Abstract
The DEAD-box RNA helicase Vasa (Vas, also known as DDX4) is required for germ cell development. In Drosophila, analysis of hypomorphic mutations has implicated maternally expressed Vas in germ cell formation and posterior embryonic patterning. vas-null females, which rarely complete oogenesis, exhibit defects in mitotic progression of germline stem cells, Piwi-interacting RNA (piRNA)-mediated transposon silencing, and translation of Gurken (Grk), an EGFR ligand. The carboxy-terminal region of Vas orthologs throughout the animal kingdom consists of several acidic residues as well as an invariant tryptophan in the penultimate or ultimate position (Trp660 in Drosophila melanogaster). Using CRISPR/Cas9 gene editing, we made a substitution mutant in this residue. Replacing Trp660 by Glu (W660E) abolishes the ability of Vas to support germ cell formation and embryonic patterning and greatly reduces Vas activity in piRNA biogenesis, as measured by transposon silencing, and in activating Grk translation. A conservative substitution (W660F) has much milder phenotypic consequences. In addition, females expressing only a form of Vas in which the seven C-terminal amino acids were replaced with the corresponding residues from Belle (Bel, also known as DDX3) show defects in perinuclear nuage assembly and transposon silencing. Oogenesis in females expressing only the chimeric Vas arrests early; however, in a vas 1 background, in which early expression of endogenous Vas supports oogenesis, the chimeric protein supports posterior patterning and germ cell specification. These results indicate that the unique C-terminus of Vas is essential for its function in piRNA biogenesis and that the conserved Trp660 residue has an important functional role.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In many organisms, the DEAD-box helicase Vasa (Vas) is essential for specification of primordial germ cells (Castrillon et al. 2000; Ikenishi and Tanaka 2000; Knaut et al. 2000; Kuznicki et al. 2000; Tanaka et al. 2000; Voronina et al. 2008). In some metazoans, Vas orthologs are also involved in the function of somatic multipotent stem cells (Alié et al. 2011; Shibata et al. 1999; Yajima and Wessel 2015). In Drosophila, Vas accumulates in the pole plasm, a specialized cytoplasmic region at the posterior pole of the oocyte and embryo that contains determinants for germ cell specification and posterior patterning (Lasko and Ashburner 1990). In the absence of functional posterior-localized Vas, determinants of germ cell specification and posterior pattern such as oskar (osk) and nanos (nos) messenger RNAs (mRNAs) are not stably localized or effectively translated in the pole plasm, and the resulting embryos lack posterior pattern elements and germ cells (Hay et al. 1988; Lasko and Ashburner 1990; Wang et al. 1994). In addition, earlier in oogenesis, Vas regulates translation of Grk, an EGFR ligand also involved in patterning of the oocyte (Styhler et al. 1998; Tomancak et al. 1998). As a result, embryos produced by females homozygous for vas PH165, a null allele, have severe defects in dorsal appendage formation (Tomancak et al. 1998). vas PH165 females produce very few eggs, because of additional roles for Vas in the progression of germline mitotic divisions and in differentiation of germline stem cells (Liu et al. 2009; Pek and Kai 2011a). Vas interacts with eIF5B, a translation initiation factor required for ribosome subunit joining (Carrera et al. 2000), and this interaction is required for Vas-mediated translational activation of grk and mei-P26 mRNAs (Johnstone and Lasko 2004; Liu et al. 2009).
Germ cells in many animals contain a unique structure termed the nuage, which is a compartmentalized site for Piwi-interacting RNA (piRNA) processing. piRNAs are small RNAs that inhibit expression of transposon-encoded genes, thereby safeguarding genome integrity against deleterious effects of excessive mobility of transposable elements (Pek et al. 2012). piRNAs can also target mRNAs for translational repression and degradation (Barckmann et al. 2015; Rouget et al. 2010). In Drosophila, vas is essential both for nuage assembly and for piRNA biogenesis (Liang et al. 1994; Xiol et al. 2014; Zhang et al. 2012). vas is involved in the transfer of piRNA precursors across the nuclear envelope and is a component of an amplifier complex participating in the ping-pong cycle that produces piRNAs (Xiol et al. 2014; Zhang et al. 2012).
RNA helicases of the DEAD-box family, including Vas, have crucial roles in a wide range of cellular processes including ribosome biogenesis, translation initiation, pre-mRNA splicing, and mRNA decay (Jankowsky 2011). DEAD-box proteins bind and hydrolyze ATP and catalyze unwinding of secondary structures in RNA, promoting strand separation (Cordin et al. 2006; Jarmoskaite and Russell 2011; Luking et al. 1998). In the helicase core, these proteins are composed of two tandemly repeated RecA-like domains, which share a set of highly conserved motifs. Belle (Bel), which belongs to a subfamily including yeast Ded1p, Xenopus An3, mouse PL10, and human DDX3, is the closest paralog of Vas (Johnstone et al. 2005). These two proteins have analogous roles in germ line and soma. Vas interacts with Aubergine (Aub) and Spindle-E (SpnE) to promote mitotic chromosome segregation in the female germ line (Pek and Kai 2011a), while Bel executes a similar role in the somatic cells through interaction with components of the endogenous siRNA (endo-siRNA) pathway, such as Ago2 (Pek and Kai 2011b).
