Abstract
Whole genome sequence analyses allow unravelling such evolutionary consequences of meso-triplication event in Brassicaceae (∼14–20 million years ago (MYA)) as differential gene fractionation and diversification in homeologous sub-genomes. This study presents a simple gene-centric approach involving microsynteny and natural genetic variation analysis for understanding SUPPRESSOR of OVEREXPRESSION of CONSTANS 1 (SOC1) homeolog evolution in Brassica. Analysis of microsynteny in Brassica rapa homeologous regions containing SOC1 revealed differential gene fractionation correlating to reported fractionation status of sub-genomes of origin, viz. least fractionated (LF), moderately fractionated 1 (MF1) and most fractionated (MF2), respectively. Screening 18 cultivars of 6 Brassica species led to the identification of 8 genomic and 27 transcript variants of SOC1, including splice-forms. Co-occurrence of both interrupted and intronless SOC1 genes was detected in few Brassica species. In silico analysis characterised Brassica SOC1 as MADS intervening, K-box, C-terminal (MIKCC) transcription factor, with highly conserved MADS and I domains relative to K-box and C-terminal domain. Phylogenetic analyses and multiple sequence alignments depicting shared pattern of silent/non-silent mutations assigned Brassica SOC1 homologs into groups based on shared diploid base genome. In addition, a sub-genome structure in uncharacterised Brassica genomes was inferred. Expression analysis of putative MF2 and LF (Brassica diploid base genome A (AA)) sub-genome-specific SOC1 homeologs of Brassica juncea revealed near identical expression pattern. However, MF2-specific homeolog exhibited significantly higher expression implying regulatory diversification. In conclusion, evidence for polyploidy-induced sequence and regulatory evolution in Brassica SOC1 is being presented wherein differential homeolog expression is implied in functional diversification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SUPPRESSOR of OVEREXPRESSION of CONSTANS 1 (SOC1) is a MADS-box type II (MIKCC) transcription factor (Lee and Lee 2010) characterised in plants. In angiosperms, SOC1 plays a crucial role in flowering time control by integrating floral input pathways (Hepworth et al. 2002). Overexpression of SOC1 hastens progression from vegetative to reproductive phase in monocots (Papaefthimiou et al. 2012), dicots (Zhong et al. 2012) and gymnosperms (Katahata et al. 2014), while downregulation of SOC1 reverses the effect (Lee et al. 2008). SOC1 is regulated by FLOWERING LOCUS T (FT), CONSTANS 1 and FLOWERING LOCUS C (FLC; Helliwell et al. 2006). SOC1 is also involved in a cross-regulatory loop with AGAMOUS-LIKE24 gene (AGL24) (Liu et al. 2008). Apart from floral transition, SOC1 mediates a variety of other biological processes in angiosperms such as floral organ senescence (Citrus sinensis, Tan and Swain 2007), floral organ identity and petal development (Gerbera hybrida, Ruokolainen et al. 2011), pathogen response (Glycine max, Sá et al. 2012), GA biosynthesis (Fragaria vesca, Mouhu et al. 2013), seed development (Hordeum vulgare, Papaefthimiou et al. 2012) and floral meristem development (Dendrobium Chao Parya Smile, Ding et al. 2013). Till date, SOC1 has not been characterised from Brassicaceae, even though it includes a large number of economically important species. Members of Brassicaceae demonstrate immense natural variation in flowering time and other life history traits.
The morphological diversity in Brassicaceae (Redden et al. 2009), comprising circa 330 genera and 3700 species, is attributed to numerous polyploidization events in the course of evolutionary history (Paterson et al. 2004; Pires and Gaeta 2011). In fact, ancestry of Brassicas traces back to angiosperm evolution and more specifically to a paleo-hexaploid ancestor that resulted from an ancient whole genome triplication event (γ) (Lysak and Koch 2011). Subsequently, angiosperm families underwent two rounds of whole genome duplications (WGDs), β and α, with the latter specific to Brassicaceae (Schranz and Mitchell-Olds 2006). The divergence of Arabidopsis lineage from Brassica circa 20–24 million years ago (MYA) (Ziolkowski et al. 2006; 14.5-20 MYA, Navabi et al. 2013) was followed by another whole genome triplication event, b, specific to Brassiceae (Lysak and Koch 2011). Repeated rounds of WGDs have caused considerable gene and genome redundancy in Brassica species (Mun et al. 2009). More recently (∼10,000 years ago (YA)), natural hybridizations of diploid species have added another layer of genome complexity in allotetraploid species (U 1935; Parkin et al. 2005). Whole genome sequence analysis of plant genomes provides evidence for differential gene fractionation within sub-genomes manifested after polyploidisation events. This evolutionary phenomenon facilitates gross genomes to re-compress to original size through large-scale gene deletions in sub-genome equivalents (Jaillon et al. 2009; Sankoff et al. 2010). For instance, recently sequenced Brassica rapa subsp. pekinensis cultivar (cv.) Chiifu-401 (The Brassica Rapa Genome Sequencing Project Consortium, Cheng et al. 2011; Wang et al. 2011) was found to contain three partially duplicated sub-genomes: least fractionated (LF), moderately fractionated (MF1) and most fractionated (MF2) sub-genomes. It is postulated that optimal gene copy number is determined by relative fitness conferred (Thomas et al. 2006). Differential expression levels of homeologous gene copies (Cheng et al. 2012) and methylation are also implicated in deciding the number of redundant gene copies (Keller and Soojin 2013).
Gene redundancy permits accumulation of mutations even in loci conferring functions that are normally conserved. Relieved of functional constraints, duplicated copies tend to undergo neo-functionalization, sub-functionalization or pseudogenisation (Sankoff et al. 2010; Cheng et al. 2012). Occasionally, mutations in regulatory sequences result in functional evolution of homeologs (Hahn 2009; Hughes et al. 2014). Brassica species are, hence, an interesting model to study polyploidy-associated genomic changes. In B. rapa, specific genes may be identified in up to three copy numbers (Cheng et al. 2011; Shivaraj et al. 2014). For instance, survey of the annotated B. rapa genome (Brassica database, BRAD) reveals three SOC1 copies, viz. Bra004928, Bra039324 and Bra000393, located in LF, MF1 and MF2 sub-genomes, respectively. Despite the criticality of role, genetic variation in Brassica homologs of SOC1 has not been described in Brassica species. The present study has thus been formulated to derive insights into SOC1 homeolog evolution in Brassicas in context to sequence and functional diversification.
Materials and methods
Plant growth conditions and nucleic acid isolation
Eighteen cultivars representing six species of Brassica (B. rapa, Brassica juncea, Brassica nigra, Brassica carinata, Brassica oleracea and Brassica napus) were grown under field conditions at TERI Gram, Haryana, India (October 2010–April 2011). Tissue samples were harvested and flash frozen in liquid nitrogen until further use. Total genomic DNA was isolated from leaf samples using modified protocol by Doyle and Doyle (1990). Total RNA from tissue samples was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA), as per manufacturers’ recommendations.
Generation of synteny maps
Sequences of three B. rapa SOC1 homeologs were retrieved from BRAD (www.brassicadb.org). In addition, genomic regions (Br-R1, Br-R2 and Br-R3, ∼300 kb) containing three B. rapa SOC1 copies were retrieved (Genome Browse, BRAD) and globally aligned (AVID, Bray et al. 2003) to Arabidopsis genome (sequence release March 2004) and visualised using gVista tool (http://genome.lbl.gov/cgi-bin/GenomeVista). Arabidopsis thaliana genes present in this region were listed along with their strandedness. Ab initio gene prediction on smaller sized genomic regions, cBr-R1 (242 kb), cBr-R2 (151 kb) and cBr-R3 (105 kb) was carried out using FGENESH (Solovyev et al. 2006; http://linux1.softberry.com/berry.phtml?topic=index&group=programs&subgroup=gfindb). Gene identities were assigned to FGENESH predicted genes by conducting homology searches (BLASTn 2.0, Altschul et al. 1990) on A. thaliana and B. rapa genomes. The predicted genes were listed maintaining the strandedness, directions and order. Synteny maps were drawn manually to depict conservation of gene content and order in cBr-R1, cBr-R2 and cBr-R3 and to analyse gene fractionation. Instances of gene inversions were indicated on synteny maps by comparing the strandedness of homologs identified for cBr-R1, cBr-R2 and cBr-R3 with At-R1.
