Abstract
The prostate tumor overexpressed-1 (PTOV1) protein was first described overexpressed in prostate cancer but not detected in normal prostate. PTOV1 expression is associated to increased cancer proliferation in vivo and in vitro. In prostate biopsy, PTOV1 detection is helpful in the early diagnosis of cancer. The purpose of this study was to analyze the relevance of PTOV1 expression to identify aggressive tumors derived from 12 different histological tissues. Tissue microarrays (TMAs) containing 182 biopsy samples, including 168 human tumors, were analyzed for PTOV1 and Ki67 expression by immunohistochemistry. Tumors of low and high histological grade were selected from lung, breast, endometrium, pancreas liver, skin, ovary, colon, stomach, kidney, bladder, and cerebral gliomas. One TMA with representative tissues without cancer (14 samples) was used as control. PTOV1 expression was analyzed semiquantitatively for the intensity and percentage of positive cells. Ki67 was evaluated for tumors proliferative index. Results show that PTOV1 was expressed in over 95% of tumors examined. Its expression was significantly associated to high-grade tumors (p = 0.014). This association was most significant in urothelial bladder carcinomas (p = 0.026). Overall, the expression of Ki67 was associated to high-grade tumors, and it was significant in several tumor types. PTOV1 and Ki67 were significantly co-overexpressed in all tumors (p = 0.001), and this association was significant in clear cell renal carcinoma (p = 0.005). In conclusion, PTOV1 expression is associated to more aggressive human carcinomas and more significantly to bladder carcinomas suggesting that this protein is a potential new marker of aggressive disease in the latter tumors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate tumor overexpressed-1 (PTOV1) was identified as a novel gene and protein during a differential display screening for genes overexpressed in prostate cancer [1]. This protein is encoded by a 12-exon gene localized in chromosome 19q13.3. The protein consists of two highly homologous domains of 151 and 147 amino acids arranged in tandem, joined by a short linker peptide. The PTOV1 domain is conserved across distant organisms and does not show significant similarities to known protein motifs [1]. A second gene, PTOV2, later identified as Arc92/MED25/ACID1, was identified in human and other organisms, coding for proteins containing a single PTOV domain [1–3]. PTOV2/Arc92/MED25/ACID1 interacts with retinoic acid receptor (RAR), the histone acetyltransferase CBP, and the mediator complex, suggesting a role for MED25 in chromatin remodeling and pre-initiation complex assembly to recruit activators to the basal transcriptional machinery [3]. PTOV1 is overexpressed in 71% of prostatic carcinomas and 80% of prostate intraepithelial neoplasia but is undetectable or expressed at low levels in normal prostate epithelium [2]. In normal endothelial cells and in neuroendocrine cells, PTOV1 is expressed at high levels [1, 2]. High levels of PTOV1 in prostatic tumors correlated significantly with Ki67 proliferative index and associated with a nuclear localization of the protein, suggesting a functional relationship between PTOV1 overexpression, proliferative status, and nuclear localization [2]. Transient overexpression of a chimeric GFP-PTOV1 protein induced proliferation and forced the entry of cells into the S phase of the cell cycle, suggesting that overexpression of PTOV1 can contribute to the biological behavior of these tumors [4]. PTOV1 interacts with the lipid raft protein flotillin-1, and both proteins colocalize at the plasma membrane and in the nucleus [4]. Depletion of PTOV1 markedly inhibited cell proliferation under basal conditions [4]. Recently, we determined that PTOV1 was significantly overexpressed in HGPIN associated with cancer but not in HGPIN lesions in prostates that do not develop cancer, indicating that PTOV1 is a useful marker to detect cancer in the absence of carcinoma lesions in the biopsy [5].
The proliferative index in human tumors is an important prognostic factor that is frequently evaluated in histological analysis of tumor specimens. The Ki67/MIB-1 antigen is a labile nuclear protein which expression is tightly linked to the cell cycle [6]. It is expressed in proliferating cells during mid-G1, increasing in level through S and G2, and peaking at the end of the M phase. Ki67/MIB-1 is not detected in resting cells [6]. Here we have analyzed the expression of PTOV1 in 12 histological types of human tumors and studied correlation with grade and Ki67 expression. We have found that PTOV1 is significantly overexpressed in the majority of the tumors analyzed, and it may be a new oncogenic factor associated with poor prognosis.