Aside from their conserved helicase domain, most DEAD-box proteins have flanking N- and C-terminal sequences, which vary significantly in their length and content. Most structural analyses have excluded these, often disordered, terminal sequences (Carmel and Matthews 2004; Caruthers et al. 2000; Kim et al. 1998; Korolev et al. 1997; Sengoku et al. 2006; Subramanya et al. 1996; Zhao et al. 2004); therefore, little is known about their contribution to the functions and structures of DEAD-box proteins. The C-terminal region of Vas contains several motifs conserved among Vas/DDX4 orthologs. Amino acids 636–646 in Drosophila melanogaster Vas are shared among some closely related Drosophila species and are required for precisely concentrating Vas in the pole plasm (Dehghani and Lasko 2015). More notably, the most C-terminal seven amino acids from even distant Vas orthologs are highly acidic. In Drosophila, deletion of the last seven amino acids (655–661), which include five acidic residues, results in severe defects in transposable element (TE) silencing, Grk translation, posterior patterning of embryos, and germ cell formation (Dehghani and Lasko 2015).
There is a highly conserved Trp (Trp660 in D. melanogaster), which in Vas orthologs is usually the penultimate amino acid but occasionally is found at the absolute C-terminus. This conserved Trp is specific to Vas/DDX4 and is absent from the sequences of most other DEAD-box proteins in Drosophila and beyond. Thus, it seems unlikely that this residue contributes to the general helicase activity of Vas orthologs; rather, it might be involved in specific interactions between Vas and the proteins or RNAs that interact with it. To gain insight into Vas functions that are dependent on Trp660, we used the CRISPR-Cas9 system to mutate this residue in endogenous vas and expressed enhanced green fluorescent protein (eGFP)-tagged Trp660-mutated Vas from transgenes, to study the consequences of these mutations in vivo. In addition, we investigated which functions of Vas could be supported by a chimeric Vas-Bel protein containing the seven C-terminal residues of Bel. Finally, we explored whether any known protein-protein interactions involving Vas were dependent on the C-terminal domain or on Trp660 specifically.
Materials and methods
Endogenous vas alleles
vas PH165 is a null allele in which the entire coding sequence of vas has been deleted (Styhler et al. 1998). vas 1 is a hypomorphic allele, generated by ethyl methanesulfonate (EMS) mutagenesis (Schüpbach and Wieschaus 1986). This allele does not carry any mutation in the coding sequence of vas; however, expression of Vas in the ovaries is undetectable after the germarium stage (Lasko and Ashburner 1990; Liang et al. 1994).
vas W660E was generated using CRISPR-Cas9-mediated homologous recombination according to the method described by Port et al. (2014). A stable transgenic line expressing the guide RNA (gRNA), targeting a sequence including the Trp660 codon (AGCAATGGGATTGAAATGTA), was generated. The transgenic males were then crossed to nos-cas9 females, and their embryos were used for injection of a donor DNA encoding the W660E mutation. The donor DNA was composed of a synthetic ssDNA oligonucleotide (Integrated DNA Technologies, Inc.) corresponding to 80 base pairs on either side of Trp660 codon and substituting that with GAA (Glu). The PAM sequence was mutated from TGG to TGA in the donor DNA to prevent its degradation by Cas9 in the cells.
Transgenic vas alleles
egfp-vas + and egfp-vas Δ15–75 have been described previously (Dehghani and Lasko 2015). egfp-vas W660E, egfp-vas W660F, and egfp-vas c.bel were generated by a PCR-based mutagenesis approach from a full-length vas cDNA.
In vivo functional assays
Functional assays including egg-laying and hatching tests as well as the statistical analysis of the data were conducted as described in Dehghani and Lasko (2015). Immunostaining, in situ hybridization, and reverse transcription quantitative PCR (RT-qPCR) tests were also described in Dehghani and Lasko (2015).
Yeast two-hybrid assays
The Matchmaker Gold Yeast Two-Hybrid System (Clontech Inc.) was used to test direct interactions between vas and other candidate genes. The coding sequences of vas +, vas Δ655–661, and vas W660E were cloned between NdeI and PstI restriction sites in the pGBKT7 vector. The entire coding sequences of gus, aret, Dynein light intermediate chain (Dlic), fsn, piwi, barr, cap-D2, spn-E, aub, ago3, csul, and vls were amplified and inserted between NdeI and BamHI restriction sites in the pGADT7 AD vector. For osk short isoform, eIF5B and qin, fragments encoding amino acids 5–468, 491–1144, and 275–1857, respectively, were cloned. Media preparation, yeast handling, and interaction assays were performed according to the manufacturer’s instructions.
Results
Vas orthologs from different species contain an invariant tryptophan in their C-terminal region
Comparison among amino acid sequences from different Vas orthologs, as far apart as sea urchins and humans, reveals that a Trp residue (W660 in Drosophila) is highly conserved within the last two amino acids of these sequences (Fig. 1a). This Trp is always flanked by several acidic residues; for example, in D. melanogaster, there are four Glu and one Asp residues preceding and following the conserved Trp, respectively. Such a conserved Trp, however, is rarely found among DEAD-box proteins other than Vas orthologs (Fig. 1b). In D. melanogaster, for example, Bel, which has the highest sequence similarity to Vas, is the only other DEAD-box protein that contains Trp in its C-terminus. Rm62 contains a phenylalanine (Phe; F), another aromatic amino acid, in a similar position. However, this Phe residue is not conserved across species; for example, DDX5 (P68), the human or mouse orthologs of Rm62, do not carry a Phe within the last five amino acids.