Informatics of primer design
Consensus primers were designed based on A. thaliana SOC1 (AT2g45660) and B. rapa SOC1 homologous sequences aligned using BioEdit version (ver.) 7.0.5.3 (Hall 1999). Oligonucleotides 5′-ATGGTGAGGGGCAAAACTCAG-3′ and 5′-TCACTTTCTTGAAGAACAAGG-3′ were employed as forward and reverse primers, respectively, for isolation of SOC1 complementary DNA (cDNA) and genomic sequences from various cultivars of Brassica. For quantification of cumulative expression levels, forward and reverse primers (5′-CGAGCAAGAAAGACTCAAGTG-3′ and 5′-CTTGAAGAACAAGGTAACCCAATG-3′, respectively) were designed based on conserved regions of B. juncea SOC1 cDNA homologs. For B. juncea homeolog-specific expression analysis, oligonucleotides 5′-GAAGGCATTGGATCATGCTCG-3′ and 5′-ACTGTCCTCGTCACCTCGTCC-3′ were used as forward and reverse primers, respectively, for analysing expression pattern of JQ845925 (MF2-specific SOC1 homeolog). Similarly, oligonucleotides 5′- GGATCGTGTCAGCACCAAACCT-3′ and 5′- TGGGCTACTCTCTTCGTCACC-3′, respectively, were employed as forward and reverse primers to analyse the expression pattern of JQ845923 representing LF sub-genome. Oligonucleotides 5′-TGAAGATCAAGGTGGTCGCA-3′ and 5′- AGAAGGCAGAAACACTTAGAAG-3′ served as forward and reverse primers, respectively, for amplification of internal control (B. juncea Actin) and were designed on Actin transcript of B. napus (GenBank ID: AF11812.1).
All oligonucleotides were synthesised by Integrated DNA Technologies (Leuven, Belgium).
Isolation of Brassica SOC1 sequences
The designed consensus pair of primers was used to isolate SOC1 homeologs from various cultivars encompassing six Brassica species through PCR amplification. The reaction mix included 50 ng of genomic DNA, 2.5 mM MgSO4, 0.2 mM dNTPs, 0.5 μM of primers, 1× reaction buffer and 1 weiss units (U) of Pyrococcus furiosus-derived DNA polymerase (Pfu) Turbo (Agilent Technologies, Santa Clara, CA, USA). Thermocycling parameters were initial denaturation of 3 min at 95 °C, 30 cycles of 94 °C for 30 s; 52 °C for 30 s and 72 °C for 2.5 min; final extension was carried out at 72 °C for 7 min. For first-strand cDNA synthesis, 1 μg of DNaseI (Fermentas, Vilnius, Lithuania)-treated RNA was reverse transcribed using RevertAid™ H Minus first-strand cDNA synthesis kit (Fermentas Vilnius, Lithuania). One microlitre of first-strand cDNA was used for reverse transcriptase-polymerase chain reaction (RT-PCR) in a 20-μl reaction mix including reaction buffer to a final concentration of 1×, 2.5 mM MgSO4, 0.2 mM dNTPs, 0.5 μM of each primer and 1 U Pfu DNA polymerase (Fermentas Vilnius, Lithuania). Thermocycling parameters were initial denaturation at 95 °C for 2 min followed by 35 cycles of 95 °C for 30 s, 54 °C for 1 min, 72 °C for 60 s and a final extension at 72 °C for 5 min. The amplicons were cloned into pGEMT-Easy vector (Promega, Madison, WI, USA) and transformed into Escherichia coli DH5α strain and sequenced (Macrogen, Seoul, Korea). At least three clones were sequenced to arrive at a consensus and submitted to GenBank for assignment of accession numbers. A nomenclature for SOC1 homologs was assigned. For example, BjuJAG1_SOC1 refers to abbreviated genus and species name followed by abbreviated cultivar name and the clone number. All sequences were suffixed with an underscore followed by SOC1 (italics).
Sequence analysis of Brassica SOC1
The Brassica SOC1 genomic and cDNA sequence dataset was further enriched by surveying prevalent databases, viz. Brassica database (BRAD, www.brassicadb.org), Phytozome v9.1 (www.phytozome.net), The Arabidopsis Information Resource (TAIR) (The Arabidopsis Genome Initiative 2000; www.arabidopsis.org) and GenBank (www.ncbi.nlm.nih.gov/genbank/). The isolated and reported Brassica SOC1 sequences were aligned using BioEdit ver. 7.0.5.3 (Hall 1999) and Clustal X ver. 2.0.12 (Larkin et al. 2007), and viewed using GeneDoc ver. 2.7 (Nicholas et al. 1999). Pair-wise percentage similarities between SOC1 gene sequences were calculated using BioEdit ver. 7.0.5.3 (Hall 1999). The number of variable sites, parsimony and nucleotide diversity (п value) was examined using DnaSp v5.10 (Librado and Rozas 2009). In silico translation of cDNA sequences were performed using ‘Translate’ tool on ExPASy server (http://web.expasy.org/translate/).
Phylogenetic analysis of SOC1 genes and cDNA sequences was carried out using Bayesian approach-based analytical pipeline available in the ‘BEAST v1.6.2’ software package. Sequence alignment file in ‘.nex’ format was generated using Clustal X ver. 2.0.12 (Larkin et al. 2007). This file was used as input for Bayesian Evolutionary Analysis Utility version 1.6.2 (BEAUti v1.6.2, Drummond and Rambaut 2007) to create an ‘.xml’ file, under default parameters, i.e. Hasegawa, Kishino and Yano (HKY) model of DNA substitution; strict clock with rate = 1.0; coalescent tree: constant size and Markov chain Monte Carlo chain length = 10,000,000. The output from BEAUti was used as input for Bayesian Evolutionary Analysis Sampling Trees version 1.6.2 (BEAST v1.6.2, Drummond and Rambaut 2007) to generate trees. The final phylogram was generated post analysis through TreeAnnotator ver. 1.6.2 and was visualised using FigTree ver 1.3.1 (Rambaut 2006).
For domain analysis of Brassica SOC1, Conserved Domain Database (Marchler-Bauer et al. 2013, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was employed. Synonymous/non-synonymous mutations in aligned cDNA sequences were identified using Highlighter for Nucleotide Sequences v2.2.1 with default parameters (Keele et al. 2008; http://hcv.lanl.gov/content/sequence/HIGHLIGHT/highlightertop.html). The d N/d S ratio (Nei and Gojobori 1986) for all sequence pairs was predicted using the Synonymous Non-synonymous Analysis Program (Korber 2000; http://hcv.lanl.gov/content/sequence/SNAP/SNAP.html). Scratch Protein Predictor (http://scratch.proteomics.ics.uci.edu/) was used to predict the tertiary structure of the K domain and cNLS Mapper (Kosugi et al. 2009; http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) was used to predict the presence and strength of any nuclear localisation signals in the amino acid sequences.