Materials and methods
Specimens
One hundred and sixty-nine human tumor specimens were retrieved from the archives of Pathology Department Vall d’Hebron University Hospital. Consent for the use of human tissues was obtained following the rules of the Spanish Biomedical Research Law and approved by the ethical committee of the hospital. Tissue microarrays (TMA) were constructed from formalin-fixed paraffin-embedded blocks of each sample. Histological grade for each tumor was determined on HE-stained slides for the same samples and was classified according to defined criteria [7]. Tumors selected were 27 lung squamous and lung adenocarcinomas (LA), 11 breast ductal, and 10 lobular carcinomas, 10 endometrial adenocarcinomas, 10 ductal pancreatic adenocarcinomas (DPA), 9 hepatocarcinomas, 11 squamous skin carcinomas, 9 malignant melanomas, 8 mucinous (MOC) and 9 serous (SOC) ovarian carcinomas, 10 gastric and 10 colon carcinomas, 11 cerebral gliomas (CG), 13 clear cell renal carcinomas (CCRC), and 11 urothelial bladder carcinomas (UBC). Tumors were classified into two groups according to their histological tumor grade following defined criteria [7]: 87 (51%) tumors were low grade (G1, G2) and 81 (48%) were high grade (G3, G4). Malignant melanomas were classified according to Breslow’s definition of staging that considers the area and the depth of invasion of cutaneous melanoma lesions [8]. Low-grade papillary urothelial carcinoma and high-grade papillary urothelial carcinoma were classified as defined [9]. Control tissues not affected by cancer (n = 14) corresponding to the analyzed histological tissue types were used for one TMA used as reference.
Generation of tissue arrays
Ten different tissue arrays were constructed using a semiautomated tissue arrayer (Chemicon International Inc., Billerica, MA, USA), containing the two major histological grades of the 169 tumors under study and one tissue array with normal control tissues. Each tissue array was assembled as a previously described [10]. Briefly, three tissue cylinders of 1.2 mm for each case were punched from morphologically representative tumor areas of each donor tissue paraffin block and brought into one recipient paraffin block (3 × 2.5 cm). Representative normal areas without tumoral pathology were included in one TMA. H&E stain for each TMA was performed to verify the histopathological findings.
Immunohistochemistry
Four-micron sections were cut from each TMA and transferred to glass slides. Antigen retrieval was carried out by autoclaving for 5 min in citrate buffer. The slides were stained on an automated DAKO stainer (Glostrup, Denmark). Endogenous peroxidase activity was blocked with 0.2% hydrogen peroxidase solution, and non-specific labeling was blocked with serum blocking solution. Sections were incubated in for 2 h at room temperature with PTOV1 antibody. As a negative control, primary antibody was omitted and replaced with phosphate-buffered saline. In addition, non-specific rabbit antibody was used and gave clean negative results in all cases tested (not shown). The reaction was revealed by the avidin–biotin complex peroxidase method (ABC Elite kit, Vector Burlingame, CA, USA) followed by staining with the peroxidase substrate 3.3 diaminobenzidine tetrachloride (Sigma-Aldrich Chemie GmbH., Deisenhofer, Germany). The slides were counterstained with 50% hematoxylin. A case was considered positive when >10% of the cells showed unequivocal staining. Both cytoplasmic and nuclear staining was recorded. A similar procedure was followed for the detection of Ki67 except that incubation with primary antibody was for 30 min at room temperature. Each slide was assessed independently by two pathologists (S.F. and I.d.T.). Discrepancies were resolved by a concurrent reexamination by both investigators using a double-headed microscope. PTOV1 expression was evaluated in a semiquantitative manner whereby the levels of expression are represented as the percentage of positive cells and the intensity of the staining \( \left[ {{\hbox{H - score}} = {1}\; \times \;\left( {\% {\hbox{ weak}}} \right) + {2}\; \times \;\left( {\% {\hbox{ moderate}}} \right) + {3}\; \times \;\left( {\% {\hbox{ intense}}} \right){\hbox{ with a ranking between 0 and 300}}} \right] \) [11]. Cases with less than 10% of cells showing positivity were considered negative. To analyze the proliferative index, the proportion of nuclear Ki67 staining over the total number of nuclei per high power field was scored. Four value ranges for Ki67 index were established (negative = 0, low = 1–5%, moderate = 6–25%, and high = >25%).