Vas orthologs share a conserved tryptophan (W) in their C-terminal end, which is not common between the other DEAD-box proteins. a Sequence alignment of the last 15 amino acids in Vas orthologs from different species indicates an invariant Trp in the penultimate or ultimate position. Acidic residues, shown in red, are also abundant in the C-terminus of Vas orthologs. b The last 15 amino acids in the C-terminal end of different DEAD-box proteins in Drosophila melanogaster are compared. This list includes at least one representative from each main group of DEAD-box proteins. The names of the human orthologs are shown in parentheses. Bel is the only other DEAD-box protein containing two consecutive Trp residues in its C-terminus, which also exhibits the highest sequence homology to Vas
Substitution of Trp660 with Glu abolishes Vas function in germ cell formation and embryonic patterning
Using CRISPR/Cas9 technology, we generated an endogenous allele of vas, vas W660E, in which Trp660 is replaced by Glu. We also generated a transgenic allele of vas W660E, which encodes a protein N-terminally tagged with eGFP, to examine how the mutant protein localizes in ovaries. Similarly, we constructed an egfp-vas W660F allele which encodes Phe as a conservative substitution for Trp660. All the mutations were confirmed through amplifying and sequencing the genomic DNA. Expression of these proteins was confirmed by western blots (Fig. 2b).
Substitution of tryptophan (W) with glutamic acid (E), which is an abundant residue in the C-terminus of Vas, is associated with severely reduced germ cell formation and impaired posterior patterning. a The percentage of stage 5–6 embryos with pole cells is compared between different genotypes carrying endogenous wild-type, vas W660E, and vas PH165 alleles (red bars) or between transgenic alleles of egfp-vas Δ15–75 (positive control), egfp-vas W660E, and egfp-vas W660F expressed in vas 1 background (green bars). One, two, or three asterisks represent P < 0.05, <0.005, or <0.0005, respectively. b The top western blot indicates expression of Vas in ovaries carrying different endogenous alleles, corresponding to the red bars in a. The bottom western blot compares expression levels of the egfp− alleles. α-Tubulin (α-Tub), Coomassie Blue staining, and endogenous (end.) Vas are used as the loading controls. c W660E, but not W660F, significantly reduces the average number of ftz stripes (illustrated on the right) in the embryos produced by the mutant females, reflecting compromised posterior patterning. Scale bar indicates 50 μm
We investigated the significance of Trp660 for Vas function in germ cell development by counting the proportion of stage 5–6 embryos from vas W660E homozygous females with pole cells. All embryos from females with two copies of wild-type vas (OrR) produce pole cells, which in number vary around an average of 34.5 (Fig. 2a). In contrast, we found that only 18 % of embryos produced by vas W660E homozygous females (subsequently referred to by the maternal genotype) form pole cells with an average of only 1.7 pole cells per embryo (n = 83). To confirm that this is specifically associated with the vas W660E mutation and to rule out second-site genetic effects, we examined vas W660E /vas PH165 embryos and observed a similarly significant decrease in the number of embryos producing pole cells compared to +/vas PH165 (7 % with pole cells, average of 0.24 pole cells per embryo, P = 5.7E−05).
We next investigated if Phe, a different hydrophobic amino acid with an aromatic ring, could replace Trp in position 660 of Vas. To address this question, we used an egfp-vas W660F transgene to rescue germ cell formation in a vas 1 background. We compared this construct with egfp-vas ∆15–75, as a positive control, since both lines expressed eGFP-Vas at similar levels (Fig. 2b) and it has been shown previously that egfp-vas ∆15–75 mimics full-length vas in these in vivo functional assays (Dehghani and Lasko 2015). Our analyses indicate that the average number of vas 1; egfp-vas W660F embryos with pole cells is 82 %, which is significantly different from only 0.5 % of vas 1; egfp-vas W660E embryos that formed pole cells (P = 4.0E−04; Fig. 2a). In contrast, egfp-vas W660F was much closer to, if still significantly lower than, egfp-vas ∆15–75 control (94 %) for its ability to induce germ cells in vas 1 embryos (p = 0.01). The average number of pole cells per embryo was 8.2 (n = 127), 0.005 (n = 393), and 9.3 (n = 90) for egfp-vas W660F, egfp-vas W660E, and egfp-vas ∆15–75, respectively.
To investigate the effects of Vas660 mutations on embryonic patterning, we examined the expression pattern of fushi tarazu (ftz), a marker for segmentation, in vas W660E embryos (Fig. 2c) (Hafen et al. 1984) . The average number of ftz stripes in stage 5–6 vas W660E embryos was 5.4, which is significantly lower than the average of 6.7 stripes in wt (P = 0.001). Our analysis using egfp transgenic alleles also indicates that unlike the W660E mutation, which decreased the number of ftz stripes from 6.5 to 5 (P = 5.5E−12), W660F did not have a significant effect on posterior patterning. We also tested the viability of vas W660E homozygous and vas W660E /vas PH165 heterozygous embryos. Our analyses show that no more than 1 % of the embryos from either of these two groups are viable, whereas +/+ or +/vas PH165 embryos have a hatching rate of 90 and 60 %, respectively (n > 10,000). We also found that the majority of vas W660E or vas W660E /vas PH165 eggs do not undergo any development, suggesting that vas W660E females have additional defects in oogenesis and many of their eggs are not fertilized.