Expression analysis of Brassica SOC1
For analysis of SOC1 expression pattern and levels in B. juncea cv. Varuna, tissue samples representing various organs (leaf, apical region, floral buds, root and pods) at critical developmental time points (35, 75 and 110 days after sowing) were harvested and quantified for SOC1 expression in real time. Quantitative real-time RT-PCR was carried out to monitor expression levels using SYBR green chemistry (SYBR Fast qPCR Master mix, Kapa Biosystems, Cape Town, South Africa). Briefly, the reaction mix contained SYBR Fast qPCR Master Mix (5 μl), forward and reverse primers (0.2 μM) and total cDNA template (1 μl); volume was made up to 10 μl with nuclease-free water (Sigma Life Science). The SOC1 primer pair designed on conserved regions was used to assess the cumulative levels of SOC1 across all spatio-temporal domains. Distinguishing primer pairs were used for homeolog-specific expression analysis of SOC1 in B. juncea cv. Varuna. B. juncea Actin was used as an internal control in all quantitative real-time (qRT) PCR experiments. PCR amplification was monitored in real time using ‘realplex’ Mastercycler (Eppendorf). The thermocycling parameters set for monitoring amplification per cycle were 50 °C for 2 min, 95 °C for 10 min, followed by 30 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 30 s, extension at 72 °C for 30 s preceding and a final extension at 72 °C for 10 min. Melting curve analysis was performed to ensure the presence of unique amplification product, with thermocycling parameters as 95 °C for 15 s, 60 °C for 15 s, followed by a 20-min gradient increase to 95 °C where it was left for 15 s. For data analysis, expression values from three technical replicates were considered. The ΔΔCt values were obtained by normalising with Actin across all samples, followed by normalisation with minima. Relative expression levels were calculated using the expression 2−ΔΔCt (Livak and Schmittgen 2001). However, 2−ΔCt was also calculated to compare expression levels of individual homeologs (Carlini et al. 2013). The PCR efficiencies of differentiating primer pairs were tested as equal. Standard deviation was calculated for mean threshold cycle number (Ct) value of replicates, and expression level differences were recorded as standard error.
Results
Analysis of microsynteny
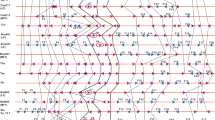
To analyse conservation in gene order and content in genomic regions containing B. rapa SOC1 homeologs, three segments Br-R1, Br-R2 and Br-R3 (∼300 kb) containing SOC1 homeologs Bra004928, Bra039324 and Bra000393 (Brassica database, BRAD) located on chromosome A05, A04 and A03, respectively, were retrieved for analysis. To compare gene fractionation status among Br-R1 (A05: 2297000-2619000), Br-R2 (A04: 18550000-18811500) and Br-R3 (A03: 10855500-11087500), two complementary approaches were employed. In the first approach, B. rapa homeologous regions (Br-R1, Br-R2 and Br-R3) were globally aligned to A. thaliana genome. This approach permitted establishing orthology of three regions to a common reference stretch spanning 77 genes (A. thaliana region (At-R); 283.5 kb) in A. thaliana located on chromosome 2 (TAIR IDs: At2g45200.t01 to At2g45970.t01; Suppl. Fig. 1) containing Arabidopsis thaliana SOC1 protein (AtSOC1). In addition, this approach permitted graphical projection (Genome Vista projections) of orthology and clearly depicted differential retention of gene content in Br-R1, Br-R2 and Br-R3 relative to Arabidopsis reference genes (Suppl. Fig. 1a–c, Suppl. Table 1). Detailed analysis revealed preservation of 53 of the 77 Arabidopsis genes (69 %, Suppl. Table 1) in Br-R1 while Br-R2 showed preservation of slightly lesser, 47 of the 77 genes (61 %, Suppl. Table 1). In contrast, Br-R3 displayed extensive gene loss with only 25 of the 77 genes (33 %, Suppl. Table 1) preserved in B. rapa. In summary, gene fractionation status of three B. rapa genomic blocks (Br-R1, Br-R2 and Br-R3) was found consistent with the reported fractionation status of respective sub-genomes of origin (LF, MF1 and MF2). Since the aforementioned strategy failed to provide complete information, viz. fractionation status of genes unique to B. rapa but absent from Arabidopsis, an alternative approach was also employed. This involved analysing synteny among predicted genes of B. rapa homeologous regions (Br-R1, Br-R2 and Br-R3) and not merely identifying the status of Arabidopsis genes in B. rapa genome. To this end, ab initio predicted genes on Br-R1, Br-R2 and BrR-R3 were assigned B. rapa gene identities (Suppl. Table 2). Since the three homeologous regions were only imperfectly overlapping, a common region was identified that was marked by co-preserved terminal genes. Here onwards, these smaller sized homeologous segments, viz. cBr-R1, cBr-R2 and cBr-R3, derived from Br-R1, Br-R2 and Br-R3, respectively, were considered as units of comparison for reliable synteny mapping. By the same logic, corresponding A. thaliana reference segment (At-R1, 58 genes, 215 kb) spanning At2g45220 and At2g45800 was identified with the said Arabidopsis genes orthologous to terminal B. rapa genes. A summary of gene preservation status in cBr-R1, cBr-R2 and cBr-R3 with reference to 58 A. thaliana genes in At-R1 is presented in Table 1. Notably, differences observed in sizes of B. rapa homeologous regions cBr-R1 (242 kb), cBr-R2 (151 kb) and cBr-R3 (108 kb) symbolised differential gene loss in B. rapa sub-genomes. As expected, these sizes correlated with the number of FGENESH predicted genes (62, 45 and 29) in cBr-R1, cBr-R2 and cBr-R3, respectively (Table 1). Similarly, the number of genes shared between At-R1 and cBr-R1; At-R1 and cBr-R2; and At-R1 and cBr-R3 was 41 (70 %), 32 (55 %) and 18 (31 %), respectively, also correlated with the fractionation status of sub-genome of origin (Table 1). This analysis further allowed synteny mapping with respect to depiction of differential gene fractionation, highlighting shared and unique genes in A. thaliana and B. rapa homeologs. Figure 1 presents the synteny map reflecting preservation of gene content and order across three B. rapa homeologous regions (cBr-R1, cBr-R2 and cBr-R3). In addition, local rearrangements such as inversions and duplications have also been indicated. Figure 1d is yet another illustration of relative gene content in cBr-R1, cBr-R2 and cBr-R3, clearly depicting differential and biased gene fractionation relative to gene content of A. thaliana in 215 kb orthologous region (At-R1), vis-à-vis hypothetical composite gene content of B. rapa. The hypothetical composite gene content of B. rapa shows 11 genes unique to B. rapa. While 6 Arabidopsis genes were found conspicuously absent from three B. rapa homeologous regions, as many as 7 Arabidopsis genes were found co-retained within the three B. rapa homeologous regions (Fig. 1d). Such asymmetric evolution of homeologous sub-genomes has also been shown in B. oleracea (Liu et al. 2014) and allotetraploid B. napus (Chalhoub et al. 2014). Overall, our results demonstrate microsyntenic conservation not only between A. thaliana and B. rapa but also among the three progressively fractionated sub-genomes within B. rapa along with highlighting rearrangements other than gene loss.
Synteny maps of B. rapa genomic regions with respect to A. thaliana. a–c Selected regions of alignment: cBr-R1 (252 kb, A05), cBr-R2 (151 kb, A04) and cBr-R3 (108 kb, A03) mapped against Arabidopsis genomic region (At-R1, 215 kb) containing a set of 58 genes. Red arrows represent Arabidopsis genes with an ortholog in the respective B. rapa genomic region, indicated in green. Purple arrows depict genes unique to Arabidopsis relative to the B. rapa region and blue arrows represent genes unique to B. rapa. FGENESH predicted genes which were identified neither in Brassica nor Arabidopsis are depicted by yellow arrows. Inversions (crossed connectors) and duplications (green connectors) are differentially indicated. d Summary of comparative gene fractionation between three homeologous regions of B. rapa containing SOC1 and (At-R1) is shown. cBr-R3 shows the highest fractionation; cBr-R2 and cBr-R1 display conservation of gene content and order. A notional master reference set of B. rapa genes has also been presented by combining the genes present on the three homeologs. The colons (:) on At-R1 reference indicate Arabidopsis genes absent in the three B. rapa regions. The asterisks (*) highlight genes present in all the three regions; SOC1 locus is boxed. The hash symbols (#) depict unique B. rapa genes
Isolation and sequence analysis of Brassica SOC1 homologs
In Arabidopsis, SOC1 (AT2g45660) is a 2.5-kb sized interrupted gene comprising 7 exons and 6 introns. Consensus primers based on Arabidopsis and reported B. rapa SOC1 sequences led to the isolation of a total of 35 SOC1 sequences comprising 8 genomic copies and 27 cDNA SOC1 variants with latter including a splice variant of SOC1 (Source: B. nigra 898 bp), retaining 6th intron. A list of cDNA and genomic DNA variants and respective GenBank accession numbers is presented in Suppl. Table 3.1.
