Abstract
We report in this paper the characterization of Dxbp-1, the Drosophila homologue of the xpb-1 gene that encodes a “bZIP”-containing transcription factor that plays a key role in the unfolded protein response (UPR), an evolutionarily conserved signalling pathway activated by an overload of misfolded proteins in the endoplasmic reticulum (ER). Dxbp-1 is ubiquitously transcribed, and high levels are found in embryonic salivary glands and in the ovarian follicle cells committed to the synthesis of the respiratory appendages. Loss of function of Dxbp-1 induced a recessive larval lethality, thus, revealing an essential requirement for this gene. The Dxbp-1 transcript was submitted to an “unconventional” splicing that generated a processed Dxbp-1s transcript encoding a DXbp-1 protein isoform, as is the case for yeast, Caenorhabditis elegans and vertebrate hac1/xbp-1 transcripts after UPR activation. However, in the absence of exogenously induced ER stress, the Dxbp-1s transcript was also detectable not only throughout embryonic and larval development but also in adults with a high level of accumulation in the male sexual apparatus and, to a lesser extent, in the salivary glands of the third-instar larvae. Using a Dxbp-1:GFP transgene as an in vivo reporter for Dxbp-1 mRNA unconventional splicing, we confirmed that Dxbp-1 processing took place in the salivary glands of the third-instar larvae. The Dxbp-1 gene appears, thus, to play an essential role during the development of Drosophila, hypothetically by stimulating the folding capacities of the ER in cells committed to intense secretory activities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The X-box binding protein-1 (Xbp-1) was first characterized as a bZIP-containing transcription factor that binds to the cis-acting X-box sequence present in the promoter region of the genes of the human major histocompatibility complex class II (Liou et al. 1990) and in the tax-dependent enhancer of HTLV1 (Yoshimura et al. 1990). Mammalian and Caenorhabditis elegans xbp-1 genes were further shown to be the orthologues of the yeast hac1 gene, a key player in the unfolded protein response (UPR; Cox and Walter 1996; Mori et al. 2000; Yoshida et al. 2001; Shen et al. 2001; for a review see Schroder and Kaufman 2005). UPR is an evolutionarily conserved signalling pathway that is activated by an over-accumulation of either misfolded or unfolded proteins in the endoplasmic reticulum (ER) lumen as the result of mutations, drug treatments or environmental stresses. UPR activation elicits several adaptative responses to maintain ER function, in particular, by restricting the import of novel proteins and by stimulating the degradation of abnormal proteins accumulated in the ER lumen. To restore ER function, UPR activation induces (1) an up regulation of genes encoding ER chaperones, folding enzymes and proteins involved in the ER-associated degradation (ERAD) system and (2) an inactivation of eiF4α mediated by PEK/PERK kinase, resulting in a global inhibition of the translational machinery to limit the import of novel proteins (for a review see Schroder and Kaufman 2005). However, when these two processes are not sufficient to restore ER homeostasis, UPR activation eventually commits cells to apoptosis through the activation of the Jun–N-term kinase pathway (Urano et al. 2000b; Nakagawa et al. 2000; Rao et al. 2001). Xbp-1 and ATF-6 are two transcription factors containing a “bZIP” domain and are key players in the UPR (Yamamoto et al. 2004). ATF6 is a transmembrane ER protein that is comprised of a lumenal-sensing domain and a cytosolic “bZIP”-containing region (Haze et al. 1999; Lee et al. 2002; Shen et al. 2005). In an early stage of UPR signalling, ATF-6 is activated by S2P-mediated cleavage of its cytosolic domain that is then transported to the nucleus (Yoshida et al. 1998; Ye et al. 2000; Lee et al. 2002; Shen et al. 2005). The activation of Xbp-1 relies upon a completely different and, as far as we know, specific post-transcriptional mechanism. The native unprocessed xbp-1 messenger RNA (mRNA), xbp-1u, encodes a transcription factor, Xbp-1u, that displays poor transcriptional activities and low stability (Yoshida et al. 2006; Tirosh et al. 2006). It has been shown that, after ER stress and UPR activation, the xbp-1u mRNA is cleaved by the site-specific endoribonuclease Ire-1 at two sites separated by 26 nucleotides (nt; in vertebrates) or 23 nt (in C. elegans), and then re-ligated by a tRNA ligase (Sidrauski and Walter 1997; Urano et al., 2000a; Yoshida et al. 2001; Lee et al. 2002). This unconventional “splicing” of 23 nt (or 26) takes place approximately in the middle of the Xbp-1u open reading frames (ORF). This induces a frame-shift in the resulting processed transcript, xbp-1s, that encodes a novel protein that displays the same N-terminal region as Xbp-1u, including the “bZIP” domain, but a distinct C-terminal region that contains a potent trans-activation domain (Yoshida et al. 2001). In the nucleus, activated ATF6 and Xbp-1s bind to the UPR and ER stress response elements (UPRE and ERSE, respectively) found in the promoter region of genes encoding ER resident proteins, such as chaperone proteins, folding enzymes and ERAD proteins, and activate their transcription (Yoshida et al. 1998; Lee et al. 2003; Yamamoto et al. 2004).