W660E substitution perturbs stable localization of Vas in the nuage and pole plasm
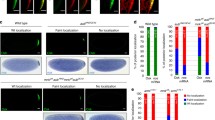
In early stages of oogenesis, Vas normally localizes to the perinuclear region of the nurse cells, also known as the nuage. At stage 10, Vas starts to be transported to the oocyte and accumulates at the pole plasm. To examine the effect of the W660E mutation on Vas localization, we used eGFP-tagged Vas fusions, as antibodies do not efficiently penetrate late-stage oocytes for immunostaining. Confocal images show that in the vas 1 background, eGFP-VasW660E not only partially localizes to the perinuclear nuage but also forms abundant cytoplasmic foci, which may indicate instability of the nuage structure (Fig. 3a). Accumulation of eGFP-VasW660E in the pole plasm is initially normal in the vas 1 background, but it is less stably maintained there, as the GFP signal was diminished in stage 14 oocytes compared to stage 13 (Fig. 3a). Consistent with this, immunostaining of vas W660E embryos shows that the majority (82 %) of stage 1–2 embryos contained no detectable Vas in their posterior region, while the others had considerably decreased levels compared to wt (n ≥ 250; Fig. 3b). In contrast, we found that, in a wild-type genetic background, eGFP-VasW660E localizes to the nuage of wild-type egg chambers comparably to eGFP-Vas+ (Fig. 3c) and persisted there stably throughout oogenesis.
eGFP-VasW660E accumulates in distinct cytoplasmic foci in the nurse cells of vas1 ovaries, and its localization to the pole plasm significantly reduces by the end of oogenesis. a eGFP-VasW660E is compared to eGFP-Vas+ in a vas1 background for its localization to the nuage (stage 3 egg chambers) and pole plasm of stage 9, 13, and 14 oocytes. Arrows indicate the cytoplasmic foci containing eGFP-VasW660E. Scale bars indicate 50 μm. b Vas immunostaining indicates posterior localization of Vas in all stage 1–2 wt (OrR) embryos, which in 70 % of the cases is strongly concentrated (n = 250). In contrast, only 18 % of vasW660E embryos maintain detectable levels of Vas at the posterior region (n = 545). DAPI staining indicates embryonic stages. c In the wt background, localization of eGFP-VasW660E mimics localization pattern of eGFP-Vas+. d, e Comparisons between transcript and protein levels of egfp-vas + and egfp-vas W660E (RT-qPCR and western blot) also indicate that VasW660E protein level is relatively stable in a vas 1 /+ background. The endogenous Vas and α-tubulin were used as the loading controls for western blot; Act5C and rp49 were used as the reference genes in RT-qPCR
We also investigated whether W660E affects protein stability by quantifying levels of egfp-vas + and egfp-vas W660E transcripts, using RT-qPCR, and comparing that with the protein levels on the western blot (Fig. 3d, e). These experiments indicate that the level of egfp-vas W660E transcript is about two times higher than egfp-vas + in the corresponding transgenic lines which were tested. As western blotting also shows a relatively high level of eGFP-VasW660E protein in the ovaries we conclude that the mutation does not have major effects on protein stability.
Mutation of Trp660 to Glu reduces Vas function in piRNA biogenesis as measured by HeT-A silencing
Vas is required for nuage assembly, and its critical role in piRNA biogenesis has been confirmed both in Drosophila and in mammals. Females homozygous for a vas-null allele, vas PH165, overexpress transcripts from several TEs including HeT-A (Zhang et al. 2012). Elevated levels of HeT-A transcripts also have been reported for most vas alleles with mutations in their conserved helicase and C-terminal domains (Dehghani and Lasko 2015). To investigate whether substitution of Trp660 with Glu affects Vas function in the piRNA pathway, and therefore transposon silencing, we compared levels of HeT-A transcript in vas W660E ovaries with wild type and found a more than tenfold increase in the mutant (P = 0.006; Fig. 4). HeT-A levels in vas W660E fall within the same range as those observed for females carrying mutations in the conserved helicase motifs of Vas or deletion of the seven C-terminal amino acids (Dehghani and Lasko 2015).
Substitution of Trp660 with Glu impairs Vas function in transposable element silencing. The retrotransposon, HeT-A, is dramatically overexpressed in vas PH165 ovaries compared to wild-type (OrR). A consistent overexpression, albeit to a lesser extent than in vas PH165, is also observed in vas W660E ovaries. egfp-vas Δ15–75 and egfp-vas W660F are able to fully suppress HeT-A overexpression in vas PH165 ovaries. On the contrary, ovaries expressing egfp-vas W660E still exhibit a significant overexpression of HeT-A. HeT-A levels in different genotypes are normalized to the wild-type level. pre-rp49 and 18 S-rRNA are used as the reference genes. Each bar represents the average from at least three biological replicates. Error bars indicate SEM
We confirmed this result by measuring the levels of HeT-A in ovaries of vas PH165 females expressing either eGFP-Vas+ or eGFP-VasW660E. In contrast to the wt construct which is able to fully suppress HeT-A overexpression, vas PH165 ovaries expressing egfp-vas W660E exhibit increased levels of HeT-A at about ten times higher than wt (P = 0.005). The W660F mutation, on the other hand, did not significantly affect the ability of Vas to suppress HeT-A overexpression in the vas PH165 background.
