Abstract
Hox and ParaHox genes are transcriptional regulators vital for many aspects of embryonic development in bilaterian animals and are considered to have originated from one ancestral proto-Hox/ParaHox cluster. Hox genes are clustered in the genome of both protostomes and deuterostomes, and there is a specific relationship between the position of a gene in the cluster and the position of its expression along the animal body axis (colinearity). It is not clear whether the ParaHox genes Gsx, Xlox, and, Cdx generally exhibit a similar phenomenon since developmental expression for all three ParaHox genes within a single species has not yet been described for any protostome animal. Here we show the spatial and temporal localization for all three ParaHox genes in the polychaete Capitella sp. I, a member of one of the morphologically most diverse and understudied groups within the Metazoa, the Lophotrochozoa. Our data demonstrate that although both CapI-Xlox and CapI-Cdx are regionally expressed in the gut, the three Capitella sp. I ParaHox genes as a group do not perfectly fit predictions of temporal or spatial colinearity. Instead, there is a conservation of expression across species associated with development of particular tissues, and the relative order of initiation of ParaHox gene expression likely reflects the relative order of species-specific tissue development during ontogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
ParaHox and Hox genes are thought to have evolved from a single ancient four-gene proto-Hox/ParaHox cluster prior to the divergence of cnidarians and bilaterians (Brooke et al. 1998; Garcia-Fernandez 2005; Kourakis and Martindale 2000). ParaHox and Hox genes are thus evolutionary sisters (or paralogues). Hox genes increased in number by tandem duplication to 8–10 genes in the postulated bilaterian ancestor (de Rosa et al. 1999) and have a clustered genomic organization in flies and vertebrates (McGinnis and Krumlauf 1992), and also amphioxus (Brooke et al. 1998). Hox genes play a role in axial patterning of animals, and there is a relationship between the relative position of a gene within the cluster and its expression domain along the anterior–posterior axis, known as spatial colinearity. That is, genes at one end of the genomic cluster are expressed at one end of the animal, and those located at the other end of the cluster are expressed at the other end of the animal. Hox genes also show a temporal colinearity that refers to the relative onset of gene expression and relative position of the gene within the cluster. When initially described for the cephalochordate Branchiostoma floridae (amphioxus; Brooke et al. 1998), the ParaHox genes were also shown to be physically linked on the chromosome, consisting of a cluster of three genes: Gsx, Xlox, and Cdx. Phylogenetic analysis revealed the following: Gsx genes show high similarity to anterior class Hox genes (lab), Xlox is similar to Hox3, and Cdx is most similar to posterior Hox genes (AbdB). In vertebrates, ParaHox genes are also clustered along the chromosome (Chawengsaksophak and Beck 1996; Fiedorek and Kay 1995; Holland 2001; Inoue et al. 1996; Pollard and Holland 2000) and exhibit both spatial and temporal colinearity, with Gsx expressed anteriorly, Xlox expressed in the middle, and Cdx expressed in a posterior position.
ParaHox genes have been less well characterized in protostomes. Only a subset of the ParaHox gene orthologues maintained in cephalochordates and vertebrates are present in ecdysozoans such as Drosophila and Caenorhabditis. Caenorhabditis has only a Cdx orthologue, pal-1, and Drosophila has Gsx (ind) and Cdx orthologues (Macdonald and Struhl 1986; Ruvkun and Hobert 1998; Weiss et al. 1998). Furthermore, in Drosophila, ind and Cdx are not linked on the chromosome. In the other major protostome supergroup, the lophotrochozoans, orthologues of the three ParaHox genes, Gsx, Xlox and Cdx, have been identified in each of two sipunculans, demonstrating that all three ParaHox genes were present in the common ancestor of the lophotrochozoans and the deuterostomes (Ferrier and Holland 2001). This result supports the prediction that three ParaHox genes were present in the bilaterian ancestor, as originally proposed by Brooke et al. (1998). Additionally, it indicates that both the Drosophila and Caenorhabditis genomes have lost ParaHox genes, and thus, ParaHox genes in these species represent a derived condition for protostomes.