Statistical analysis
Cross-tabulations were used to establish the presence of associations among variables. The Fisher’s exact test was used to evaluate associations among variables and the Cramer’s V coefficient to measure the strength of relationships [12]. The non-parametric method of Kruskal–Wallis was used to test equality of population medians among groups [13]. A p value <0.05 was considered significant in all analyses. For the multivariate analysis, we used the multiple correspondence analysis (MCA) technique to represent the distances among the categories of the qualitative variables and between the observations [14, 15]. This technique allows to handle large and complex datasets and to represent the pattern of relationships of several categorical dependent variables. MCA can be seen as a generalization of principal component analysis when the variables to be analyzed are categorical instead of quantitative. That is, MCA can be understood as a reduction of the dimensionality of a dataset with a large number of interrelated variables into a smaller number of independent variables (known as new dimensions). First dimension retains as much as possible of the inertia (or variability) present in the data, and similarly each succeeding dimension retains the remaining inertia that it is always less than the previous dimension. Data analyses were performed with the open source R statistical software package [16].
Results
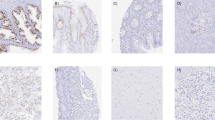
A total of 182 biopsies that included 168 malignant tumors and 14 normal tissues were evaluated for the expression of PTOV1 in a semiquantitative manner. PTOV1 expression, quantified by the H-score value, was absent or very low (less than 10%) in the cytoplasm and nucleus of all normal tissues examined, including lung, breast, endometrium, pancreas, liver, skin, ovary, stomach, colon, kidney, bladder, and brain (Fig. 1). Blood vessels and red blood cells showed positive expression for PTOV1, confirming previous observations [1, 2].
Expression of PTOV1 protein in tumors. Representative images of immunohistochemistry performed with anti PTOV1 antibody. Six histological tumor types of high and low grade were selected together with six representative no cancerous tissues (normal). Insets are magnifications showing details of PTOV1 protein localization. Faint or absence of staining was observed in no cancerous tissues. Nuclear staining was detected in several tumor types, including ductal pancreatic adenocarcinomas (DPA), cerebral gliomas (CG), clear cell renal carcinomas (CCRC), and urothelial bladder carcinomas (UBC). Strong association of PTOV1 with mitotic figures was observed in colon adenocarcinoma (CA) and breast ductal carcinomas (BDC)
The expression of PTOV1 was detected in 162 of 168 tumors (97%) with a minimum median H-score of 100 and a maximum of 200 (Table 1). The mean H-score of PTOV1 expression in tumors of low grade was 120, ranging between 0 and 240, whereas in high-grade tumors it was 134, with a range between 0 and 300. These data suggested possible differences in the distribution of PTOV1 staining among the tumors analyzed. To confirm this observation, the Kruskal–Wallis test was applied and the results confirmed that the medians of PTOV1 score among tissues are different (p = 0.006984). Therefore, in order to compare the levels of PTOV1 expression in the different tumors, the levels of protein detection were defined as follows: No expression was labeled “0”, H-score lower or equal to the median for a specific tissue was labeled “low”, and H-score greater than the median for a specific tissue was labeled “overexpression”. The expression of PTOV1 showed a significant association with tumor grade in UBC (Fisher’s exact test p = 0.026; Cramer’s V coefficient = 0.841), where the H-score for PTOV1 expression in the high-grade tumors group is clearly not overlapping with the values encountered in low-grade tumors (Fig. 2). This observation, also put into evidence by the high Cramer’s coefficient value, suggests that although a low number of cases are included in the group, the detection of PTOV1 is able to distinguish in a significant manner high-grade bladder tumors from low grade. PTOV1 has been previously shown to shift from the cytoplasm to the nucleus [2]. Overexpression of the exogenous PTOV1 is nuclear-localized and promotes cell entry into the S phase of the cell cycle, and in addition, nuclear PTOV1 is significantly associated to higher Ki67 index in prostate tumors [2]. Thus, we have analyzed PTOV1 subcellular localization in relation with tumor grade. PTOV1 nuclear localization was strongly associated with high-grade UBC (p = 0.015; Cramer’s V coefficient = 0.833) suggesting that its presence in the nucleus is important for the proliferation and progression of this type of cancer (see representative images in Fig. 1).