The genomic DNA copies isolated in this study are classified into two types. The first category comprises typical eukaryotic interrupted genes, viz. BnaGSC1_SOC1 (2446 bp, GenBank ID: JQ845912), BniIC1_SOC1 (2445 bp, GenBank ID: JQ845911) and BraRAG17_SOC1 (2446 bp, GenBank ID: JQ845910) isolated from B. rapa cv. Ragini, B. nigra cv. IC247 and B. napus cv. GSC-1, respectively (Suppl. Table 3.1). The second category comprises extremely small-sized (642 bp) intronless gene copies, identified from B. carinata cv. Pusa Swarnim (GenBank IDs: JQ973096 and JQ973097), B. oleracea cv. PO5 (GenBank ID: JQ973098), B. juncea cv. Varuna (GenBank ID: JQ973094) and B. rapa cv. Ragini (JQ973093). Full-length SOC1 gene sequences of Brassicaceae members retrieved from Brassica database (BRAD) and NCBI were also included in the data set (Suppl. Table 3.2).
To analyse average nucleotide polymorphism within SOC1 genomic DNA sequences for comparison of levels of sequence divergence within and between introns and exons, sliding window analysis was carried out. As expected, high levels of sequence conservation were observed in exons compared to introns (Fig. 2a); the introns displayed significantly high Pi values and size variation implying the absence of purifying selection. Interestingly, within the large sized first intron, two microregions of high conservation (Pi∼0.15) were identified implying regulatory significance.
Sequence conservation in genes and predicted proteins of SOC1 homologs. a Nucleotide polymorphism between Brassica SOC1 genomic DNA sequences. Pi values (Dnasp v5.10.01) plotted along the length of gene (consensus sequence). The window and step-size were taken as 100 and 25 nt, respectively. Low Pi values correspond to 7 exons of SOC1. The first intron showed regions with unusually low Pi values (asterisk). The MIKC domains of predicted SOC1 protein are presented. b Alignment of Brassica SOC1 proteins predicted from cDNA homologs. The MADS-box, intervening, K-box and C-terminal domains of proteins predicted isolated from SOC1 homologs are underscored. A conserved MADS-box (57 aa) domain includes a conserved arginine (marked with arrow) at position 24 across in all homologs. The predicted nuclear localisation signal (NLS) is marked in the MADS domain. The three α-helices, K1, K2 and K3 in K-box region of Brassica SOC1, are indicated with brackets
Genetic relationships between Brassica SOC1 homologs
The isolated SOC1 cDNA sequences were sequence characterised along with the intronless gene copies. An additional set of 29 Brassica SOC1 coding sequence (CDS) retrieved from various databases, viz. BRAD, and GenBank, TAIR and Phytozome v9.1 were also included for sequence analysis (Suppl. Table 3.2). Multiple alignments of these sequences revealed high sequence identity ranging from 91 to 100 % (Suppl. Table 4.1). In contrast, SOC1 gene sequences exhibit a low sequence identity, ranging between 25 and 64 % mainly due to divergence in introns. As an exception, homolog pairs including sequences derived from B. rapa and B. napus SOC1 were found to share a very high sequence identity of 95 and 99 % (Suppl. Table 4.2).
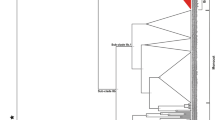
To trace ancestry and derive evolutionary relationships among Brassica SOC1 homologs, phylograms were generated using gene (Fig. 3a) as well as cDNA (Fig. 3b) sequences. Since the homeologs from B. rapa (LF, MF1 and MF2) and B. oleracea (LF, MF1 and MF2) sub-genomes were known a priori, the phylograms allowed us to infer the relationship of homeologs from hitherto uncharacterised Brassica genomes. The separation of SOC1 sequences (Bra004928, Bra039324 and Bra000393) from the three sub-genomes of B. rapa (Brassica diploid base genome A (AA); LF, MF1 and MF2) and SOC1 sequences (Bol030200 and Bol021742) from two sub-genomes of B. oleracea (Brassica diploid base genome C (CC); LF and MF1) into separate clades, and their grouping with corresponding putative homeologs of other Brassica species is indicative of existence of similar sub-genomes in Brassica diploid base genome B (BB) and CC base genomes. Such groupings may be reflective of shared ancestry of homeologous blocks that may have existed in the last common ancestor to Brassica lineage. In other words, it is possible to deduce orthologous relationships between homeologs of diploid base genomes merely through phylogenetic studies if the homolog sampling is adequately representative.
Phylogram based on BEAST derived from SOC1 sequences. a Phylogram based on gene sequences. Three major clades are indicated (I, II.A and II.B), representing homeologous groups of SOC1 sequences drawn from MF1, LF and MF2 sub-genomes, respectively. Identity of each clade has been determined on the basis of prior known sequences marked with an asterisk (*). Sub-clades of I and II.A represent divergence of AA and CC base genomes. Clade II.B contains an obvious AA-MF2 sub-clade, a B. napus sequence (BnaSOC1-Cnn_random) of unknown origin and a single divergent B. nigra sequence (BB base genome). Orthologs appear to be closely related in comparison to paralogs. Out-group (O) consists of A. thaliana, A. lyrata and C. rubella SOC1 gene sequences, while T. halophila SOC1 is not a part of the out-group. b Phylogram based on cDNA sequences. Two major clades are indicated (I and II). Clade I contains MF1-derived Brassica SOC1 coding sequences (IA) from AA (AA-MF1) and CC (CC-MF1) base genomes, along with distantly related T. halophila SOC1. Clade II splits into LF-derived (II.A) and MF2-derived (II.B) sub-clades. Within II.A, sequences show base genome-specific grouping apparent as AA-LF, CC-LF and BB-LF sub-clades. Within II.B, the AA and CC orthologous clades have been marked clearly. Dotted sub-clades diverging from this clade contain B. juncea and B. carinata sequences with unresolved ancestry (X and Y). Homeologs appear more divergent than orthologs. Out-group (O) consists of A. thaliana, A. lyrata and C. rubella SOC1 cDNA sequences. Annotated B. rapa cv. Chiifu-401 and B. oleracea sequences present in each clade have been marked (asterisk)
For instance, the phylogram based on genomic sequences categorises Brassica SOC1 sequences into three major clades (Fig. 3a). The first clade (I) consists of sequences isolated from B. rapa (AA), B. oleracea (CC) and B. napus (AACC). We infer that this clade represents respective MF1 sub-genome-specific sequences owing to the presence of MF1-specific Bra039324. Similarly, the two major clades that separate as (II.A and II.B) comprise putative LF and MF2 sub-genome-derived sequences, respectively. The LF-specific clade (II.A) further splits into sub-clades which represent sequences derived from AA and CC genomes. A putatively MF2-specific clade (II.B) is observed to branch out into two sub-clades comprising sequences from AA genome. A B. nigra-specific sequence was found to diverge out as a clade suggesting its origin from MF2 sub-genome of BB diploid genome. Remarkably, divergence of sequences derived from BB diploid genome in all major clades was marked well before the divergence of AA and CC diploid base genomes. SOC1 sequences from A. thaliana, Arabidopsis lyrata and Capsella rubella diverged out clearly from Brassica sequences while Thellungiella halophila SOC1 sequence was found to be evolutionarily closer to Brassica sequences.
The phylogenetic tree based on coding sequences of Brassica SOC1 (Fig. 3b) recapitulated evolutionary relationships depicted in genomic DNA-based phylogram in a more robust manner. Using the earlier logic, MF1 sub-genome derivatives of AA and CC genomes were found to constitute clade I which was phylogenetically distant to their MF2 and LF counterparts in clades II.A and II.B. Within clade II.A representing putatively MF2-derived SOC1 sequences, clades specific to AA and CC base genomes could be discerned. A minor clade specific to BB diploid genome was also observed within II.A. The clade II.B comprised AA-LF- and CC-LF-derived homologs assorted into sub-clades. However, two B. juncea (BjuSOC1_JQ906706.1 and BjuRH8_SOC1_JQ845919) and two B. carinata SOC1 (BcaPS10_IntronlessSOC1_JQ973096 and BcaNCP12_SOC1_JQ845941) did not belong to any major clade and were marked as sub-clades X and Y (Fig. 3b) since the ancestry of these clades could not be inferred clearly from phylogeny. T. halophila, A. thaliana, A. lyrata and C. rubella SOC1 cDNA sequences were included as out-groups (O) even as T. halophila was found to group along with Brassica sequences (I.B, Fig. 3b).