The presence of three ParaHox genes in lophotrochozoans raises several questions. Are the ParaHox genes clustered along the chromosome in lophotrochozoans as they are in cephalochordates and vertebrates? Do lophotrochozoan ParaHox genes exhibit spatial and temporal colinearity? If so, we can infer that the common ancestor of the lophotrochozoans and deuterostomes exhibited these features. Expression patterns for all three ParaHox genes have not yet been reported within a single lophotrochozoan species, and there is no available genomic linkage data. Thus far, expression for a Cdx orthologue has been described in the mollusc Patella vulgata (Le Gouar et al. 2003), the oligochaete Tubifex tubifex (Matsuo et al. 2005), and the polychaete Platynereis (de Rosa et al. 2005), and Xlox orthologues have been described in the two leech species, Helobdella triserialis and Hirudo medicinalis (Wedeen and Shankland 1997; Wysocka-Diller et al. 1995). Given the gene loss in ecdysozoans and the incomplete data from lophotrochozoans, it has been difficult to determine ancestral bilaterian features of the ParaHox genes.
To gain insight into the evolution of the ParaHox genes in the Bilateria, we report the sequence as well as temporal and spatial expression patterns for all three ParaHox orthologues by whole mount in situ hybridization from embryonic through juvenile stages in the segmented polychaete annelid Capitella sp. I. Although we do not yet have genetic linkage data for the Capitella sp. I ParaHox genes, we can predict the appearance of a colinear expression pattern based on data from chordates; Gsx expression would be initiated first in the most anterior tissues, Xlox expression would be initiated second and localized to tissues in the middle of the body, and Cdx would appear last in posterior tissues. Our data show that each ParaHox gene displays a distinct spatiotemporal expression pattern, and together, they are observed in derivatives of all three germ layers. Our results only partially support expected spatial and temporal colinear expression relationships predicted from chordate examples.
Methods
Collection of developmental stages of Capitella sp. I
Embryos and larvae were collected as previously described (Seaver et al. 2005). To obtain juveniles, competent St. 9 larvae were induced to undergo metamorphosis by addition of mud to a finger dish with filtered seawater (FSW). Following relaxation in 0.19 M MgCl2 in FSW, juveniles were collected by sifting them through a fine mesh sieve, and then transferred to fresh dishes of FSW without food for 2 days to empty their digestive tracts. For the collection of older juveniles, animals were kept in their original dishes, fed with fresh mud weekly until the desired age was reached, and then transferred to fresh dishes for 2 days to empty their digestive tracts prior to fixation.
Cloning of ParaHox genes from Capitella sp. I and riboprobe synthesis
Degenerate primers corresponding to the conserved homeodomain were designed to isolate fragments of Capitella sp. I ParaHox orthologues. Fragments of Cdx and Xlox were recovered in a degenerate Hox screen using the primers AQLVELEKE (5 -GCB CAR YTN GTH GAR YTV GAR AAR G-3') and WFQNRR (5'-CKN CKR TTY TGR AAC CA-3'). A Gsx fragment was recovered using the primers QLLELER (5'-CAR YTK YTD GAR YTH GAR MGR G-3') and WFQNRR. Additional sequence for each fragment was obtained by rapid amplification of cDNA ends (RACE) using the SmartRACE Kit (BD Biosciences) and gene-specific primers (sequences available upon request) with subsequent cloning into the pGEM-Teasy vector (Promega). A linear template for probe synthesis was generated by the polymerase chain reaction (PCR) with SP6 and T7 oligonucleotides. Digoxigenin-labeled riboprobes were transcribed in vitro with the MEGAscript High Yield Transcription Kit (Ambion, Austin, TX) in the presence of 11-dig-UTP (Roche). Riboprobes were diluted to a working concentration of 1 ng/μl for CapI-Cdx, 3 ng/μl for CapI-Gsx, and 3 ng/μl for CapI-Xlox. The CapI-Cdx riboprobe was 1,546 bp in length and included 822 bp of open reading frame (ORF) and 547 bp 5'UTR, CapI-Gsx was 612 bp with 236 bp ORF and 199 bp 3'UTR, and CapI-Xlox was 1,121 bp including 594 bp ORF and 350 bp 5'UTR.
Phylogenetic analyses
The identity of cloned sequences was initially evaluated by BLASTX searches of the GenBank database from NCBI. Gene orthology analysis was performed by first generating amino acid alignments in CLUSTALW of homeodomain regions using the MacVector software (version 7.2.2, Accelyrs Inc.), with default alignment parameters (alignment available upon request). Sequences included the recovered Capitella sp. I clones and ParaHox sequences retrieved from the databases on NCBI. Neighbor Joining analysis was performed in PAUP*4.0b10 (Swofford 2002) with 1,000 replicates. Bayesian analysis was performed with MrBayes V3.0 (Huelsenbeck and Ronquist 2001) using the wag model for protein evolution. Four independent chains were run for 1,000,000 generations, and a majority consensus tree was generated in PAUP*4.0b10 from 9,500 trees representing 950,000 stable generations.