In C. elegans, xbp-1 RNAi-depleted worms are highly sensitive to ER stress, but these mutants are viable under physiological conditions, thus, showing that this gene does not play an essential role during development (Shen et al. 2001). Nevertheless, pek-1(ok275); xbp-1(RNAi) double mutant worms arrest their development at or before the L2 larval stage, suggesting that the two genes mediate redundant pathways that are essential for survival. In contrast, Reimold et al. (2000) and Zhao et al. (2003) have shown that the functions of vertebrate xbp-1 are crucial during development, and that it is essential for the viability of embryos both in mice and frogs. In mouse, xbp-1 is required for cardiac myocyte survival (Masaki et al. 1999), liver development (Reimold et al. 2000), skeletal formation and terminal differentiation of immature plasma cells into immunoglobulin secreting B-lymphocytes (Iwakoshi et al. 2003a,b; for a review see Wu and Kaufman 2006). In addition, these data suggest that xbp-1 does not play a crucial role during early embryonic development in mammals. On the other hand, in Xenopus, xbp-1 is essential during the early stages of embryo development. It forms a regulatory loop with bone morphogenetic protein-4 (BMP-4), which is required for mesoderm and neuroderm differentiation (Zhao et al. 2003; Cao et al. 2006). All together, these data suggest that xbp-1 has been recruited during vertebrate evolution to play several essential roles during development, particularly in cells committed to intense secretory activities in mammals and in the BMP-4 pathway in frogs.
In this present research, we report the first study of the function of Dxbp-1, the Drosophila xbp-1 orthologue. Our results show that, like in vertebrates, this gene plays an essential role during development. We observed that, in the absence of exogenously induced ER stress, the unconventional splicing of Dxbp-1 takes place throughout development and in adult tissues, particularly in secretory organs such as the larval salivary glands and the male accessory glands. Moreover, using an UAS-Dxbp-1:GFP transgene as a reporter for Dxbp-1 processing, we confirmed that putative Ire-1 dependent unconventional splicing took place in the salivary glands of the third-instar larvae. All together, our data support the hypothesis that, as in mammals, Dxbp-1 plays an essential role during the development of Drosophila, particularly in tissues that show high levels of secretory activity.
Materials and methods
Drosophila stocks
Drosophila melanogaster stocks were raised on a standard cornmeal, yeast, agar medium at 25°C. The Dxbp-1 k13803, Df(2)Exel 6042, Df(2) CC2, da-Gal4, en-Gal4 and puc e69 -lacZ lines are described in FlyBase, and relevant fly stocks are available from the Bloomington Stock Centre at Indiana University. The pDI:GFP protein trap line and the lio-Gal4 driver line were kindly provided by Alain Debec (Bobinnec et al. 2003) and Jean-Maurice Dura (Taillebourg et al. 2005), respectively. Dxbp-1 k13803 has been shown to contain an insertion of the P[w + ; lacZ] enhancer trap transposon (Bier et al. 1989) in the 5′-UTR region of the Drosophila xbp-1 gene. In all experiments, the w 1118 stock was used as a wild-type control.
UAS–Dxbp-1:GFP construct
In the UAS-Dxbp-1:GFP construct, a genomic DNA fragment, corresponding to the 5′ region of the Dxbp-1 gene and extending 488 bp downstream from the unconventional intron and, thus, containing the entire Dxbp-1u ORF, was amplified by PCR with additional 5′ BamHI and 3′ KpnI restriction sites and cloned as an XbaI-KpnI fragment into the pUAS P -GFP vector (Januschke et al. 2002) in-frame with the ORF of the Dxbp-1u RNA. Transcription of the UAS-Dxbp-1:GFP construct, which was verified by sequencing, was predicted to encode a fluorescent DXbp-1s:GFP protein after the removal of the 23-bp intron. We generated UAS-Dxbp-1:GFP transgenic lines as previously described, using a w 1118 strain as a recipient stock (Rubin and Spradling 1982).
RNA in situ hybridisation
For in situ hybridisation, RNA probes were synthesized with T7 or T3 RNA polymerase and a Dxbp-1 fragment as template, extending from position +511 to +1,380. We amplified this DNA fragment by PCR, using two pairs of primers designed to include either a T7 promoter sequence within the 3′ backward oligonucleotide or a T3 promoter within the 5′ forward primer.