Boxplot of PTOV1 H-score per tumor grade and tissue. Bar diagrams and boxplots are used to show distributions of protein expression. The graph shows the H-score for PTOV1 (vertical axis) and the tumor grade (1 for low-grade and 2 for high-grade) for the different histological tumors. Yellow and red dots show the median values of PTOV1 H-score for each tumor subtype
A significant association with high-grade tumors was also observed considering all tumors (excluding UBC; p = 0.045; Cramer’s V coefficient = 0.201). The proliferative status of tumors was evaluated by staining for Ki67 (MIB-1). The expression of Ki67 was evaluated considering the proportion of nuclear staining over the total number of nuclei (Ki67 index). Ki67 was absent in 43 (25%) tumors, low in 27 (16%), and high or overexpressed in 96 (56%) tumors. A higher Ki67 index was significantly associated with high-grade tumors in UBC, LA, SOC–MOC, and DPA. Overall, this association was also significant (p = 5.3947e−07), and it was stronger than the one observed with PTOV1 (Cramer’s coefficient = 0.461).
The association between PTOV1 and Ki67 expression was significant in CCRC (p = 0.001; Cramer’s V coefficient = 0.557) and considering all tumors (excluding CCRC) (p = 0.004; Cramer’s V coefficient = 0.268), again suggesting that higher PTOV1 expression levels are associated with more aggressive proliferative tumors, in agreement with previous findings in prostate cancer [2].
Integrating information collected from various sources provides a better interpretation of the underlying biological processes than data from a single source. To get further insight of the biological relevance of our findings and to discover associations among different groups of variables (tissue, tumor grade, Ki67, PTOV1, and nuclear or no-nuclear localization), we performed a multivariate analysis based on a MCA. The results in Fig. 3 show a representation for the first two dimensions: PTOV1 covers quiet well the map representation with the three levels of expression defined. Different groups of tissues are clearly defined and show association with the histological grade of tumors. Tumors overexpressing PTOV1 (labeled PTOV1.2 in Fig. 3) are found in very close proximity with high-grade tumors and UBC (bladder cancer: bladder), confirming our previous results. These parameters are also in close proximity to breast (breast) and pancreas (pancreas) cancer. Lower levels of PTOV1 (PTOV1.1) appear more related to low-grade tumors and specifically to ovary (ovary) and CNS (CNS) cancer. Interestingly, tumors not expressing PTOV1 (PTOV1.0) are quite distant from all the other variables. This unsupervised analysis integrates the five variables studied and confirms our previous observations that higher PTOV1 expression is significantly associated to aggressive bladder carcinomas and high-grade tumors.
MCA analysis representation of the variables studied in the first two dimensions. This analysis permits to discover associations among the different groups of variables studied: tissue, tumor grade, Ki67 levels, PTOV1 levels, and PTOV1 nuclear localization. These variables are indicated by different colors. Red PTOV1 levels, indicated as PTOV1.2 = overexpression, PTOV1.1 = low expression (lower or equal to the median for a specific tissue), PTOV1.0 = no expression. Yellow PTOV1 localization, nuclear is indicated by 1 and no-nuclear is indicated by 0. Blue cancer tissues. Green tumor grade LG and HG. Pink Ki67 levels, indicated by Ki67.3 = overexpression, Ki67.2 = moderate, Ki67.1 = low, Ki67.0 = no expression. Quadrant III, for example, suggests an association among the variables: high-grade (HG) tumors, tumors with overexpression of Ki67, tumors with high levels of PTOV1, as well as bladder, breast, and pancreas tumors
Discussion