vas W660E females produce a similar number of eggs to wild type, but these eggs have dorsal appendage defects
Vas has a critical role in oogenesis by regulating multiple processes including germline mitotic divisions, stem cell differentiation, and dorsal ventral patterning of the oocytes (Liu et al. 2009; Pek and Kai 2011a; Tomancak et al. 1998). In vas PH165, females all of these pathways are perturbed; these mutants produce very few eggs and their eggs do not form normal dorsal appendages. In addition, our previous analyses indicate that several mutations in the conserved motifs of Vas that are critical for its enzymatic activity do not abolish female fecundity, even though these residues are indispensable for other functions of Vas such as piRNA biogenesis or germ cell formation (Dehghani and Lasko 2015).
To examine if the W660E mutation, in addition to its effects on germ cell formation, embryonic patterning, and transposon silencing, affects female fecundity, we counted the number of eggs laid by individual females within the first 3 days from eclosion (Fig. 5a). vas W660E homozygous or hemizygous (over vas PH165) produces eggs in numbers comparable to wild-type females. Expression of either egfp-vas W660E or egfp-vas W660F in the vas PH165 background also restored fecundity, although rescue by egfp-vas W660E was not as robust as egfp-vas W660F and the control. We conclude that the W660E mutation has little effect on female fecundity.
vas W660E females produce a wild-type number of embryos. This mutation, however, has a significant effect on Grk expression and dorsal appendage formation. a Females carrying two copies of wild-type vas (OrR), two copies of vas W660E, or only one copy of vas W660E (over vas PH165) produce a comparable number of embryos (red bars). Similarly, expression of either egfp-vas W660E or egfp-vas W660F could largely restore female fecundity to vas PH165, although egfp-vas W660E still had significantly lower fecundity compared to egfp-vas Δ15–75 control. b The percentage of embryos with two separate or semi-fused dorsal appendages is significantly reduced in vas W660E compared to wild-type; this effect is even more pronounced when females are hemizygous for vas W660E (over vas PH165). Expression of egfp-vas Δ15–75 or egfp-vas W660F increases the number of embryos with two dorsal appendages from 0 % in vas PH165 to 68 or 84 %, respectively. On the contrary, such embryos produced from vas PH165; egfp-vas W660E females comprise only 34 % of the total. n (the number of females tested) > 20; error bars represent SEM. c Representative images of Grk immunostaining in stage 7–8 egg chambers of homozygous vas W660E compared to OrR (left) and vas PH165 egg chambers expressing egfp-vas Δ15–75, egfp-vas W660F, or egfp-vas W660E (right). Scale bar indicates 50 μm
We, however, observed that 23 % of eggs from vas W660E homozygous females had only a single fused dorsal appendage, a phenotype that we did not observe in wild-type eggs (P = 1.7E−08; Fig. 5b). This phenotype was aggravated in hemizygous vas W660E/vas PH165 females in that 74 % of the eggs had only one dorsal appendage versus 0.2 % in +/vas PH165 (P = 1.3E−20). This indicates that VasW660E has a significantly decreased activity in establishing the dorsal-ventral axis. Again, our analysis using egfp transgenic alleles confirmed these results and as well showed that the conservative substitution, W660F, does not reduce Vas activity in dorsal-ventral patterning. To determine whether these effects were correlated with decreased Grk expression, we immunostained ovaries with an antiserum recognizing Grk. This experiment showed that the defects in dorsal appendages of vas W660E or vas PH165; egfp-vas W660E embryos are indeed correlated with decreased amounts of Grk in stage 7–8 oocytes (Fig. 5c). The dorsal appendage defects observed in vas W660E embryos could also account for the large number of these embryos which remain unfertilized.
Direct binding of Vas to its known interacting partners is independent of Trp660
Previous studies, using different in vitro and in vivo assays, have identified a number of proteins that are associated with Vas in ribonucleoprotein complexes (Table S1). To investigate possible correlations between these protein interactions and the functional defects observed for vas W660E and vas ∆655–661 (Dehghani and Lasko 2015), we tried to co-immunoprecipitate Vas-protein complexes from ovaries or embryos. With this method, however, known interacting partners of Vas, such as Aubergine (Aub), Oskar (Osk), and Valois (Vls), were not detected in the IP eluates from ovaries or embryos expressing either eGFP-Vas or endogenous Vas. We reasoned that this might be due to weak or transient interactions or to low abundance or instability of these proteins in the whole ovary/embryo lysates. As an alternative, we used a yeast two-hybrid assay to compare VasW660E and Vas∆655–661 with wild-type Vas for their interactions with 16 other proteins that had been identified as Vas interactors by various means in other studies. Our experiments confirmed interactions between Vas and Osk, Gus, and eIF5B in this assay, which have been previously reported (Breitwieser et al. 1996; Carrera et al. 2000; Styhler et al. 2002). These interactions remained unchanged for VasW660E and Vas∆655–661 (Fig. S1). For the other 13, which were mostly tested as full-length proteins, we did not find an interaction with Vas as measured by this assay.