Total and polyA+ RNA isolation and RT-PCR
Total RNA was isolated using the RNeasy kit (Qiagen), and polyA+ RNA was further purified using the Oligotex mRNA kit (Qiagen). For RT-PCR experiments, 10 μg of total RNA or 1 mg of polyA+ RNA were reverse transcribed with random hexamers and the first-strand cDNA synthesis kit for RT-PCR (Roche). To visualize the processing of the 23-bp intron, we carried out PCR amplifications of the newly synthesized complementary DNA (cDNA) templates using the forward primer 5′-CAGATGCATCAGCCAATCCAAC-3′ and the backward primer 5-GAGTGAGACCTTTCAACAC-3′ that were predicted to amplify fragments of 191 bp (Dxbp-1u RNA) or 168 bp (the Dxbp-1s RNA). PCR products were separated by electrophoresis in 4% metaphor (FMC) gels. All RNA samples were tested for a possible contamination by genomic DNA through RT-PCR, using a couple of primers flanking the 64-bp conventional intron common to both Dxbp-1 RNA transcripts.
Results
Structure of the Dxbp-1 gene and transcripts
The structure of the Drosophila xbp-1 gene was first deduced from the annotated Drosophila genome sequence determined by the Berkley Drosophila genome project (release 3.2 of the annotated D. melanogaster genome) and from the partial or complete sequencing of numerous expressed sequence tag (EST) clones carried out by the Berkley Drosophila genome project. However, the predicted structure of Dxbp-1 transcripts did not fit with the sequence of several EST clones. The annotated Drosophila genome sequence predicted two Dxbp-1 RNA species, Dxbp1-RA and Dxbp1-RB, as a result of the alternative splicing of a 53-nt intron in Dxbp1-RB. Although sequencing data from 16 EST clones confirmed the structure of the Dxbp1-RA transcript, none of the sequenced EST corresponded to the predicted Dxbp1-RB transcript isoform. Three ESTs corresponded to an alternatively spliced transcript distinct from the predicted Dxbp1-RB by the splicing of 23 nt instead of 53 (Fig. 1a). Because of this inconsistency, we sequenced a set of PCR fragments encompassing the complete Dxbp-1 genomic region, and we performed RT-PCR experiments using several couples of primers flanking the predicted or observed introns and RNA extracted from various tissues or at various developmental stages. The amplified RT-PCR products were also sequenced to determine intron lengths and sequences with precision.
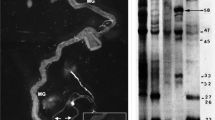
Structure and expression of Dxbp-1 transcripts. a The P[lac; w] K13803 transposon is shown inserted into the 5′-UTR-region of the Dxbp-1 gene. Intergenic sequences are depicted as single lines. Untranslated exonic sequences are represented by light grey boxes and open reading frames are shown as dark grey (common Dxbp-1u/Dxbp-1s ORF), black (Dxbp-1s-specific ORF) and striped (Dxbp-1u/Dxbp-1s overlapping ORFs) boxes. The intron is represented as an empty box, and the unconventional intron (uc. i.) is indicated by two convergent stem-loop structures. Arrow shows the direction of transcription. The translation initiation codon (ATG) and the two termination codons (TAG1 and TAG2) are indicated above the Dxbp-1 transcribed region. The structure of the Dxbp-1u and Dxbp-1s transcripts is shown below. Untranslated sequences are represented by single lines. The Dxbp-1u ORF and that shared by the Dxbp-1u and Dxbp-1s transcripts are indicated as red boxes, with the basic domain coloured in yellow, and the “leucine zipper” as a hatched box. The Dxbp-1s-specific ORF is shown as a blue box. Unprocessed and spliced unconventional introns are depicted by two convergent stem-loops and by a single stem-loop, respectively. b RT-PCR analysis of the expression of the Dxbp-1u and Dxbp-1s transcripts. PCR amplifications were performed using genomic DNA (Gen) as control or reverse-transcribed polyA+ RNA extracted from 0 to 1 h, 2 to 4 h, 6 to 12 h and 12 to 24 h embryos or total RNA extracted from the first (L1), second (L2) and third (L3) instar larvae or dissected third-instar larval salivary glands (SG), ovaries (Ov) or male sexual apparatus (Test), as templates. The length of the two RT-PCR products was determined by the direct sequencing of the amplified fragments. c The sequence alignment of the basic domain of D. melanogaster (D. mel), Caernorhabditis elegans (C. eleg), Mus Musculus (M. mus), Homo Sapiens (H. sap), Rattus norvegicus (R. norv) and Brachydanio rerio (D. rer) Xbp-1 proteins
Our data confirmed that the Dxbp-1 gene gave rise to the transcription of two RNA isoforms. The major transcript that corresponded to the predicted Dxbp1-RA RNA was composed of two exons, 591 and 1552 nt in length, separated by a 64-nt intron flanked by canonical 5′, GUAAGU, and 3′, CAG, consensus sequences. The predicted translation product of this transcript was a 307-aa protein, whose amino-terminal region contained a “bZIP” domain; a 35-aa region rich in basic amino acids followed by seven evenly spaced leucine residues forming a “leucine zipper” dimerization domain (see Figs. S1 and 1a).
