis remains elusive and their role in metastasis is subject of debate. Previous observations suggest that miR-200c expression can suppress migration of carcinoma cells [18, 25, 28, 29]. However, in melanoma cell lines, miR-200 family members do not inhibit cell migration but have profound effects on melanoma cell morphology and mode of melanoma cell invasion in 3D matrices [30]. MiR-200c overexpression drives the nonmesenchymal amoeboid form of invasion in type I collagen gels [30]. A different study demonstrated that melanoma cells can express markers of amoeboid invasion at the invasive front of melanomas, which most often consists of cells without a spindled morphology [6]. In that study, miR-200c expression was not analyzed. Our data, however, demonstrate that loss of miR-200c expression occurs at the invasive front and argues that miR-200c-mediated ameboid invasion in 3D culture models with its known limitations [31] is not representative for melanoma invasion in tissue. A recent study using a breast cancer cell line suggests a biphasic role of miR-200 family members in metastasis in which their expression can hinder tumor cell dissemination and entry into circulation but that it can support outgrowth at a secondary site [32]. Our data, however, demonstrate that loss of miR-200c expression occurs at the invasive front and that its expression in metastases is significantly lower compared to primary lesion from which it originated. This may suggests that the metastases originated from the miR-200-negative cells at the invasive front. The discrepancies between our data, miR-200c-mediated mode of invasion in 3D culture models, and the possible biphasic effect of the expression of miR-200 family members in melanoma warrants further investigations.
The contribution of miR-200 a and c to tumor progression most likely extends beyond its ability to modulate invasion via the ZEB/miR-200/E-cadherin axis. Our data demonstrate a very strong correlation between miR-200a, c, and miR-203 expressions. ZEB1, in addition to repressing E-cadherin transcription, also represses miR-203 expression [17]. It has been proposed that miR-203 is a key molecule repressing expression of epithelial differentiation by ‘stemness’-related genes [33, 34], such as, the stem cell-related transcription factors BMI1 [35] and SOX2 [17]. Expression of Sox2 has been documented in melanoma cells with presumed stem cell or tumor-initiating characteristics, in particular, at the invasive front of primary cutaneous melanoma [36, 37]. A stem cell-like phenotype is associated with the cell’s ability to grow in an anchorage-independent manner. Loss of miR200a and c may, therefore, be important for cells to acquire this ability. Although we were not able to maintain long-term down regulation of miR200c expression using anti-miR-200c that allowed us to test if loss of miR-200c loss increased colony formation in soft agar (data not shown), others have demonstrated that forced expression of miR-200c in A375 melanoma cells reduced their capacity to grow in soft agar [27]. This suggests that loss of miR-200c could indeed promote anchorage-independent growth. The observations that ZEB1 expression is necessary for TrkB-mediated suppression of anoikis in rat intestinal epithelial and kidney cells [38] and that TrkB expression is predominantly observed in the invasive growth component of melanoma [39] lend further support to additional effects of disturbing the ZEB-miR200 equilibrium beyond that of modulating E-cadherin-mediated cell adhesion in melanoma cells.
Our results demonstrate a clear and highly significant correlation between loss of miR-200a, c, and miR-203 expression at the invasive front of melanoma, which is casually related to loss of E-cadherin expression. Expression of these miRNAs is strongly reduced in melanoma metastases compared to the matched primary tumors. Several lines of research point to a variety of effects of miR-200 activity in different tumor types and cell lines. If and how these are relevant for melanoma progression warrants further investigations.
References
Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial–mesenchymal transitions in development and disease. Cell 139:871–890
Na YR, Seok SH, Kim DJ, Han JH, Kim TH, Jung H, Lee BH, Park JH (2009) Bone morphogenetic protein 7 induces mesenchymal-to-epithelial transition in melanoma cells, leading to inhibition of metastasis. Cancer Sci 100:2218–2225
Nurmenniemi S, Sinikumpu T, Alahuhta I, Salo S, Sutinen M, Santala M, Risteli J, Nyberg P, Salo T (2009) A novel organotypic model mimics the tumor microenvironment. Am J Pathol 175:1281–1291
Yang J, Price MA, Li GY, Bar-Eli M, Salgia R, Jagedeeswaran R, Carlson JH, Ferrone S, Turley EA, McCarthy JB (2009) Melanoma proteoglycan modifies gene expression to stimulate tumor cell motility, growth, and epithelial-to-mesenchymal transition. Cancer Res 69:7538–7547
Hsu MY, Wheelock MJ, Johnson KR, Herlyn M (1996) Shifts in cadherin profiles between human normal melanocytes and melanomas. J Investig Dermatol Symp Proc 1:188–194
Sanz-Moreno V, Gaggioli C, Yeo M, Albrengues J, Wallberg F, Viros A, Hooper S, Mitter R, Feral CC, Cook M, Larkin J, Marais R, Meneguzzi G, Sahai E, Marshall CJ (2011) ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell 20:229–245
Jaeger J, Koczan D, Thiesen HJ, Ibrahim SM, Gross G, Spang R, Kunz M (2007) Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res 13:806–815
Tucci MG, Lucarini G, Brancorsini D, Zizzi A, Pugnaloni A, Giacchetti A, Ricotti G, Biagini G (2007) Involvement of E-cadherin, beta-catenin, Cdc42 and CXCR4 in the progression and prognosis of cutaneous melanoma. Br J Dermatol 157:1212–1216
George E, Polissar NL, Wick M (2010) Immunohistochemical evaluation of p16INK4A, E-cadherin, and cyclin D1 expression in melanoma and Spitz tumors. Am J Clin Pathol 133:370–379
Katoh M (2011) Network of WNT and other regulatory signaling cascades in pluripotent stem cells and cancer stem cells. Curr Pharm Biotechnol 12:160–170
