Abstract
EGFR mutation testing in non-small cell lung cancer (NSCLC) is a novel and important molecular pathological diagnostic assay that is predictive of response to anti-epidermal growth factor receptor (EGFR) therapy. A comprehensive compilation of a large number of EGFR mutation analyses of the German Panel for Mutation Analyses in NSCLC demonstrates (a) a higher than previously reported mutation frequency outside the conventionally tested exons 19 and 21 and (b) an overall superiority of sequencing based assays over mutation-specific PCR. The implications for future diagnostic EGFR mutation testing are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epidermal growth factor receptor (EGFR)-targeted therapy has been shown to significantly improve progression-free and overall survival in a subgroup of non-small cell lung cancer (NSCLC) patients carrying “activating” mutations of the EGFR gene in their tumors, predominantly representing those NSCLC with adenocarcinoma differentiation. Targeted therapy against EGFR using gefitinib has obtained approval by the European Medicines Agency as first-line therapy in non-resectable NSCLC under the premise that the presence of an “activating” (presumably gefitinib-sensitive) mutation has been demonstrated prior to treatment [1]. Thus, the detection and correct reporting of “activating” mutations in the tumor tissue is essential for gefitinib therapy and has gained significant impact in diagnostic molecular pathology. In order to initiate and supervise large-scale and nationwide quality-controlled testing for EGFR mutations in NSCLC in Germany, a steering panel consisting of six institutions was implemented. Besides quality management, central questions addressed by the panel were the frequency and potential therapeutic relevance of EGFR mutations as well as the critical evaluation of testing modalities. The latter aspect concentrated on the evaluation of the two methodologies available for diagnostic testing, i.e., sequencing of the exons in question (18–21) and mutation-specific PCR.
Methodology
All panel institutions performed mutation analyses by bidirectional Sanger sequencing of exons 18–21 of the EGFR gene from formalin-fixed paraffin-embedded tissues obtained by biopsy or resection. A complete compilation of the applied technical procedures of all panel institutes including DNA extraction methods, primer sequences, and PCR conditions is available online (www.dgp-berlin.de/downloads/public/protocols/EGFR_Mutations_Protocols_engl.pdf). Internal quality assessment of the panel institutes was carried out as following: A series of 40 NSCLC (20 mutated, 20 non-mutated) were initially analyzed by two panel institutes (20 cases contributed by each institute). All 40 cases were cross-tested in a blinded manner by the respective other institutes; there was no disagreement on the analysis results between the two institutes. From this collective, a test set of ten cases (five mutation-positive) was composed and circulated among the four other panel institutes. Positive quality assessments were given to those institutes which correctly analyzed nine of ten cases within a turnaround time of ten working days. All panel institutes participating in this analysis identified all test cases correctly.
Mutation data derived exclusively from routine diagnosis (n = 1,047) from the six panel institutes were collected and analyzed for the distribution of mutations among the different exons. The compiled collective consisted of biopsies (between 25% and 40% of the cases, depending on the respective center) and resection specimen; cytology specimens were not among the analyzed cases. Microscopically guided dissection (by an experienced morphologist, no laser microdissection) was performed in almost all cases in order to increase tumor content above 50% and was only omitted in those resection cases that contained almost exclusively tumor in the analyzed material. Cases without tumor in the preanalytical HE section (∼4%) were not analyzed and not included in the compilation. Cases with <50% tumor content after dissection were analyzed and counted valid when mutations were detected; the non-mutated cases (∼5%) among them were reported with the necessary precautions and were included in the compilation. All new mutations (n = 14; not present in the Catalogue of Somatic Mutations in Cancer Database, www.sanger.ac.uk/genetics/CGP/cosmic/) were confirmed by a second independent diagnostic analysis of the same DNA source. Furthermore, all mutations reported were carefully reviewed in regard to sequence pattern potentially resulting from formalin fixation artifacts [2]. The detectability of these mutations by the commercially available mutation-specific PCR detection kit (TheraScreen® EGFR29 Mutation Kit; DXS Ltd., Manchester, UK) was assessed based on the detailed description of its composition by the supplier. Evaluation of “activation” status (presumed gefitinib responsiveness) was based on the data published by Sharma et al. [3] and, if not addressed in this publication, by the Somatic Mutation-EGFR database [4]. Additional relevant parameters (costs, turnaround time, aspects of quality management and sensitivity, and innovative potential) affecting the choice of assay were addressed.
Results
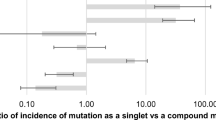
Compilation of all reported mutation data (Supplementary Table) resulted in 163 mutation-positive tumors among 1,047 analyses representing a mutation rate of 15.6% (status 1st December 2009). This frequency is unlikely to reflect exactly the true mutation frequency in German NSCLC since clinical practice of ordering the diagnostic assay preselected patient populations with higher prevalence of EGFR mutations (adenocarcinoma/non-smoker or light smokers) to various extents. Nevertheless, it is likely to reflect the relative distribution of the mutations in regard to the different exons. The overall frequency of exon 20 mutations in this collective may be slightly underestimated since two of the six institutions did not perform bidirectional sequencing of exon 20 in all cases. Mutation frequency in exon 18 (10.4%) was significantly higher compared to Sharma et al. [3], at the relative expense of exon 21 mutations (Table 1). Frequencies of exon 18 and exon 20 mutations were remarkably different between some of the panel centers.