To clarify ambiguous phylogenetic relationships, another criterion was employed. The cDNA sequences were aligned on the basis of total number of silent and non-silent mutations (Highlighter for Nucleotide Sequences v2.2.1, HCV). The highlighter output arranged the sequences in an ascending order of divergence relative to B. rapa cv. YTS151 (GenBank ID: JQ845914) master sequence (Suppl. Fig. 2). Groupings emerging from pattern of shared mutations between aligned sequences recapitulated groupings observed in the phylograms and provided additional information on sub-clades and sequences with ambiguous ancestry as observed in the phylogram.
Figure 4 represents manual rearrangement of sequences highlighting groups based on the pattern of shared mutations. Nine groups could be clearly discerned marked as AALF, AAMF2, AAMF1, CCLF, CCMF2, CCMF1, BBMF2, BBLF and BB* based on inferred ancestry from the phylograms. These groups were highlighted near identical pattern of polymorphism within members belonging to a particular group but were remarkably distinct between members of different groups. For example, sequences derived from AA and CC base genomes can be easily categorised into three sub-groups based on sub-genome of origin of the sequences (LF, MF1 and MF2) in accordance with the clustering observed in phylogram. Interestingly, the ambiguities relating to homolog ancestry prevailing in the phylograms were easily resolved based on this criterion involving analysis of shared pattern of silent and non-silent mutations. For example, B. juncea sequences (BjuSOC1_JQ906706.1 and BjuRH8_SOC1_JQ845919, indicated as clade X in Fig. 3b) were found to have obvious similarity in mutation pattern with sequences belonging to AALF-specific group, whereas this inference was not apparent in the phylograms. Further, the sequences derived from BB base genome were seen to resolve into three sub-groups in the highlighter output even though these appeared monophyletic in dendrogram. The group marked BB-MF2 in highlighter output corresponds to the cluster in the cDNA phylogram that was putatively inferred to be of MF2 origin thereby resolving the ancestry of these homologs. Given the distinctness of mutation pattern of B. carinata sequences (BcaPS10_IntronlessSOC1_JQ973096 and BcaNCP12_SOC1_JQ845941, indicated as sub-clade Y in Fig. 3b), we speculate that these represent BBLF-specific clade. Another pair of BB-derived sequences (BcaPS5_SOC1_JQ845939 and BniIC4_SOC1_JQ845933) was seen to form a distinct group (BB*). In the absence of whole genome information for BB, it may at best be inferred that such BB-genome-specific orthologs are not derived from MF2 or LF sub-genomes. The AA and CC genomes show existence of three homeologous sub-genomes, while only two were determined in BB base genome. Overall, both phylogram and highlighter plot reveals sub-genomic organisation in the three Brassica base genomes with the presence of multiple homeologs of SOC1.
Distribution and pattern of silent and non-silent mutations highlighted on aligned SOC1 cDNA homologs. Highlighter v.2.2.3 output (modified) of SOC1 cDNA homologs showing divergence relative to reference master sequence (B. rapa YTS151_SOC1, JQ845913). The original output has been rearranged to highlight 9 groups of orthologous based on the extent of shared pattern of silent/non-silent mutations. Groups coded in green are suggestive SOC1 homologs originating from MF2, LF and MF1 sub-genomes specific to base genome AA. Similarly, groups coded in red suggest paralogous groups specific to BB while groups coded in blue suggest sub-genomes (CC-MF2, CC-LF and putative CC-MF1) specific to CC
Structure analysis of SOC1
The Brassica SOC1 cDNA sequences, upon in silico translation, predicted 213 aa protein differing with AtSOC1 with respect to the absence of a threonine residue at the 187th position. Domain analysis of in silico-translated cDNA sequences revealed a characteristic MIKCC type of organisation with clearly discernible MADS, I, K-box and C-terminal domains in Brassica cDNA homologs (Suppl. Table. 5). A higher sequence conservation was observed in MADS and I domains compared to the K-box and C-terminal domains (Suppl. Table. 6).
The domain structure of Brassica SOC1 is reminiscent of type II (MIKC) MADS-box transcription factors with MADS-box, intervening region, K-box and C-terminal domains comprising 57, 16, 97 and 43 aa, respectively (Suppl. Table 5). Further, Brassica SOC1 was inferred to belong to MIKCC type MADS-box family since MADS domain was encoded by first exon, while the I domain was encoded partly by the first exon in addition to exons 2 to 3. Exons 4, 5 and 6 encoded K-box while the C-terminal domain was encoded by exon 7 (Fig. 2a).
Protein sequence alignment (Fig. 2b) revealed a 99 % sequence conservation in α-helices of MADS and I domains across all Brassica homologs. There was a strong illustration of a nuclear localization signal of moderate strength in the MADS domain. The amino acid signatures imparting DNA-binding propensities such as an invariant arginine (position 24th; cited by Lee et al. 2010) were identified within MADS domain (Fig. 2b). A 33 aa-sized nuclear localization signal was also found conserved within the MADS domain. The characteristic heptad repeat pattern was distinctly visible in the three α-helices in K-box (K1, K2 and K3; Fig. 2b). The MADS domain was found devoid of any variation in sequence (Fig. 2b) and exhibited over 99 % sequence conservation, while the K-box was found to be divergent with sequence conservation ranging from 86.5 to 95 %. The C-terminal domain was even more divergent (5 to 23.5 % dissimilarity). Interestingly, the intron-retaining B. nigra transcript (JQ845937), a putative splice variant, encoded a truncated protein (175 aa) due to a premature stop codon. The predicted protein was found to contain 5 aa peptide following a K-box, as opposed to the 43 aa C-terminal extension.
Expression analysis of SOC1 paralogs
In order to analyse expression domain and cumulative expression levels of SOC1 in B. juncea, quantitative real-time expression analysis was carried out using consensus primers targeting SOC1 homologs isolated in the present study. SOC1 expression was detected across all tissues and developmental stages (Fig. 5a). The highest expression levels were observed in apical buds (post-flowering) with 2700-fold higher relative expression compared to the floral buds, where least expression was observed. In leaf and root samples drawn at post-flowering stage, SOC1 transcript accumulation was 563-fold and 111-fold higher, respectively, than corresponding pre-flowering stages. SOC1 levels, however, declined as the plant reached maturity. For instance, tissue samples harvested at 110 days after sowing (DAS) demonstrated lower levels of SOC1 accumulation. Significantly high SOC1 levels (1054-fold) were, however, detected in pods implying additional role of SOC1 in Brassicas (Fig. 5a).
Expression pattern of Brassica juncea SOC1. a Real-time monitored cumulative expression levels of SOC1 quantified with consensus primers in various tissue samples representing vegetative stages 30 DAS (days after sowing), post-flowering stages (75 DAS) and maturity (110 DAS). Error bars represent standard deviation in technical replicates for each sample. b Differential expression pattern of B. juncea SOC1 homeologs (GenBank accession nos. JQ845925 and JQ845923) representing MF2 (black bars) and LF (shaded bars) sub-genomes, respectively, of base genome ‘AA’
To examine the possibility of differential expression pattern of transcripts originating from distinct SOC1 homeologs of B. juncea, specific primers were designed for Brassica SOC1 sequences (GenBank IDs: JQ845925 and JQ845923) inferred as representing sub-clades AAMF2 and AALF, respectively, through phylogenetics. The PCR efficiencies of the primer pairs were validated as equal. For unbiased comparison of homeolog expression levels, relative expressions of both were also calculated as a function of ΔCt (CtSOC1 –CtActin ) (Suppl. Table 7). Relative expression levels of the two transcripts were quantified across the tissue sample set. The two transcripts displayed quantitative differences in expression levels. Our analyses established predominant expression of JQ845925 (MF2) over the JQ845923 (LF) (Fig. 5b) with JQ845925 exhibiting 560-fold higher than minima (30-day-old root samples) and JQ845923 displaying only 43-fold higher expression than minima (Fig. 5b). Our results indicate a greater functional relevance of the former. Also, JQ845925 and not JQ845923 recapitulated the expression patterns observed for cumulative SOC1 levels. Besides, JQ845925 (MF2) exhibited significant expression (21-fold) in pods.