The PTOV1 protein, first found overexpressed in prostate carcinomas, is involved in the mechanisms leading to the progression of this neoplasia, as reported by previous findings showing that the exogenous overexpression of a recombinant PTOV1 protein activates cell proliferation [1, 2, 4]. To evaluate whether its overexpression is a more general feature associated to neoplastic cells, we have analyzed the expression of PTOV1 in a cohort of 168 malignant tumors of 12 different histological types, each one comprising low-grade and high-grade tumors. Many types of human neoplasias can be found within a given organ. This heterogeneity and the great variability of oncogenic alterations that have been described to trigger tumor growth are an indication of the numerous regulating circuits and cellular targets that may exist in cancer. It is essential to study several hundreds or several thousands of human tumors to identify critical genetic and biochemical factors associated with the tumor progression and prognosis [10, 17–19]. Our results show that PTOV1 is expressed in 97% of all tumors and, in general, it is overexpressed in more aggressive disease. In our series of 168 tumors, PTOV1 immunoreactivity was cytoplasmic or mixed cytoplasmic nuclear in all primary carcinomas, in contrast with the normal epithelial counterpart lacking significant reactivity. Because the median of expression of PTOV1 was significantly different among the various cancers, the values above the median were taken as “overexpression” levels in each type. This allowed us to compare the levels of the immunoreactive protein in neoplasias of different types. Considering all 168 tumors, those with high grade significantly overexpressed the protein PTOV1. In particular, the association between stronger expression of PTOV1 and high grade of malignancy was very significant in urothelial bladder carcinomas. UBCs are also very significantly associated with nuclear localization of PTOV1, suggesting an action of the protein at this location in promoting UBC cancer progression. These observations would suggest a function for PTOV1 similar to the one described for the related protein PTOV2 in transcription as part of the Mediator complex [3]. PTOV2/MED25 was described to associate with RA-bound RAR and increase RAR/RXR-mediated transcription [20]. The intrinsic transcriptional activity of MED25 was found in its PTOV domain, likely accomplished by direct association with the general activator CBP. In that work, PTOV1 competed with MED25 function, possibly by competition of the co-activator CBP, and blocked RAR response [20].
Several carcinoma types among those studied here showed a higher Ki67 index in association with a high grade of malignancy, in agreement with previous descriptions [21–25]. The proliferative activity of CCRC, measured by Ki67 immunoreactivity, was among the best prognostic tools [21]. However, in our series of CCRC, neither Ki67 index nor PTOV1 expression alone were significantly associated with high-grade tumors. However, when the expression levels of PTOV1 and Ki67 index were considered, a significant association was found, suggesting that assessment of both parameters is useful to indicate tumor aggressiveness. This finding is in part supported by our previous in vitro studies where the exogenous PTOV1 expression induced proliferation of different cell types, including PZHPV7 and PC3 prostate cancer cells and COS7 fibroblasts [2]. In addition, in prostate carcinomas, PTOV1 protein was significantly associated with tumors with higher Ki67 index and increased PSA blood levels [4].
Our previous finding that PTOV1 overexpression is detected in HGPIN lesions specifically associated with cancer but not in HGPIN that are not associated with cancer suggests that high levels of PTOV1 can identify premalignant lesions committed to develop into carcinomas [5]. This observation indicates the implication of PTOV1 in the early changes that lead to full cancer development. Our present findings indicate that the detection of PTOV1 expression might be helpful in the analysis of tumor aggressiveness for UBC and CCRC. Although further studies using a larger number of cases are needed to confirm the usefulness of PTOV1 as a potential therapeutic target in a number of carcinomas from different origin, the results obtained from the present pilot study predict the involvement of this protein in tumor progression, particularly in the bladder.