Poser I, Dominguez D, de Herreros AG, Varnai A, Buettner R, Bosserhoff AK (2001) Loss of E-cadherin expression in melanoma cells involves up-regulation of the transcriptional repressor Snail. J Biol Chem 276:24661–24666
Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y (2009) Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 15:195–206
Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, Eggermont AM, Lenoir G, Sarasin A, Tursz T, van den Oord JJ, Spatz A (2006) Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst 98:472–482
Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12:247–256
Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O’Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, Urban N, Knudsen BS, Tewari M (2010) Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol 116:117–125
Tryndyak VP, Beland FA, Pogribny IP (2009) E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int J Cancer 126:2575–2583
Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur HA, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T (2009) The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11:1487–1495
Park SM, Gaur AB, Lengyel E, Peter ME (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22:894–907
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Kraft P, Schadt E, Aten J, Horvath S (2003) A family-based test for correlation between gene expression and trait values. Am J Hum Genet 72:1323–1330
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
van Kempen LC, Rijntjes J, Mamor-Cornelissen I, Vincent-Naulleau S, Gerritsen MJ, Ruiter DJ, van Dijk MC, Geffrotin C, van Muijen GN (2008) Type I collagen expression contributes to angiogenesis and the development of deeply invasive cutaneous melanoma. Int J Cancer 122:1019–1029
Alexaki VI, Javelaud D, Van Kempen LC, Mohammad KS, Dennler S, Luciani F, Hoek KS, Juarez P, Goydos JS, Fournier PJ, Sibon C, Bertolotto C, Verrecchia F, Saule S, Delmas V, Ballotti R, Larue L, Saiag P, Guise TA, Mauviel A (2010) GLI2-mediated melanoma invasion and metastasis. J Natl Cancer Inst 102:1148–1159
van Muijen GN, Cornelissen LM, Jansen CF, Figdor CG, Johnson JP, Brocker EB, Ruiter DJ (1991) Antigen expression of metastasizing and non-metastasizing human melanoma cells xenografted into nude mice. Clin Exp Metastasis 9:259–272
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10:593–601
Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M (2008) MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18:549–557
Xu Y, Brenn T, Brown ER, Doherty V, Melton DW (2012) Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer 106:553–561
Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 9:582–589
Korpal M, Lee ES, Hu G, Kang Y (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283:14910–14914
Elson-Schwab I, Lorentzen A, Marshall CJ (2010) MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS One 5
Sabeh F, Shimizu-Hirota R, Weiss SJ (2009) Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol 185:11–19
Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y (2012) Direct targeting of Sec23a by miR-200 s influences cancer cell secretome and promotes metastatic colonization. Nat Med 17:1101–1108
Lena AM, Shalom-Feuerstein R, Rivetti DV, Aberdam D, Knight RA, Melino G, Candi E (2008) miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ 15:1187–1195
Yi R, Poy MN, Stoffel M, Fuchs E (2008) A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452:225–229
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD (2011) miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 39:D163–D169
Laga AC, Lai CY, Zhan Q, Huang SJ, Velazquez EF, Yang Q, Hsu MY, Murphy GF (2011) Expression of the embryonic stem cell transcription factor SOX2 in human skin: relevance to melanocyte and merkel cell biology. Am J Pathol 176:903–913
Laga AC, Zhan Q, Weishaupt C, Ma J, Frank MH, Murphy GF (2011) SOX2 and nestin expression in human melanoma: an immunohistochemical and experimental study. Exp Dermatol 20:339–345
Smit MA, Peeper DS (2011) Zeb1 is required for TrkB-induced epithelial–mesenchymal transition, anoikis resistance and metastasis. Oncogene 30:3735–3744
Innominato PF, Libbrecht L, van den Oord JJ (2001) Expression of neurotrophins and their receptors in pigment cell lesions of the skin. J Pathol 194:95–100
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
In situ detection of miR-200 and E-cadherin in a desmoplastic melanoma. Consecutive tissue sections of a desmoplastic melanoma (A, H&E staining) were subjected to Ecadherin immunohistochemistry (B,C) and miR-200c in situ hybridization (D,E). E-cadherin and miR-200c expression was not observed in the tumor (B and D, respectively), but the overlying epidermis was strongly positive for membranous E-cadherin (C, brown staining) and cytoplasmic miR-200c (E, blue staining; arrows indicate a few of many positive cells) expression (PDF 246 kb)
Table S1
List of 23 patients with primary melanomas and their pertinent clinical and histological features, used in the discovery phase of the study (PDF 44 kb)
Table S2A
List of 15 patients with primary melanomas and their pertinent clinical and histological features, used in the first validation phase of the study (PDF 40 kb)
Table S2B
List of 16 patients with melanomas metastases and 6 nevi used in the first validation phase of the study (PDF 33 kb)
Table S3
Pertinent clinical data and Ct values of the set of 11 paired primary-metastatic melanomas used in the second validation study (PDF 34 kb)
Table S4
Spread sheets of all miRNAs analysed, raw Ct values and calculated values (PDF 966 kb)
Rights and permissions
About this article
Cite this article
van Kempen, L.C., van den Hurk, K., Lazar, V. et al. Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch 461, 441–448 (2012). https://doi.org/10.1007/s00428-012-1309-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-012-1309-9