Discussion
Polyploid genomes of Brassica provide a lucid understanding on sequence and functional diversification of duplicated genes as a consequence of genome redundancy. In fact, the meso-triplication event specific to Brassiceae, post Arabidopsis–Brassica divergence (14.5–20 MYA, Mun et al. 2009; Navabi et al. 2013), underpins the evolutionary bases for observed natural variation in Brassicas. Extensive gene fractionation events that followed WGDs have resulted in partially redundant sub-genomes within Brassica. With this perspective, the current study was framed to understand evolution of sub-genomes and its gene components within a species using SOC1 as a case in point. To this end, we mapped conservation in gene content and order in three microsyntenic homeologous blocks (300 kb) of B. rapa (Br-R1, Br-R2 and Br-R3) containing SOC1 to demonstrate evolutionary changes defining genome divergence in the two genera. Subsequently, SOC1 sequences from diverse species of Brassica were isolated and characterised to understand dynamics of sequence and functional diversification in gene copies after WGD and gene fractionation events specific to Brassicas.
Since the genome structure of ancestral Brassicaceae genome, the true reference remains unknown; the extensively annotated member of Brassicaceae (A. thaliana) genome was taken as a reference. Since A. thaliana did not undergo meso-hexaploidization (b), it was expected to display semblance to the last common ancestor of Brassiceae and Arabidae.
Two complementary comparative genomics-based approaches were employed to deduce robust inferences. The first approach exploited availability of A. thaliana whole genome sequence to establish orthology between three genomic segments of B. rapa carrying SOC1 homologs to a common segment in A. thaliana, thus providing evidence for existence of three SOC1 homeologs in AA (B. rapa) base genome, and amphidiploid Brassica species (B. napus, B. juncea) that contain AA base genome. Establishment of homeology among the three B. rapa regions allowed studying the extent of preservation of gene content and order. In addition, conformation of gene fractionation status of individual homeologous B. rapa fragments with respective sub-genomes of origin was established. The second approach was more refined as it involved studying fate of the entire predicted gene list in B. rapa homeologs rather than analysing only a subset found co-preserved in A. thaliana, as was the case in previous analysis. Secondly, synteny analysis was undertaken after establishing a priori and precise correspondence between the three B. rapa homeologous regions (cBr-R1, cBr-R2 and cBr-R3) and A. thaliana ortholog. Broad findings of these two approaches were consolidated as synteny maps which clearly illustrated differential gene loss in triplicated sub-genomes in B. rapa. Biased and differential nature of gene fractionation was observed in cBr-R1, cBr-R2 and cBr-R3, in accordance with the overall status of respective sub-genome of origin (LF, MF and MF2, respectively) corroborating the findings of Sankoff et al. (2010) and Cheng et al. (2012). Despite widespread gene fractionation, at least 7 genes in the homeologous regions were found co-retained as three homeologs in B. rapa, B. rapa SOC1 being one of these. The decision on retention of gene copies, as opposed to fractionation, is influenced by relative fitness or sensitivity conferred to the species (Birchler and Veitia 2010; Thomas et al. 2006). Therefore, maintenance or deletion of all three homeologs of a few particular genes is an indicator of selection. Comparative mapping also revealed conspicuous absence of 6 distinct A. thaliana genes from the three homeologs of B. rapa syntenic regions, while 11 genes in B. rapa did not have orthologs in A. thaliana. However, it is difficult to ascertain if the apparent deletion in one of the genera is reflective of gene insertion in the other subsequent to split of Arabidopsis–Brassica lineages.
To understand the extent of gene diversification as a consequence of polyploidy, genomic and cDNA sequences of Brassica SOC1 were isolated and characterised from diverse species. Full-length genomic SOC1 copies showed expected divergence and conservation in sequences of introns and exons, respectively. However, the presence of conserved regions in the first intron of SOC1 implied a regulatory function.
Interestingly, our study also led to the discovery of intronless SOC1 gene sequences from at least four Brassica species. Similar intronless FLOWERING LOCUS C (FLC) gene copies have also been discovered by our group in Brassicas (Shivaraj and Tyagi, personal communications). Co-occurrence of intronless gene copies along with full-length genes may be attributed to retroposition events (Long et al. 2003), though the presence of conserved UTRs, direct repeats and poly-adenosine tail (Kaessmann 2010) is required to be sequence validated. Surprisingly, neither SOC1 nor FLC intronless gene copies could be identified through in silico screen of whole genome sequence of B. rapa cv. Chiifu-401 (BRAD). The detection of intronless Brassica SOC1 in eukaryotic genomes provides important insights into the evolution of gene structure as well as retroposition (Yan et al. 2014).
The coding sequences of SOC1 homologs were largely conserved and a low d N/d S ratio (=0.21) was indicative of purifying selection (Yang and Bielawski 2000). However, small but detectable variation provided important insights into phylogeny and function.
Phylogenetic analysis illustrated ancestry of SOC1 sequences in concert with the evolution of diploid and amphidiploid species of Brassica. The phylogram depicted ortholog-specific groupings with paralogs separating out as major clades. Similar inferences have been made by phylogenetic analysis of genomic sequences from Brassicas (Kumari et al. 2013; Shivaraj et al. 2014). Most importantly, our study showed the application of simple phylogenetics as a tool for identification of sub-genome-specific copies in polyploid genomes of Brassicas for the first time. Until now, sub-genome-specific copies in polyploids have only been studied in the context of whole genome sequence (Krasileva et al. 2013; Hughes et al. 2014).
The major clades in the dendrogram of SOC1 sequences isolated from diverse species of Brassica represented homeolog diversification after meso-hexaploidy event. Within each clade, groupings were reminiscent of lineages that diversified after the speciation of B. rapa (AA), B. nigra (BB) and B. oleracea (CC) base genomes with variation retained in the allotetraploids. Based on grouping of sequences observed in phylogram, evidence could be obtained for the presence of three sub-genomes in CC similar to three sub-genomes in AA, viz. LF, MF1 and MF2. Our analysis shows that all three base genomes are likely to contain three sub-genome equivalents indicating a hexaploid ancestor before they diverged recently (Navabi et al. 2013). This sub-genome structure is also possibly maintained in allotetraploid Brassicas, as is the case of B. napus (Chalhoub et al. 2014). Phylogenetic analysis also provided molecular evidence for early divergence of B. nigra (B genome)-specific lineage from B. rapa (A) and B. oleracea (C) lineages. Navabi et al. (2013) have timed the split of B. nigra lineage from B. rapa (A) and B. oleracea (C) lineages (6.2 MYA), based on physical mapping of B. nigra BACs against B. rapa and B. oleracea genomes. SOC1 sequences from other members of Brassicaceae (A. thaliana, A. lyrata, C. rubella, T. halophila) which did not undergo the whole genome triplication (b) in Brassica are obviously divergent from Brassica SOC1. T. halophila SOC1 is relatively closer owing to the phylogenetic proximity of tribes Brassicaceae and Eutremeae (Franzke et al. 2011). Groupings emerging from the pattern of shared mutations (Highlighter for Nucleotide Sequences v2.2.1, HCV) between aligned sequences provided additional information on sub-clades and sequences with ambiguous ancestry observed in phylogram.