Whole-mount in situ hybridization
The whole-mount in situ hybridization protocol used was according to that of Seaver and Kaneshige (in press; detailed protocol available upon request). After rehydration, fixed larvae and juveniles were treated with 0.01 mg/ml proteinase K in PTw [1× phosphate-buffered saline (PBS), 0.1% Tween 20] for 3 or 5 min, respectively, and stopped by washing twice with 2 mg/ml glycine in PTw, and then postfixed in 3.7% formaldehyde in PTw. Specimens were hybridized in hybridization buffer [50% formamide; 5× saline sodium citrate (SSC), pH 4.5; 50 μg/ml heparin; 0.1% Tween 20; 1% sodium dodecyl sulfate (SDS); 100 μg/ml denatured sheared salmon sperm DNA] with either a sense or antisense riboprobe (0.5–3 ng/μl) at 65°C for 72 h, and then gradually washed to 0.05× SSC. Riboprobes were detected using an alkaline-phosphatase-coupled anti-digoxigenin Fab fragment (1:5,000; Roche) in 1% blocking buffer (Roche blocking powder in 100 mM maleic acid, 150 mM NaCl, pH 7.5) overnight at 4°C and visualized by incubation in NBT/BCIP (Amersham-Pharmacia Biotech) in alkaline phosphatase (AP buffer) [100 mM NaCl, 50 mM MgCl2, 100 mM Tris (pH 9.5), 0.1% Tween 20]. Specimens were mounted in 80% glycerol supplemented with 1 μg/ml Hoechst 33342 (Sigma) to stain nuclei and were analyzed with a Zeiss Axioskop plus. Digital photomicrographs were taken with a Nikon Coolpix 4,500 digital camera (4.0 megapixel).
Results
Fragments of the three ParaHox orthologues for Capitella sp. I were isolated by degenerate PCR using primers designed to complement the conserved regions of the homeodomain. Additional sequence information for the putative Gsx, Xlox, and Cdx orthologues was recovered by RACE PCR using a template generated from mixed stage larval RNA. The composite sequences of the cDNA clones were tentatively named CapI-Gsx, CapI-Xlox, and CapI-Cdx. Examination of the predicted amino acid sequence for the homeodomain reveals the presence of diagnostic residues (Ferrier and Holland 2001) for each of the three Capitella sp. I ParaHox genes (not shown), and complete open reading frames (ORF) were recovered for CapI-Xlox, and CapI-Cdx. To determine the orthology assignments for each gene, we performed phylogenetic analysis using multiple methods. Both Neighbor Joining and Bayesian analyses strongly support the orthology assignments of CapI-Gsx, CapI-Xlox, and CapI-Cdx in the three distinct ParaHox gene families, rather than clustering with Hox genes (Fig. 1).
Neighbor Joining consensus tree of ParaHox genes. The Capitella sp. I sequences group robustly with their respective ParaHox orthologues. Numbers above the branches represent bootstrap values for Neighbor Joining analysis and those below the line represent posterior probabilities for Bayesian analysis. Species abbreviations: Artemia franciscana (Afr), Branchiostoma floridae (Bfl), Bombyx mori (Bmo), Capitella sp. I (CapI), Chaetopterus variopedatus (Cva), Drosophila melanogaster (Dme), Hirudo medicinalis (Hme), Mus musculus (Mmu), Nephasoma minuta (Nmi), Nereis virens (Nvi), Perionyx excavatus (Pex), Ptychodera flava (Pfl), Phascolion strombus (Pst), Patella vulgata (Pvu), Schistocerca gregaria (Sgr), Strongylocentrotus purpuratus (Spu), Tubifex tubifex (Ttu), Xenopus laevis (Xla). Accession numbers are as follows: CapI-Gsx, DQ132894; CapI-Xlox, DQ102390; CapI-Cdx, DQ102389
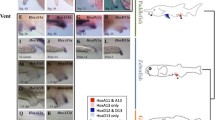
CapI-Gsx is transiently expressed during early stages of brain formation in a subset of anterior neuroectoderm cells at St. 4 (Fig. 2b). Cerebral ganglia formation is initiated as localized thickening of the ectoderm in bilateral areas (Fig. 2c), which results from continued cell divisions in the brain neuroectoderm (Seaver et al. 2005). CapI-Gsx is expressed prior to the internalization and separation of the brain from the ectoderm epithelia and is not observed in any other tissue, or at any other larval or juvenile stage.