References
Benedit P, Paciucci R, Thomson T, Valeri M, Nadal M, Càceres C, de Torres I, Estivill X, Lozano JJ, Morote J, Reventós J (2001) PTOV1, a novel protein overexpressed in prostate cancer containing a new class of protein homology blocks. Oncogene 20:1455–1464
Santamaría A, Fernández P, Farré X, Benedit P, Reventós J, Morote J, Paciucci R, Thomson TM (2003) PTOV1, a novel protein overexpressed in prostate cancer, shuttles between the cytoplasm and the nucleus and promotes entry into the S phase of the cell division cycle. Am J Pathol 162:897–905
Yang F, DeBeaumont R, Zhou S, Näär AM (2004) The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A 101(8):2339–2344
Santamarıía A, Castellanos E, Gómez V, Benedit P, Renau-Piqueras J, Morote J, Reventós J, Thomson TM, Paciucci R (2005) PTOV1 enables the nuclear translocation and mitogenic activity of flotillin-1, a major protein of lipid rafts. Mol Cell Biol 25:1900–1911
Morote J, Fernández S, Alaña L, Iglesias C, Planas J, Reventós J, Ramón Y Cajal S, Paciucci R, de Torres IM (2008) PTOV1 expression predicts prostate cancer in men with isolated high-grade prostatic intraepithelial neoplasia in needle biopsy. Clin Cancer Res 14(9):2617–2622
Tan PH, Bay BH, Yip G, Selvarajan S, Tan P, Wu J, Lee CH, Li KB (2005) Immunohistochemical detection of Ki67 in breast cancer correlates with transcriptional regulation of genes related to apoptosis and cell death. Mod Pathol 18(3):374–381
American Joint Committee on Cancer (2002) AJCC cancer staging manual, 6th edn. Springer, New York
Breslow A (1970) Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann Surg 172:902–908
Eble JN, Epstein JI, Sesterhenn I (eds) (2004) World Health Organization classification of tumors. Pathology and genetics of tumors of the urinary system and male genital organs. IARCC, Lyon, p 114
Kononen J, Budendorf L, Kallioiemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP (1998) Tissue microarrays for high-throughput molecular profiling of tumors specimens. Nat Med 4:844–847
Detre S, Saclani Jotti G, Dowsett M (1995) A “quickscore” method for immunohistochemical semiquantitation: validation for estrogen receptor in breast carcinomas. J Clin Pathol 48(9):876–878
Quinn GP, Keough MJ (2008) Experimental design and data analysis for biologists. Cambridge University Press, Cambridge
Sprent P, Smeeton NC (2001) Applied nonparametric statistical methods, 4th edn. Chapman & Hall, Boca Raton
Dray S, Dufour AB (2008) The ade4 package: implementing the duality diagram for ecologists. J Stat Software 22(4):1–20
Greenacre MJ, Blasius J (2006) Multiple correspondence analysis and related methods. Chapman & Hall, Boca Raton
Maindonald J, Braun J (2007) Data analysis and graphics using R, 2nd edn. Cambridge University Press, Cambridge
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Bild AH, Yao G, Chang JT et al (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439(7074):353–357
Lu Y, Yi Y, Liu P, Wen W, James M, Wang D, You M (2007) Common human cancer genes discovered by integrated gene-expression analysis. PLoS ONE 2(11):e1149
Lee HK, Park UH, Kim EJ, Um SJ (2007) MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J 26:3545–3547
Cindolo L, Cantile M, Galasso R, Marsicano M, Napodano G, Altieri V (2001) Not traditional prognostic factors in human conventional renal carcinoma. Minerva Urol Nefrol 53(4):211–219
van Diest PJ, van der Wall E, Baak JP (2004) Prognostic value of proliferation in invasive breast cancer: a review. J Clin Pathol 57(7):675–681
Dudderidge TJ, Stoeber K, Loddo M, Atkinson G, Fanshawe T, Griffiths DF, Williams GH (2005) Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clin Cancer Res 11(7):2510–2517
Rindi G, D’Adda T, Froio E, Fellegara G, Bordi C (2007) Prognostic factors in gastrointestinal endocrine tumors. Endocr Pathol 18(3):145–149
Ohsie SJ, Sarantopoulos GP, Cochran AJ, Binder SW (2008) Immunohistochemical characteristics of melanoma. J Cutan Pathol 35(5):433–444
Acknowledgments
This study was supported by the Health Ministry (FIS-PI020764; FIS-PI050818), Science and Education Ministry (SAF05-05848; MTM2008-00642), Science and Innovation Ministry (SAF2008-03936), TV3 Marathon (052610; 052710), and Urologic Research Foundation (01/2004).
Conflict of interest statement
The authors declare to have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Inés de Torres and Rosanna Paciucci equally contributed to the work.
Rights and permissions
About this article
Cite this article
Fernández, S., Mosquera, J.L., Alaña, L. et al. PTOV1 is overexpressed in human high-grade malignant tumors. Virchows Arch 458, 323–330 (2011). https://doi.org/10.1007/s00428-010-1018-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-010-1018-1