Analysis of predicted protein sequences classified Brassica SOC1 as MADS-box type II transcription factor. These were further classified as MIKCC and not MIKC*following the criteria proposed by Gramzow and Theissen (2010). Since the I domain of Brassica SOC1 was found encoded by the second exon only, Brassica SOC1 proteins were classified as MIKCC type transcription factor. The four domains of Brassica SOC1 (MIKC) were differentially conserved implying distinct selection pressure correlated to criticality of function. While MADS and I domains (DNA site recognition, DNA binding and nuclear localisation) were found highly conserved, K and C domains were slightly divergent. It has been reported earlier that K-box of MIKC type II transcription factors mediates protein–protein interactions and hetero-dimerisation (Yang et al. 2003). However, the MADS-box and I domains of SOC1 have been shown to be necessary and sufficient for interaction with AGL24, the only known interacting partner of SOC1 in A. thaliana (Lee et al. 2008). It may thus be hypothesised that K-box and C-terminal domains of Brassica SOC1 homeologs in polyploid Brassicas may have undergone sequence divergence on being relieved of their conventional function of mediating protein–protein interactions. Nevertheless, tertiary structure of K-box containing three characteristic α-helices still indicates possibility of protein–protein interaction via K-box of Brassica.
Another interesting aspect of the study was the discovery of a SOC1 splice variant from B. nigra that was found to retain the 6th intron. The in silico translated polypeptide of Brassica SOC1 splice-form obtained from B. nigra cv. SRB98 is predicted to retain MADS, I and K-box domains but lack C-terminal domain. Functional characterisation of splice variants is anticipated to provide comprehensive insights. Alternative splicing is common in plant genomes (Reddy et al. 2013; Rosloski et al. 2013). Partially spliced transcript of SOC1 has been reported in mutant background of A. thaliana ecotype Wassilewskija (Song et al. 2009).
While sequence conservation among SOC1 homologs implied functional conservation, expression pattern of Brassica SOC1 was analysed to understand possible novel roles implied in expression domains. Expression analysis was also undertaken for two inferred SOC1 homeologs of B. juncea. Cumulative expression levels of SOC1 were quantified in the genetic background of B. juncea in diverse tissue and developmental samples. Elevated Brassica SOC1 expression in diverse tissue samples harvested post-flowering implicated the role of SOC1 in flowering. Interestingly, the detection of significant expression levels in pods suggested the expansion of expression domains via regulatory sub-functionalisation. Such observations have been made in several plant species wherein SOC1 expression has been detected in tissue samples other than flowering (Tan and Swain 2007; Ruokolainen et al. 2011; Papaefthimiou et al. 2012). Homeolog-specific expression analysis revealed that AA-MF2-specific homeolog was found to be expressed in pods. More importantly, this homeolog consistently displayed higher expression levels compared to JQ845923 (AA-LF homeolog) indicating its significant contribution to cumulative SOC1 expression levels across all the expression domains analysed. Epistatic dynamics and sub-genome dominance has been reported in paralogs of B. rapa (Cheng et al. 2012). However, our results are in contradiction with Cheng and co-workers who showed dominant expression of genes present in LF sub-genome relative to homeologs from more fractionated sub-genomes. This discrepancy may be attributed to complex regulatory evolution occurring in allotetraploids such as B. juncea. Sub-genome dominance could also have been obscured by genome fusion events.
Conclusion
In this study, we have demonstrated sub-genome evolution and biased gene fractionation in B. rapa and discussed the significance of differential selection of retained homeologs. Taking SOC1 as an example, we have analysed sequence and functional conservation among homeologous genes. We have introduced a novel approach to determine the lineages of homologous sequences, which involves phylogenetic analysis combined with analysis of shared pattern of mutations (silent/non-silent). Our results depict sub-genomic organisation in Brassica genomes. Further, we have shown evidence for regulatory evolution in SOC1 that may contribute to functional divergence among homeologs. Our results highlight that B. rapa microsyntenic regions containing SOC1 homeologs show differential gene fractionation in accordance with respective sub-genomes of origin. Also, while most genes have lost at least one of the homeologs to genome fractionation, maintenance or deletion of all three homeologs indicates selection. High level of sequence conservation is present within coding regions in B. rapa SOC1 homeologs implying functional conservation. Further, it was observed that phylogenetic reconstruction of Brassica species using SOC1 sequences retraces the occurrence of major events during Brassica evolution, which include meso-hexaploidization, early B base genome divergence and A/C base genome split. In addition, ancestry of all isolated SOC1 homolog sequences could be ascertained. Shared patterns of silent and non-silent mutations revealed the base genome origins of homologs from allotetraploid species. Identification of respective ancestries of SOC1 homologs indicated the presence of three groups of homeologs in each diploid genome.
Hence, we conclude that while homeologous gene copies are present in all Brassica genomes, B. juncea SOC1 homeologs have undergone expression domain evolution, most likely via regulatory diversification. This was apparent as their differential contribution to overall molecular phenotype.
Abbreviations
- A. thaliana :
-
Arabidopsis thaliana
- AA:
-
Brassica diploid base genome A
- AGL24 :
-
AGAMOUS-LIKE24 gene
- Arg:
-
Arginine
- AtSOC1:
-
Arabidopsis thaliana SOC1 protein
- At-R:
-
A. thaliana region
- B. juncea :
-
Brassica juncea
- B. napus :
-
Brassica napus
- B. oleracea :
-
Brassica oleracea
- B. rapa :
-
Brassica rapa
- BB:
-
Brassica diploid base genome B
- BRAD:
-
Brassica database
- CC:
-
Brassica diploid base genome C
- CDS:
-
coding sequence
- Ct :
-
Threshold cycle number
- CTD:
-
Carboxy-terminal domain
- cv.:
-
Cultivar
- DAS:
-
Days after sowing
- FLC:
-
FLOWERING LOCUS C gene
- FT :
-
FLOWERING LOCUS T gene
- LF:
-
Least fractionated
- MF1:
-
Moderately fractionated
- MF2:
-
Most fractionated
- MIKC:
-
MADS, intervening, K-box, C-terminal
- MY:
-
Million years
- MYA:
-
Million years ago
- NLS:
-
Nuclear localisation signal
- Pfu :
-
Pyrococcus furiosus-derived DNA polymerase
- qRT:
-
Quantitative real-time
- RT-PCR:
-
Reverse transcriptase-polymerase chain reaction
- SOC1/SOC1:
-
SUPPRESSOR of OVEREXPRESSION of CONSTANS 1 gene/protein
- U:
-
Weiss units
- ver.:
-
Version
- WGD:
-
Whole genome duplication
- YA:
-
Years ago
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Birchler JA, Veitia RA (2010) The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol 186:54–62
Bray N, Dubchak I, Pachter L (2003) AVID: a global alignment program. Genome Res 13:97
Carlini DB, Satish S, Fong DW (2013) Parallel reduction in expression, but no loss of functional constraint, in two opsin paralogs within cave populations of Gammarus minus (crustacea: amphipoda). BMC Evol Biol 13(1):89
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X (2014) Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345(6199):950–953
Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Wang X (2011) BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11(1):136
Cheng F, Wu J, Fang L, Sun S, Liu B, Lin K, Wang X (2012) Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 7(5):e36442
Ding L, Wang Y, Yu H (2013) Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant Cell Physiol 54(4):595–608
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7(1):214
Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K (2011) Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci 16(2):108–116
Gramzow L, Theissen GA (2010) Hitchhiker’s guide to the MADS world of plants. BMC Genome Biol 11:214
Hahn MW (2009) Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered 100(5):605–617
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high‐molecular‐weight protein complex. Plant J 46(2):183–192
Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering‐time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21(16):4327–4337
Hughes TE, Langdale JA, Kelly S (2014) The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole genome duplication in maize, Genome Res. gr-172684