The second Dxbp-1 RNA differed from the major transcript only by the absence of a 23-nt sequence located 304 nt downstream from the common 64-nt intron. The predicted 498-aa protein encoded by this transcript displayed the same 188-aa-long amino-terminal region as the protein encoded by the unprocessed Dxbp1-RA transcript, but showed a distinct carboxy-terminal domain because of the frame shift induced by the splicing of this 23-nt intron (see Figs. S1 and 1a). This 23-nt intron was not flanked by canonical splice site consensus sequences, and visual and in silico analyses revealed the presence of complex secondary structures in the flanking sequences. These included a long stem and two evolutionarily conserved stem-loop structures showing strong structure and sequence similarities with the Ire-1 endoribonuclease cleavage sites identified in worms and vertebrates (see Fig. S2). Thus, although the Dxbp-1 gene did not display any significant sequence similarities with the fish, mammal or worm xbp-1 genes outside the region coding for the basic domain (64% identity and 80% similarity; Fig. 1c), the presence of these conserved features indicates that this gene is the Drosophila xbp-1 orthologue. These data also suggested that the Dxbp-1 transcript was submitted to an Ire-1-mediated unconventional “splicing” similar to that of the yeast hac1, and worm and vertebrate xbp-1 transcripts after UPR signalling. In agreement with the nomenclature used for worms and vertebrates, we will refer to Dxbp-1u as the unprocessed transcript and to Dxbp-1s as the spliced Dxbp-1 transcript.
We showed that the level of Dxbp-1s was increased in flies fed for 24 h with tunicamycine, a drug that inhibits protein glycosylation and folding, and consequently induces UPR signalling (see Fig. S3). Because we also detected Dxbp-1s in the absence of exogenously induced ER stress (see Fig. S3), we determined the stage and tissue specificities of the two Dxbp-1 transcripts by RT-PCR, using either total or polyA+ RNA extracted from embryos at various developmental stages (first, second and third instar larvae), the dissected third instar larval salivary glands, and adult ovaries and testes. Both transcripts were detected in all samples analysed, but the levels of accumulation of the xbp-1s transcript compared to those of the xbp-1u form differed in the various stages and tissues analysed. Although levels of Dxbp-1s were low or barely detectable when compared to those of Dxbp-1u in embryos and in ovaries, its relative accumulation was significantly increased in the salivary glands of the third-instar larvae and, even more so, in the sexual apparatus of adult males (Fig. 1b).
Dxbp-1 has an essential function during development
The l(2) k13803 line (Torok et al. 1993) has been shown to contain an insertion of the P[w; lacZ] enhancer trap transposon (Bier et al. 1989) in the 5′-UTR region of the Dxbp-1 gene (Fig. 1a). We used PCR experiments to confirm that this line contains an insertion of the P[w; lacZ] transposon at position +156 respective to the predicted start site of the transcription of the Dxbp-1 gene. The lack of complementation between l(2) k13803 and Df(2)CC 2 or Df(2)Exel 6042 , two deficiencies that delete Dxbp-1, was a first indication that the recessive lethal phenotype in line l(2) k13803 was due to the P[w; lacZ] insertion in the gene. Then, we generated 100 l(2) k13803; w - revertant lines following Δ[2-3] transposase-induced mobilization of the transposon, and we isolated 43 homozygous viable and fertile lines. In all these lines, we mapped the excision event by PCR analysis and subsequent sequencing of the amplified fragments. Precise excision of the transposon was detected in 28 of the 43 lines, therefore, demonstrating (1) that the recessive lethality of line l(2) k13803 was due to the insertion of the P[w; lacZ] transposon leading to a mutation of the Dxbp-1 gene and (2) that line l(2) k13803 did not contain a second recessive lethal or sterile mutation. We, therefore, refer to this mutation as Dxbp-1 k13803.
We observed that 95% of Dxbp-1 k13803 homozygous animals hatched and, at that time, were indistinguishable from control. However, as individuals aged, they remained at the first instar stage, became sluggish and sick before dying after two to four days. A very similar phenotype was observed in hemizygous Dxbp-1 k13803 individuals obtained with either Df(2)CC 2 or Df(2)Exel 6042 , showing that Dxbp-1 k13803 is either an amorphic or a strong hypomorphic mutation. All together, these data suggest that the Dxbp-1 gene has an essential function during development.
Detection in situ of xbp-1 transcripts during oogenesis and embryogenesis
The pattern of accumulation of Dxbp-1 mRNA during oogenesis and embryogenesis was established by whole-mount in situ hybridisation. Data showed that Dxbp-1 transcripts accumulated almost ubiquitously throughout embryogenesis and oogenesis (data not shown). However, we detected significantly higher levels of Dxbp-1 RNAs superimposed on this ubiquitous pattern of expression: (1) during oogenesis in follicle cells that are committed to the synthesis of the respiratory appendages (Fig. 2a), (2) during embryogenesis, in the labial segment of stage 11 embryos (Fig. 2b) and (3) in the salivary glands of stages 14 to 17 embryos (Fig. 2c). In addition, a segmented pattern of Dxbp-1 transcript accumulation was detectable in stage 10-11 embryos (Fig. 2b).