Next, the panel addressed the question which mutations detected by the sequencing approach would have been identified by the commercially available mutation-specific PCR test. Compilation of the data (Table 2) demonstrated that 67 of the 169 mutations (40%) would have been missed by the mutation-specific PCR analysis. For the different exons, the results were as follows: exon 18, 17/21 (81%); exon 19, 33/82 (40%); exon 20, 9/9 (100%); exon 21, 8/57 (14%). Based on Sharma et al. [3] and the Somatic Mutation-EGFR-Database [4], 47 of the 67 mutations (70%) missed by mutation-specific PCR represented “activating” mutations. Evaluating the different exons, the distribution of these mutations was as follows: exon 18, 11/17 (65%); exon 19, 30/33 (91%); exon 20, 3/9 (33%); exon 21, 3/8 (38%). In summary, 47 of all 149 activating mutations (32%) detected by routine diagnostic sequencing of exons 18–21 by the panel institutes would have been missed by sole application of mutation-specific PCR testing.
Discussion
The panel evaluation represents the first assessment of daily routine diagnostic application of EGFR mutation testing in NSCLC on a quality-controlled multicenter and on a nationwide basis. All panel centers had agreed to perform bidirectional sequencing of exons 18–21 and had previously submitted their diagnostic testing to standardized internal quality control (round-robin testing). Compilation of the data obtained between 01 January 2009 and 1 December 2009 provided a sufficient basis for combined analyses and generated several unexpected results with significant impact on diagnostic testing.
Interestingly, the panel evaluation demonstrated that the distribution of mutations between the different exons varied from published data. Especially, the frequency of mutations in exons 18 and 20 that were not uniformly addressed in general and by several previous trials was found to represent 16% of all detected EGFR mutations, which was a higher rate compared to previous reports. Based on the available information, 69% (18/26) of these mutations were judged as activating (gefitinib-responsive). This result suggests the reevaluation of recommendations and studies that were based only on the analysis of exons 19 and 21. Thus, based on these findings, the panel recommends including exons 18 and 20 into routine diagnostic testing. Importantly, frequencies of exon 18 and exon 20 mutations varied remarkably between the different centers. The most likely explanation are differences in patient collectives, e.g., due to patient background or exposure bias. Etiology-specific mutation pattern are a well-documented phenomenon in other solid adulthood malignancies [5] and are likely to be present also in NSCLC, as suggested by the correlation of smoking habits and EGFR mutation frequency [6]. Thus, comprehensive analyses of mutation distribution may provide an important source for molecular epidemiology and unraveling of so far undetected etiological factors of NSCLC. This will be addressed by future studies and further comprehensive compilation of EGFR mutations by the panel.
An important aspect of the evaluation was the question which assay(s) should be implemented in diagnostic settings. All panel institutes used bidirectional sequencing of exons 18–21. Exact central reporting of all mutation data allowed their evaluation in regard to detectability by mutation-specific PCR. Overall, 32% of tumors carrying “activating” mutations as detected by sequencing would have been missed by the commercially available PCR test; notably, 38% of the complex exon 19 mutations would have been missed. This is especially relevant since there is some indication that tumors carrying exon 19 mutations may respond especially favorably to gefitinib treatment [7]. Overall, the percentage of missed mutations is too high to recommend the use of mutation-specific PCR for diagnostic application. It should be mentioned that none of the 38 institutions that had passed the first nationwide quality assessment in Germany for EGFR mutation testing in NSCLC had performed mutation-specific PCR testing as the sole diagnostic test. In all institutes, final reporting was based on sequencing analyses. Furthermore, among the 169 mutations detected by the panel institutes in this study, 14 had not been reported before, underscoring that only sequencing-based assays will further enhance our knowledge about EGFR mutations. Considering the fact that well over 200 different EGFR mutations are reported meanwhile (and the vast majority being considered “activating”), a mutation-specific approach appears to be unsuited for diagnostic EGFR mutation analyses for technological reasons. Further evaluation by the panel of other aspects, such as costs, turnaround time, and quality management, failed to demonstrate favorable aspects of the current mutation-specific PCR testing. In conclusion, the panel emphasizes the disadvantages of the current mutation-specific PCR assays for the detection of EGFR mutations in NSCLC. Instead, the use of sequencing-based detection is recommended. Currently, the analysis of exons 19 and 21 is considered mandatory, but the compiled results of the panel institutes strongly suggest including the analysis of exons 18 and 20 into the diagnostic setting.
References
Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M (2009) Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361:947–957
Marchetti A, Felicioni L, Buttitta F, Tsao MS, Kamel-Reid S, Shepherd FA (2006) Assessing EGFR mutations. N Engl J Med 354:526–528
Sharma SV, Bell DW, Settleman J, Haber DA (2007) Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 7:169–1681
Somatic Mutations in Epidermal Growth Factor Receptor DataBase (2008) Department of Molecular Pathology and Translational Oncology at Metropolitan Hospital, Athens, Greece. www.somaticmutations-egfr.info/index.html
Bressac B, Kew M, Wands J, Ozturk M (1991) Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 350:429–431
Rosell R, Moran T, Queralt C et al (2009) Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 361:958–967
Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Jänne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV, Johnson BE (2009) Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res 15:5267–5273
Conflict of interest
Round-robin testing of the panel institutes was supported by Astra Zeneca.
Author information
Authors and Affiliations
Corresponding author
Additional information
R. Penzel, C. Sers, Y. Chen, U. Lehmann-Mühlenhoff, S. Merkelbach-Bruse, A. Jung, T. Kirchner, R. Büttner, H.H. Kreipe, I. Petersen, M. Dietel, P. Schirmacher are members of the German Panel for Mutation Testing in NSCLC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM. 1
(PDF 9.08 kb)
Rights and permissions
About this article
Cite this article
Penzel, R., Sers, C., Chen, Y. et al. EGFR mutation detection in NSCLC—assessment of diagnostic application and recommendations of the German Panel for Mutation Testing in NSCLC. Virchows Arch 458, 95–98 (2011). https://doi.org/10.1007/s00428-010-1000-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-010-1000-y