The Bel C-terminus cannot substitute for the corresponding region of Vas
Among other DEAD-box proteins, Bel has the highest sequence similarity to Vas, and its C-terminal end contains two conserved Trp residues in its third and fourth from last positions. Given the significance of the C-terminal domain and its conserved Trp660 for several functions of Vas, we explored whether the last seven amino acids of Bel could replace those from Vas by testing an eGFP-Vasc.bel chimeric protein (Fig. 6a, b).
Replacing the last seven amino acids of Vas with the corresponding residues from Bel results in complete oogenesis failure; however, expression of this protein in the embryos from vas 1 females significantly rescues abdominal patterning. a The last 15 amino acids of Vasc.bel composed of fragments from Vas and Bel. b Expression level of eGFP-Vasc.bel is compared to eGFP-VasΔ15–75, as a positive control used in the subsequent experiments. End. Vas and Actin serve as the loading controls. c In contrast to its normal localization to the nuage in a wild-type background, eGFP-Vasc.bel expressed in vas 1 ovaries accumulates in perinuclear and cytoplasmic foci. Localization of this protein to pole plasm, however, is maintained in fully developed oocytes (stage 14) in both wild-type and vas 1 backgrounds. Scale bar indicates 50 μm. d eGFP-Vasc.bel fails to support oogenesis progression. e Unlike vas Δ15–75 control, which fully represses HeT-A overexpression in vas PH165 ovaries, expression of egfp.vas c bel does not restore transposon silencing. f Surprisingly, abdominal patterning is significantly restored upon expression of egfp.vas c.bel in vas 1 embryos. About 15 % of these embryos also produced germ cells (g). One, two, or three asterisks indicate P < 0.05, <0.005, or <0.0005, respectively
Our experiments using this construct show that, similar to eGFP-VasW660E, in the vas 1 background, eGFP-Vasc.bel exhibits a granular localization in the nuage as opposed to a smooth perinuclear ring in the wt background (Fig. 6c). However, unlike eGFP-VasW660E, localization of eGFP-Vasc.bel in the pole plasm of vas 1 oocytes is maintained until stage 14. Surprisingly, eGFP-Vasc.bel failed to restore any fecundity in vas PH165 females, which is usually at least partially rescued by different Vas mutants (Fig. 6d; Dehghani and Lasko 2015). As well, the few embryos produced by vas PH165; egfp-vas c.bel females, like vas null, exhibited severe defects in dorsal-ventral patterning (data not shown). We also observed that vas PH165 ovaries expressing eGFP-Vasc.bel transcribe high levels of HeT-A similar to vas null (Fig. 6e), which is consistent with the defects in localization of this mutant protein to the nuage, and indicates defects in piRNA biogenesis.
Despite its failure to support oogenesis in vas PH165, when expressed in vas 1 background, eGFP-Vasc.bel could increase the average number of ftz stripes from 3.4 to 5.5 (P = 1.8E−10; Fig. 6f). These numbers still remained significantly lower than the average number of ftz stripes supported by egfp-vas Δ15–75 control; nevertheless, about 10 % of these embryos hatched (n = 5000). Interestingly, 15 % of vas 1; egfp-vas c.bel embryos also produced germ cells ranging in number between 2 and 15 (n = 480 embryos). These results indicate that replacement of the seven C-terminal residues of Vas with those of Bel has its most profound effects during oogenesis and specifically on piRNA biogenesis, while Vas functions in events that occur later in development, namely posterior patterning and germ cell specification, are less severely affected.
Discussion
Our previous analysis revealed that the most C-terminal seven amino acids (655–661) of Vas are critical for many of its developmental functions (Dehghani and Lasko 2015). In the current study, we found that a single missense mutation, W660E, behaves similarly to the deletion of motif 655–661, in that it severely compromises most functions of Vas including germ cell specification, oocyte axis determination, and transposon silencing. This mutation alone, however, did not decrease the number of eggs produced by females, implying that the acidic residues that are adjacent to the conserved Trp are essential for oogenesis. In addition, we found that the C-terminal domain cannot be replaced with terminal seven amino acids from Bel, which contains Trp residues but is otherwise different from Vas.
The vas W660E phenotype is intermediate between vas 1 and vas-null
In this study, for the first time, we edited the endogenous allele of vas, and this allowed us to examine the effect of a point mutation by direct comparisons with wild type. Our analyses indicate that vas W660E resembles vas 1 in that females produce a normal number of embryos, but these embryos do not form germ cells and are not viable because of posterior patterning defects. However, vas W660E ovaries resemble vas-null ovaries in that they produce high levels of HeT-A transcripts, indicating that piRNA biogenesis is compromised. Also, like vas-null mutations, vas W660E oocytes display defects in their dorsal appendages, suggesting that this mutant form of Vas does not optimally activate Grk translation. In addition, vas W660E females produce more eggs than do females expressing only a catalytically dead form of Vas (Dehghani and Lasko 2015), which suggests that VasW660E still retains some enzymatic activity.