Accumulation pattern of the Dxbp-1 transcript during oogenesis and embryogenesis. (a–c) Whole-mount in situ hybridisations with a Dxbp-1 anti-sense RNA probe on a early stage 14 ovarian follicle, b stage 11 and c stage 17 embryos. d Whole-mount in situ hybridisation with a Dxbp-1 sense RNA probe on a stage 11 embryo. Note that the intensely labelled cells observed in the labial segment of stage 11 embryo likely correspond to the salivary gland anlagen. Dorsal appendage-secreting cells (dasc), salivary glands (sg). Anterior is to the left
Unconventional splicing of Dxbp-1s occurs in the salivary glands of the third-instar larvae
Because the structure of the two Dxbp-1 transcripts made it impossible to design a Dxbp-1s-specific probe to establish the pattern of accumulation of this RNA isoform by in situ hybridisation, we constructed an UAS-Dxbp-1:GFP transgene as a tool to visualize Dxbp-1 unconventional splicing. In this construct, a genomic fragment encompassing the 5′-UTR of the Dxbp-1 gene, the entire Dxbp-1u ORF, including the unconventional intron and 488 bp of 3′ downstream sequences, was cloned upstream from the reporter green fluorescent protein (GFP) gene in the pUASp:GFP vector (Januschke et al. 2002). The construct was designed to reveal unconventional splicing through the production of a fluorescent DXbp-1s:GFP fusion protein after the splicing of the 23-nt intron (see Fig. 3a and Materials and methods). A similar transgenic model has been designed in the mouse for monitoring ER stress (Iwawaki et al. 2004). We first tested various ubiquitous Gal4 drivers, such as act5C-Gal4, arm-Gal4 or da-Gal4, as well as more specific drivers, such as en-Gal4. However, none of these drivers allowed us to detect fluorescent cells at any stage, even in larvae fed with drugs known to induce the UPR pathway such as tunicamycine, dithiothreitol or thapsigargin. Because the Dxbp-1s transcript appeared to accumulate at a relatively high level in salivary glands of the third-instar larvae (Fig. 1b), we analysed the expression of the UAS-Dxbp-1:GFP construct using the lio-Gal4 driver line (Taillebourg et al. 2005) that leads to high levels of expression of the Gal4 transcription factor in the salivary glands of the third-instar larvae. In lio-Gal4; UAS-Dxbp-1:GFP third-instar larvae, we observed a strong GFP fluorescent staining in the salivary glands (Fig. 3b). Consistent with the fact that the DXbp-1u and DXbp-1s proteins are transcription factors, this GFP staining was restricted to salivary gland nuclei. Moreover, an increase in the GFP staining was observed in larvae fed for 24 h with tunicamycine. This demonstrates that putative Ire-1-dependent splicing does indeed occur in salivary glands (Fig. 4). These results, which are also consistent with our RT-PCR data, confirmed that unconventional splicing of Dxbp-1 occurred at relatively high levels in salivary glands of third-instar larvae.
In vivo detection of Dxbp-1 RNA processing. a Structure of the UAS-Dxbp-1:GFP transgene. Dxbp-1 sequences shown as light grey (5′-UTR), dark grey (Dxbp-1u and Dxbp-1s shared ORF), empty (common intron), hatched (Dxbp-1u/Dxbp-1s overlapping ORFs) and black (3′-truncated Dxbp-1s specific ORF) boxes were inserted between UAS sequences (dotted box), and the GFP-encoding gene (green box) in the pUAS P -GFP vector (Januschke et al. 2002) in phase with the Dxbp-1u ORF. An arrow points in the direction of transcription. Translation initiation (ATG) and termination (TAG Dxbp-1u and TAG GFP ) codons are indicated above the transcribed sequence. Underneath is depicted the predicted structure of the encoded transcripts. Whereas, the native RNA should encode the bona fide DXbp-1u protein (colours as in Fig. 1a), the processed Dxbp-1s:GFP transcript resulting from the splicing of the unconventional intron (uc. i.) is predicted to encode a fluorescent DXbp-1s:GFP fusion protein (colours as in Fig. 1a with the GFP encoding sequences indicated as a green box). b Salivary glands of lio-Gal4; UAS-Dxbp-1:GFP third-instar larvae showing strong specific nuclear fluorescent staining
ER stress increases processing of Dxbp-1:GFP transcripts. Feeding lio-Gal4; UAS-Dxbp-1:GFP larvae tunicamycine increases fluorescent labelling of salivary gland nuclei. All salivary glands were photographed in a single shot to fully visualize the increased fluorescent staining of the salivary glands of lio-Gal4; UAS-Dxbp-1:GFP third instar larvae fed with 50 μg/ml of tunicamycine for 24 h (left) compared to that of lio-Gal4; UAS-Dxbp-1:GFP control larvae (right)
Discussion
In this study, we have analysed the structure, expression pattern and requirement during the development of a novel Drosophila gene encoding a “bZIP” protein. This gene was identified as being the Drosophila orthologue of the xbp-1 gene, as deduced from (1) the strong sequence similarities of the basic domain of the encoded protein with that of vertebrate and nematode Xbp-1 protein, (2) the conservation of both structure and sequences of putative Ire-1 cleavage sites in the Dxbp-1 transcript, (3) and the unconventional splicing of a 23-nt intron, which so far has been detected in xbp-1 transcripts only.