Jaillon O, Aury JM, Wincker P (2009) “Changing by doubling”, the impact of whole genome duplications in the evolution of eukaryotes. Compt Rendus Geosci 332(2):241–253
Kaessmann H (2010) Origins, evolution, and phenotypic impact of new genes. Genome Res 20(10):1313–1326
Katahata SI, Futamura N, Igasaki T, Shinohara K (2014) Functional analysis of SOC1-like and AGL6-like MADS-box genes of the gymnosperm. Cryptomeria Jpn Tree Genet Genome 10(2):317–327
Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C et al (2008) Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci 105(21):7552–7557
Keller TE, Soojin VY (2013) DNA methylation and evolution of duplicate genes. Proc Natl Acad Sci 111(16):5932–5937
Korber B (2000) HIV signature and sequence variation analysis. Comput Anal HIV Mol Seq 4:55–72
Kosugi S, Masako H, Masaru T, Hiroshi Y (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci 106(25):10171–10176
Krasileva KV, Buffalo V, Bailey P, Pearce S, Ayling S, Tabbita F, Dubcovsky J (2013) Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol 14(6):R66
Kumari G, Kusumanjali K, Srivastava PS, Das S (2013) Isolation and expression analysis of miR165a and REVOLUTA from Brassica species. Acta Physiol Plant 35(2):399–410
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Higgins DG (2007) Clustal W and Clustal X version 20. Bioinformatics 23(21):2947–2948
Lee J, Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61(9):2247–2254
Lee J, Oh M, Park H, Lee I (2008) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J 55(5):832–843
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452
Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135(8):1481–1491
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA et al (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 5
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Long M, Betrán E, Thornton K, Wang W (2003) The origin of new genes: glimpses from the young and old. Nat Rev Genet 4(11):865–875
Lysak MA, Koch MA (2011) Phylogeny, genome, and karyotype evolution of crucifers (Brassicaceae). In: Schmidt R, Bancroft I (eds) Genetics and genomics of the Brassicaceae. Springer, New York, pp 1–31
Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41(Database Issue):D348–52
Mouhu K, Kurokura T, Koskela EA, Albert VA, Elomaa P, Hytönen T (2013) The Fragaria vesca homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 represses flowering and promotes vegetative growth. Plant Cell Online 25(9):3296–3310
Mun JH, Kwon SJ, Yang T-J et al (2009) Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes. Genome Biol 10(10):RIII
Nagaharu U (1935) Genome analysis in Brassica with special reference to the experimental formation of Brassica napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Navabi ZK, Huebert T, Sharpe AG, O’Neill CM, Bancroft I, Parkin IA (2013) Conserved microstructure of the Brassica B genome of Brassica nigra in relation to homologous regions of Arabidopsis thaliana, B. rapa and B. oleracea. BMC Genomics 14(1):250
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3(5):418–426
Nicholas KB, Nicholas HB Jr, Deerfield DW (1999) II (1997) GeneDoc: analysis and visualization of genetic variation. Embnew news 4(14):370
Papaefthimiou D, Kapazoglou A, Tsaftaris AS (2012) Cloning and characterization of SOC1 homologs in barley (Hordeum vulgare) and their expression during seed development and in response to vernalization. Physiol Plant 146(1):71–85
Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171(2):765–781
Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A 101(26):9903–9908
Pires JC, Gaeta RT (2011) Structural and functional evolution of resynthesized polyploids. In: Schmidt R, Bancroft I (eds) Genetics and genomics of the Brassicaceae. Springer, New York, pp 195–214
Rambaut A (2006) FigTree: tree fig drawing tool version 131. Inst Evol Biol Univ Edinb
Redden R, Vardy M, Edwards D, Raman H, Batley J (2009) Genetic and morphological diversity in the Brassicas and wild relatives, 16th Australian Research Assembly on Brassicas Ballarat, Victoria
Reddy ASN, Marquez Y, Kalyna M, Bartab A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25:3657–3683
Rosloski SM, Singh A, Jali SS, Balasubramanian S, Weigel D, Grbic V (2013) Functional analysis of splice variant expression of MADS AFFECTING FLOWERING 2 of Arabidopsis thaliana. Plant Mol Biol 81(1-2):57–69
Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH (2011) Over-expression of the Gerbera hybrida At-SOC1-like 1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann Bot 107(9):1491–1499
Sá MELD, Lopes MJC, Campos MDA, Paiva LV, Santos RMAD, Beneventi MA, Sá MFGD (2012) Transcriptome analysis of resistant soybean roots infected by Meloidogyne javanica. Genet Mol Biol 35(1):272–282
Sankoff D, Zheng C, Zhu Q (2010) The collapse of gene complement following whole genome duplication. BMC Genomics 11(1):313
Schranz ME, Mitchell-Olds T (2006) Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18:1152–1165
Shivaraj SM, Dhakate P, Mayee P, Negi MS, Singh A (2014) Natural genetic variation in MIR172 isolated from Brassica species. Biol Plant 58(4):627–640
Solovyev V, Kosarev P, Seledsov I, Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7(1):101–1012
Song HR, Song JD, Cho JN, Amasino RM, Noh B, Noh YS (2009) The RNA binding protein ELF9 directly reduces SUPPRESSOR OF OVEREXPRESSION OF CO1 transcript levels in Arabidopsis, possibly via nonsense-mediated mRNA decay. Plant Cell Online 21(4):1195–1211
Tan FC, Swain SM (2007) Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol Plant 131(3):481–495
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814):796
Thomas BC, Pedersen B, Freeling M (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose sensitive genes. Genome Res 16:934–946
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S et al (2011) The genome of the mesopolyploid crop species Brassica Rapa. Nat Genet 43(10):1035–1039
Yan H, Jiang C, Li X, Sheng L, Dong Q, Peng X et al (2014) PIGD: a database for intronless genes in the Poaceae. BMC Genomics 15(1):832
Yang Y, Fanning L, Jack T (2003) The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J 33(1):47–59
Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol Pers Ed 15(12):496–503
Zhong X, Dai X, Xv J, Wu H, Liu B, Li H (2012) Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Mol Biol Rep 39(6):6967--6974
Ziolkowski PA, Kaczmarek M, Babula D, Sadowski J (2006) Genome evolution in Arabidopsis/Brassica: conservation and divergence of ancient rearranged segments and their breakpoints. Plant J 47(1):63–74
Acknowledgments
This work was supported by grants (BT/PR10071/AGR/36/31/2007 and BT/PR628/AGR/36/674/2011) received from the Department of Biotechnology, Govt. of India. The authors thank Dr. S.B. Tripathi for offering valuable suggestions and Mr. M.S. Negi for the technical assistance. The contribution of Mr. Hari Ram Gupta in field-related activities is duly recognised. The authors thank Dr. Sandip Das (Department of Botany, Delhi University) for his kind guidance on synteny and phylogenetic analysis. Contributions from Shivaraj SM and Shikha Tyagi (Dept. of Biotechnology, TERI University) towards consolidating this paper are acknowledged. Financial assistance as JRF to Sri, T. from University Grants Commission, Govt. of India is gratefully acknowledged. Infrastructural support from TERI and TERI University are duly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Sureshkumar Balasubramanian
Tanu Sri and Pratiksha Mayee contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Fig 1
Global alignment of the three genomic regions of B. rapa containing SOC1 and A. thaliana genome. Vista output shows regions of alignment between B. rapa and A. thaliana. The three regions (Br-R1, Br-R2 and Br-R3) on B. rapa chromosomes A05, A04 and A03, respectively, map to a genomic region At-R (283.5 kb) comprising of a stretch of 77 genes on chromosome 2 of A. thaliana; the blue coloured peaks indicate coding regions. The boxed regions indicate SOC1 locus (a–c). (GIF 362 kb)
Suppl. Fig 2
Distribution and pattern of silent and non-silent mutations among SOC1 cDNA homologs: Highlighter v.2.2.3 output (original) of SOC1 cDNA homologs showing divergence relative to reference B. rapa YTS151SOC1 JQ845913 sequence. (GIF 147 kb)
Suppl. Table 1
(DOCX 22 kb)
Suppl. Table 2
(DOCX 39 kb)
Suppl. Table 3
(DOCX 23 kb)
Suppl. Table 4
(DOCX 24 kb)
Suppl. Table 5
(DOCX 17 kb)
Suppl. Table 6
(DOCX 18 kb)
Suppl. Table 7
(DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Sri, T., Mayee, P. & Singh, A. Sequence and expression variation in SUPPRESSOR of OVEREXPRESSION of CONSTANS 1 (SOC1): homeolog evolution in Indian Brassicas. Dev Genes Evol 225, 287–303 (2015). https://doi.org/10.1007/s00427-015-0513-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-015-0513-4