A vas transgene encoding Phe instead of Trp660 supports Vas functions
Our analysis of egfp-vas W660F indicates that Vas functions in oogenesis and germ cell formation generally could tolerate this conservative substitution. This was surprising since Trp660 has remained invariant through evolution. It remains possible that W660F results in small changes in Vas functions that reduce fitness in natural environments. An endogenous allele of vas W660F would facilitate more accurate quantitative analyses to investigate this.
The nature of interactions between Trp660 and other parts of Vas and its possible role in protein conformation remain to be investigated through crystal structure analyses that would include the C-terminal region. We observed, however, that the non-conservative substitution to Glu, despite its severe effects on most Vas functions, had mild or no effect on female fecundity, suggesting that the mutant protein still retains a conformation that supports some function. Yet, it is possible that a subtle change in Vas folding affects certain functions more than the others.
The acidic C-terminus of Vas is distinct among DEAD-box proteins and essential for its function
The closest paralog of Vas, Bel, in its core region shows 68 % sequence similarity to Vas, and its long glycine-rich N-terminus, like Vas, contains multiple RGG motifs. The conserved helicase core in Bel is followed by a C-terminal domain, about three times longer than Vas C-terminus, which exhibits some sequence similarity to Vas toward the end, including the presence of two Trp residues. Phylogenetic analyses suggest that vas-like genes emerged from PL10-like genes, including bel, through a gene duplication occurring after diversion of plants but before appearance of sponges (Mochizuki et al. 2001). In Drosophila, strong bel alleles are recessive lethal, consistent with the essential role of Bel in mitotic chromosome segregation in somatic cells (Johnstone et al. 2005; Pek and Kai 2011b). A similar function in the germline is carried out by Vas (Pek and Kai 2011a). Bel is also expressed in Drosophila ovaries and is required for female fertility (Johnstone et al. 2005), but most likely in different pathways than Vas, as we found that overexpression of Bel in vas PH165 ovaries does not rescue any oogenesis defects (data not shown).
vas c.bel is among the very few non-null alleles of vas that completely abolish female fecundity (Dehghani and Lasko 2015; Liang et al. 1994; Tomancak et al. 1998). In contrast to severe defects in oogenesis progression and transposon silencing, Vasc.bel supports many of the later functions of Vas. It localizes in the pole plasm of late-stage oocytes and early embryos, and it even partially supports posterior patterning and germ cell formation. This raises the possibility that the acidic motif is involved in specific interactions with the components of piRNA pathway. The divergent C-terminal regions of Bel and Vas could explain why Bel interacts with components of the siRNA pathway while Vas interacts with components of the piRNA pathway. Further analysis of protein-protein interactions involving Vas and Bel, and of how these interactions are affected by C-terminal mutations, will be required to test this hypothesis.
References
Alié A, Leclere L, Jager M, Dayraud C, Chang P, Le Guyader H, Queinnec E, Manuel M (2011) Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Dev Biol 350:183–197
Anand A, Kai T (2012) The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J 31:870–882
Anne J (2010) Targeting and anchoring Tudor in the pole plasm of the Drosophila oocyte. PLoS One 5:e14362
Barckmann B, Pierson S, Dufourt J, Papin C, Armenise C, Port F, Grentzinger T, Chambeyron S, Baronian G, Desvignes JP, Curk T, Simonelig M (2015) Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep 12:1205–1216
Breitwieser W, Markussen FH, Horstmann H, Ephrussi A (1996) Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev 10:2179–2188
Carmel AB, Matthews BW (2004) Crystal structure of the BstDEAD N-terminal domain: a novel DEAD protein from Bacillus stearothermophilus. RNA 10:66–74
Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P (2000) Vasa mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell 5:181–187
Caruthers JM, Johnson ER, McKay DB (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A 97:13080–13085
Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP (2000) The human vasa gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci USA 97:9585–9590
Cordin O, Banroques J, Tanner NK, Linder P (2006) The DEAD-box protein family of RNA helicases. Gene 367:17–37
Dehghani M, Lasko P (2015) In vivo mapping of the functional regions of the DEAD-box helicase Vasa. Biol Open 4:450–U457
Hafen E, Kuroiwa A, Gehring WJ (1984) Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell 37(3):833–841
Hay B, Ackerman L, Barbel S, Jan LY, Jan YN (1988) Identification of a component of Drosophila polar granules. Development 103:625–640
Ikenishi K, Tanaka TS (2000) Spatio-temporal expression of Xenopus vasa homolog, XVLG1, in oocytes and embryos: the presence of XVLG1 RNA in somatic cells as well as germline cells. Dev Growth Differ 42:95–103
Jankowsky E (2011) RNA helicases at work: binding and rearranging. Trends Biochem Sci 36:19–29
Jarmoskaite I, Russell R (2011) DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA 2:135–152
Johnstone O, Lasko P (2004) Interaction with eIF5B is essential for Vasa function during development. Development 131:4167–4178
Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P (2005) Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol 277:92–101
Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89–100
Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C (2000) Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol 149:875–888
Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G (1997) Major domain swiveling revealed by the crystal structures of complexes of E-coli Rep helicase bound to single-stranded DNA and ADP. Cell 90:635–647