The UPR pathway and its major components such as the per/perk, ire-1 and hac1/xbp-1 genes have been conserved during the evolution from yeast to mammals (for a review see Ma and Hendershot 2001), but in higher eukaryotes, the xbp-1 gene has also been shown to possess functions that are essential during development, independently from pathological or drug-induced ER stresses (for a review see Wu and Kaufman 2006).
Conserved requirement for xbp-1 in secretory cells
In mammals, the first indication of a possible link between the xbp-1 gene and secretory activities was provided by an in situ hybridisation analysis that revealed an abundant transcription of the gene in exocrine glands and in osteoblasts during skeleton formation (Clauss et al. 1993). It was further shown that, at later stages, xbp-1 is required for terminal differentiation of plasma cells into antibody-secreting B cells (Reimold et al. 2001; Gass et al. 2002; Iwakoshi et al. 2003a,b). Indeed, whilst xbp-1 -/- hematopoietic cells underwent normal B-cell activation, germinal centre differentiation and class-switch recombination, they failed to secrete immunoglobulins, which is the final step in B-cell differentiation (Reimold et al. 2001). In addition, an increased accumulation of xbp-1s mRNA has been observed upon the induction of the differentiation of the mouse B-cell lymphoma CH12 cells into antibody-secreting B-cells by lipopolysaccharide. The level of xbp-1s transcript started to increase after 8-12 h of stimulation and then peaked at 32 h, whereas a sharp increase in the accumulation of Xbp-1s protein was detected after 24 h of stimulation, just before an increased translation of Ig chains (Gass et al. 2002). These data seem to corroborate the hypothesis that xbp-1 plays a key role during the very last stage of lymphocyte B-cell differentiation, that is, Ig secretion (Gass et al. 2002). More recently, Lee et al. (2005) designed xbp-1 -/- mouse embryos whose early lethality, which is due to severe impairment of hepatocyte development (Reimold et al. 2000), was rescued by targeting the expression of an xbp-1 transgene in liver cells. These mice showed defects that were restricted to secretory organs, such as exocrine pancreas and salivary glands, and they died at an early post-natal stage due to a lack of pancreatic digestive enzymes (Lee et al. 2005). These data all argue in favour of the postulate that xbp-1 plays a key role in secretory organs in mammals.
Our results also suggest that Dxbp-1 plays an important role in cells showing an intense secretory activity in Drosophila. Firstly, in situ hybridisation data revealed that this gene is expressed in two groups of 50 follicle cells that will secrete in less than 2 h the large quantity of proteins required for the synthesis of the two respiratory appendages at late stages of oogenesis. However, because it is not possible to design a Dxbp-1s-specific probe to detect Dxbp-1s by in situ hybridisation, we were not able to establish which of the Dxbp-1 mRNA isoforms, Dxbp-1u or Dxbp-1s, had accumulated in these follicle cells. Secondly, our RT-PCR experiments revealed that the Dxbp-1s transcript is comparatively more abundant in two types of tissue: (1) the paragonia, which are two accessory glands of the male sexual apparatus that secrete a large quantity of proteins that are transferred with the sperm during copulation and, (2) to a lesser extent, in the salivary glands of third-instar larvae. Thirdly, in agreement with this latter observation, we have confirmed with the Dxbp-1:GFP transgene used to detect Dxbp-1 unconventional splicing that significant levels of presumptive Ire-1-dependent processing of the Dxbp-1 transcript take place in the salivary glands of third-instar larvae, a tissue that is committed to the synthesis of large quantities of glue proteins right at the end of the third larval instar. These data suggest that, like the mouse gene, the Dxbp-1 gene is active in secretory cells.
Because secretory cells display intense translational and post-translational activities such as covalent modifications and protein folding, their requirement for an active Xbp-1 protein likely reflects their important needs in ER resident proteins, such as chaperones proteins and modifying enzymes, that are encoded by genes directly activated by the Xbp-1s protein. In this context, it would be interesting to determine whether ATF-6, the second “bZIP” transcription factor of the UPR pathway, also plays a role in secretory cells. It could be proposed that this “physiological” aspect of the UPR pathway, i.e. the maintenance of ER homeostasis in secretory cells, is the ancestral function of the xbp-1/hac1 and ire-1 genes.
xbp-1 plays an essential role during development
During the early stages of the Xenopus development, both the xXBP-1 protein and the BMP-4 protein are components of a regulatory loop that is essential for mesoderm and neuroderm differentiation (Zhao et al. 2003; Cao et al. 2006). During this process, xXBP-1 acts both as an activator of BMP-4 and a repressor of Xvent-2 genes through direct binding to their promoter sequences. This dual activity of xXBP-1 depends upon its association with various co-factors such as c-Jun, p300 and Smad1 to form transcriptional activator complexes, or with Smad6 and Smad7 in the case of inhibitory complexes (Cao et al. 2006).