Kugler JM, Woo JS, Oh BH, Lasko P (2010) Regulation of Drosophila Vasa in vivo through paralogous cullin-RING E3 ligase specificity receptors. Mol Cell Biol 30:1769–1782
Kuznicki KA, Smith PA, Leung-Chiu WMA, Estevez AO, Scott HC, Bennett KL (2000) Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C-elegans. Development 127:2907–2916
Lasko P, Ashburner M (1990) Posterior localization of Vasa protein correlates with, but is not sufficient for pole cell development. Genes Dev 4:905–921
Lerit DA, Gavis ER (2011) Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr Biol 21:439–448
Liang L, Diehljones W, Lasko P (1994) Localization of Vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120:1201–1211
Liu NK, Han H, Lasko P (2009) Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev 23:2742–2752
Luking A, Stahl U, Schmidt U (1998) The protein family of RNA helicases. Crit Rev Biochem Mol Biol 33:259–296
Megosh HB, Cox DN, Campbell C, Lin H (2006) The role of Piwi and the miRNA machinery in Drosophila germline determination. Curr Biol 16:1884–1894
Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T (2001) Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol 211:299–308
Patil VS, Kai T (2010) Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 20:724–730
Pek JW, Kai T (2011a) A role for Vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol 21:39–44
Pek JW, Kai T (2011b) DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci U S A 108:12007–12012
Pek JW, Patil VS, Kai T (2012) piRNA pathway and the potential processing site, the nuage, in the Drosophila germline. Dev Growth Differ 54:66–77
Port F, Chen HM, Lee T, Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A 111:E2967–E2976
Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467:1128–1132
Schüpbach T, Wieschaus E (1986) Maternal-effect mutations altering the anterior-posterior of the Drosophila embryo. Rouxs Arch Dev Biol 195:302–317
Sengoku T, Nureki O, Nakamura A, Satoru KI, Yokoyama S (2006) Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell 125:287–300
Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K (1999) Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol 206:73–87
Styhler S, Nakamura A, Swan A, Suter B, Lasko P (1998) vasa is required for Gurken accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125:1569–1578
Styhler S, Nakamura A, Lasko P (2002) Vasa localization requires the SPRY-domain and SOCS-box containing protein, Gustavus. Dev Cell 3:865–876
Subramanya HS, Bird LE, Brannigan JA, Wigley DB (1996) Crystal structure of a DExx box DNA helicase. Nature 384:379–383
Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T (2000) The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 14:841–853
Tomancak P, Guichet A, Zavorszky P, Ephrussi A (1998) Oocyte polarity depends on regulation of gurken by Vasa. Development 125:1723–1732
Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G (2008) Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol 314:276–286
Wang C, Dickinson LK, Lehmann R (1994) Genetics of Nanos localization in Drosophila. Dev Dynam 199:103–115
Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM (1997) Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev 11:2510–2521
Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Coute Y, Conn S, Kadlec J, Sachidanandam R et al (2014) RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157:1698–1711
Yajima M, Wessel GM (2015) Essential elements for translation: the germline factor Vasa functions broadly in somatic cells. Development 142:1960–1970
Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD et al (2012) UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151:871–884
Zhao R, Shen JP, Green MR, MacMorris M, Blumenthal T (2004) Crystal structure of UAP56, a DExD/H-Box protein involved in pre-mRNA splicing and mRNA export. Structure 12:1373–1381
Acknowledgments
We are grateful to Beili Hu for microinjection into the embryos. All the images were taken using the Cellular Imaging and Analysis Network (CIAN) facility at McGill University. We would also like to thank Fillip Port for his technical support during CRISPR gene editing and for sharing some of his unpublished observations. This work was supported by NSERC Discovery grant RGPIN-2014-06340 to P. L.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Angelika Stollewerk
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Interactions of Vas with Osk, Gus and eIF5B are not abolished by ∆655-661 or W660E. A. A β-Galactosidase filter assay shows similar results for vas +, vas ∆655-661 and vas W660E in terms of interactions with Osk, Gus and eIF5B. B. Positive interactions would enable yeast to grow on the quadruple drop-out (-Trp, -Leu, -His and –Ade) media. This test also confirmed that the interactions with Osk, Gus and eIF5B are not notably different among vas +, vas ∆655-661 and vas W660E. (GIF 169 kb)
Table S1
The list of the proteins that have been previously found to associate with Vas through different assays, including yeast two-hybrid (Y2H), GST pull-down or co-immunoprecipitation (co-IP). The latter has been performed using the endogenous or tagged proteins from Drosophila ovaries or Bombyx mori BmN4 cell lines. We expressed each one of these candidates in yeast, either as a full length protein or a fragment. Our Y2H assays only showed a direct interaction between full-length Vas and Osk, Gus and eIF5B, consistent with the previous studies. 1: (Breitwieser et al. 1996), 2: (Anne 2010), 3: (Styhler et al. 2002), 4: (Kugler et al. 2010), 5: (Carrera et al. 2000), 6: (Webster et al. 1997), 7: (Lerit and Gavis 2011), 8: (Patil and Kai 2010), 9: (Megosh et al. 2006), 10: (Pek and Kai 2011a), 11: (Anand and Kai 2012), 12: (Xiol et al. 2014) (GIF 78 kb)
Rights and permissions
About this article
Cite this article
Dehghani, M., Lasko, P. C-terminal residues specific to Vasa among DEAD-box helicases are required for its functions in piRNA biogenesis and embryonic patterning. Dev Genes Evol 226, 401–412 (2016). https://doi.org/10.1007/s00427-016-0560-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-016-0560-5