Mouse XBP-1−/− embryos died between E12.5 and E13.5 after organogenesis was completed and showed growth retardation, liver hypoplasia and reduced blood cell production (Reimold et al. 2000). The lethality in these mutant embryos could be rescued by targeting the expression of xbp-1 in hepatocytes only (Lee et al. 2005). Thus, the absolute requirement of xXBP-1 during the early stages of embryonic development in frogs appears to be absent in mammals. This suggests that xbp-1 was recently recruited in amphibians to play a role in the early patterning of the embryo. Alternatively, this function of xbp-1 in early frog development might have disappeared or became masked during mammal evolution by the function of a yet unidentified gene. In this context, it would be interesting to use morpholino-mediated gene knockdown to determine whether xbp-1 also plays an essential role during early embryonic development in fishes such as the zebrafish.
In Drosophila, the segmented pattern of the expression of Dxbp-1 in fully elongated germ-band embryos suggested a role for this gene during embryonic development, particularly in embryo patterning. However, the lack of detectable phenotype of Dxbp-1 homozygous or hemizygous mutant embryos did not support this hypothesis. Alternatively, this lack of embryonic phenotype might be due to either residual, albeit sufficient, Dxbp-1 function in available mutants or from maternal Dxbp-1 contribution that provided sufficient Dxbp-1 function for the completion of embryonic development. All attempts to isolate a molecularly null allele of Dxbp-1 that would allow us to perform a germ line clone analysis and generate embryos devoid of maternal Dxbp-1 contribution have failed so far. Whether Dxbp-1 is essential for embryonic development, thus, remains to be established.
Whereas Dxbp-1 mutants hatched normally, homozygous or hemizygous larvae hardly grew at all and did not moult. They lived for 2 to 4 days as sluggish L1s and then died. Thus, in contrast to C. elegans in which xbp-1 null mutants were fully viable and fertile (Shen et al. 2001; Calfon et al. 2002), our data demonstrate an essential requirement for Drosophila xbp-1 during larval development. Because [35S] methionine incorporation experiments suggested that protein synthesis was not significantly decreased in mutant larvae (data not shown); this larval lethality might be due to impaired secretory processes. The growth defects might reflect a lack, or at least a severe reduction, in the secretion of digestive enzymes, and the moult inhibition is possibly the consequence of defects in ecdysone secretion.
Whatever the function of Dxbp-1 during embryogenesis is, this gene appears to play an essential and evolutionarily conserved role in Drosophila, hypothetically, by stimulating the folding capacities of the ER in cells committed to intense secretory activities.
References
Bier E, Vaessin H, Shepherd S, Lee K, Mc Call K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, Jan LY, Jan YN (1989) Searching for pattern and mutation in the Drosophila genome with a p-lacZ vector. Genes Dev 3:1273–1287
Bobinnec Y, Marcaillou C, Morin X, Debec A (2003) Dynamics of the endoplasmic reticulum during early development of Drosophila melanogaster. Cell Motil Cytoskelet 54:217–225
Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002) IRE-1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96
Cao Y, Knöchel S, Oswald F, Donow C, Zhao H, Knöchel W (2006) XBP1 forms a regulatory loop with BMP-4 and suppresses mesodermal and neural differentiation in Xenopus embryos. Mech Dev 123:84–96
Clauss IM, Gravallese EM, Darling JM, Shapiro F, Glimcher MJ, Glimcher LH (1993) In situ hybridization studies suggest a role for the basic region-leucine zipper protein Hxbp-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev Dyn 197:146–156
Cox JS, Walter P (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391–404
Gass JN, Gifford NM, Brewer JW (2002) Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem 277:49047–49054
Haze K, Yoshida H, Yanagi H, Yura T, Mori K (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10:3787–3799
Iwakoshi NN, Lee AH, Glimcher LH (2003a) The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev 194: 29–38
Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH (2003b) Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol 4:321–329
Iwawaki T, Akai R, Kohno K, Miura M (2004) A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med 10:98–102
Januschke J, Gervais L, Dass S, Kaltschmidt JA, Lopez-Schier H, St. Johnston D, Brand AH, Roth S, Guichet A (2002) Polar transport in the Drosophila oocyte requires dynein and kinesin I cooperation. Curr Biol 12:1971–1981
Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ (2002) IRE-1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16:452–466
Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459
Lee AH, Chu GC, Iwakoshi NN, Glimcher LH (2005) XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24:4368–4380
Liou HC, Boothby MR, Finn PW, Davidon R, Nabavi N, Zeleznik-Le NJ, Ting JP, Glimcher LH (1990) A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science 247:1581–1584
Ma Y, Hendershot LM (2001) The unfolding tale of the unfolded protein response. Cell 107:827–830
Masaki T, Yoshida M, Noguchi S (1999) Targeted disruption of CRE-binding factor TREB5 gene leads to cellular necrosis in cardiac myocytes at the embryonic stage. Biochem Biophys Res Commun 261:350–356
Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T (2000) mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci U S A 97:4660–4665
Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J (2000) Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403:98–103
Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM, Bredesen DE (2001) Coupling endoplasmic reticulum stress to the cell death program. Mechanism of caspase activation. J Biol Chem 276:33869–33874
Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH (2000) An essential role in liver development for transcription factor XBP-1. Genes Dev 14:152–157
Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimsher LH (2001) Plasma cell differentiation requires the transcription factor XBP-1. Nature 412:300–307
Rubin GM, Spradling AC (1982) Genetic transformation of Drosophila with transposable element vectors. Science 218:348–353
Schroder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74:739–789
Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107:893–903
Shen X, Ellis RE, Sakaki K, Kaufman RJ (2005) Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet 1:355–368
Sidrauski C, Walter P (1997) The transmembrane kinase Ire-1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90:1031–1039
Taillebourg E, Moreau-Fauvarque C, Delaval K, Dura JM (2005) In vivo evidence for a regulatory role of the kinase activity of the linotte/derailed receptor tyrosine kinase, a Drosophila Ryk ortholog. Dev Genes Evol 215:158–163
Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL (2006) Rapid turnover of unspliced xbp-1 as a factor that modulates the unfolded protein response. J Biol Chem 281:5852–5860
Torok T, Tick G, Alvarado M, Kiss I (1993) P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: isolation of lethals with different overgrowth phenotypes. Genetics 135:71–80
Urano F, Bertolotti A, Ron D (2000a) IRE-1 and efferent signaling from the endoplasmic reticulum. J Cell Sci 113:3697–3702
Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000b) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE-1. Science 287:664–666
Wu J, Kaufman RJ (2006) From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ 13:374–384
Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K (2004) Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem (Tokyo) 136:343–350
Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL (2000) ER stress induces cleavage of membrane-bound ATF 6 by the same proteases that process SREBPs. Mol Cell 6:1355–1364
Yoshida H, Haze K, Yanagi H, Yura T, Mori K (1998) Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins; involvement of basic-leucine zipper transcription factors. J Biol Chem 273: 33741–33749
Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE-1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891
Yoshida H, Oku M, Suzuki M, Mori K (2006) pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol 172:565–575
Yoshimura T, Fujisawa J, Yoshida M (1990) Multiple cDNA clones encoding nuclear proteins that bind to the tax-dependent enhancer of HTLV-1: all contain a leucine zipper structure and basic amino acid domain. EMBO J 9:2537–2542
Zhao H, Cao Y, Grunz H (2003) Xenopus X-box binding protein 1, a leucine zipper transcription factor, is involved in the BMP signaling pathway. Dev Biol 257:278–291
Acknowledgements
We thank Jean Maurice Dura for kindly providing the lio-Gal4 driver line and all the members of the laboratory for helpful discussions throughout the course of this work. We also thank the two anonymous reviewers for their insightful comments and suggestions. S.S. was supported by a predoctoral fellowship from the Fondation pour la Recherche Médicale.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Desplan
Electronic supplementary material
Below is the image is a link to a high resolution version
Fig. S1
Deduced sequences of DXbp-1u and DXbp-1s proteins. Amino acids of the amino-terminal region shared by the two proteins are depicted in black. Within this region, the basic domain is underlined and the leucine residues constituting the “leucine zipper” are indicated in green. Residues of the carboxy-terminal specific domains of the DXbp-1u and DXbp-1s proteins are represented in blue and red, respectively (GIF 12 841 kb)
Fig. S2
Hypothetical structures of Ire-1 cleavage sites. Secondary structure of Ire-1 cleavage sites (red arrowheads) and adjacent sequences of D. melanogaster, C. elegans, Brachydanio rerio and Mus musculus xbp-1 transcripts, as deduced from visual and in silico examinations of xbp-1 sequences (GIF 6 767 kb)
Fig. S3
ER stress induces putative Ire-1-dependent processing of Dxbp-1 RNA. RT-PCR analysis of Dxbp-1s accumulation after the activation of the UPR pathway in adults. PCR amplifications were performed using genomic DNA (Gen) or reverse transcribed RNA extracted from non-treated adults (0) or from flies fed with 20, 40 or 60 μg/ml tunicamycine (GIF 32 523 kb)
Rights and permissions
About this article
Cite this article
Souid, S., Lepesant, JA. & Yanicostas, C. The xbp-1 gene is essential for development in Drosophila . Dev Genes Evol 217, 159–167 (2007). https://doi.org/10.1007/s00427-006-0124-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-006-0124-1