ParaHox gene expression during Capitella sp. I development. a Schematic of embryonic and larval stages for Capitella sp. I. b–l (except c) Whole-mount in situ hybridizations. Anterior is to the left in all panels. b Expression of CapI-Gsx in the forming brain (arrowheads) of a St. 4 larva, dorsal view. c Epidermal thickening (arrowheads) during early brain formation (St. 4), visualized with the nuclear stain Hoechst. d CapI-Xlox expression in the gut (bracket; St. 8), lateral view. e–l Expression of CapI-Cdx. Expression domains are marked as follows: brain (black arrows), peristome region/pharynx/oesophagus (white arrows), pygidium/hindgut (black arrowheads), belly plate mesoderm (white bracket), and coelomesoblasts (white arrowhead). e Initial expression in coelomesoblasts (white arrowheads) shortly after blastopore (white ring) closure in St. 3, ventral view. f Same embryo and view as in e, nuclear staining with CapI-Cdx expression superimposed. g Brain expression (early St. 4), ventral view, midbody focal plane. h CapI-Cdx is expressed in brain, belly plate mesoderm, peristome, and pygidium of the same larva as in g. i Lateral view of a St. 6 larva, midbody focus plane, showing expression in the brain, peristome, segmental mesoderm, and pygidium. j Continued expression in domains shown in h (ventral view, St. 7.5). k Lateral view of a juvenile 3 days post-metamorphosis. Highest expression levels are in the hindgut, pharynx/oesophagus, and ventral nerve cord (broad black arrowhead). Note the weak brain expression. l Expression in the hindgut after addition of new juvenile segments (3 weeks post-metamorphosis). Abdominal ganglia are labeled A1–A10. Asterisk marks the stomodeum, and brackets mark the position of the posterior growth zone
CapI-Xlox also shows limited spatiotemporal expression and is found only in endodermal cells that form the midgut epithelium during gut morphogenesis at late larval stages (St. 8; Fig. 2d). In Capitella sp. I gut development, endodermal cells are initially scattered within the yolk and later migrate to the yolk boundaries and form the definitive midgut (Eisig 1899). CapI-Xlox expression has a patchy appearance, reflecting CapI-Xlox expression in midgut progenitor cells that have not yet formed a continuous epithelium. Once the midgut is fully formed, CapI-Xlox expression is no longer detected. CapI-Gsx expression is initiated prior to that of CapI-Xlox and has a more anterior localization, thus the relative expression of these two genes exhibit characteristics of both spatial and temporal colinearity.
CapI-Cdx displays a spatially and temporally dynamic expression pattern that, unlike both CapI-Gsx and CapI-Xlox, includes tissues derived from multiple germ layers. Soon after closure of the blastopore, CapI-Cdx is expressed in a pair of bilaterally symmetric lateral cells in the posterior half of the embryo at St. 3 (Fig. 2e), located beneath the ectoderm, with large nuclei relative to surrounding cells (Fig. 2f). The position of these cells coincides with that of 4d-derived mesendodermal stem cells, or coelomesoblasts (Eisig 1899), characteristic of spiral-cleaving lophotrochozoans. Soon after, two ventrolateral bands of Cdx-expressing mesodermal cells appear (early St. 4), extending anteriorly from the coelomesoblasts (which produce the body wall mesoderm; Fig. 2h). Over time, the mesodermal bands expressing CapI-Cdx enlarge in area and reflect the circumferential expansion of the midbody mesoderm (Seaver et al. 2005; Fig. 2i). CapI-Cdx becomes broadly expressed throughout the segmental mesoderm (Fig. 2j, St. 7), as it is in other lophotrochozoans (Le Gouar et al. 2003; Matsuo et al. 2005).
CapI-Cdx is also expressed in the anterior nervous system and the developing gut. Soon after initial expression in larval coelomesoblasts (early St. 4), CapI-Cdx is up-regulated in the anterior neuroectoderm (Fig. 2g,h) of the brain primordia. As the cerebral ganglia internalize and separate from the overlying ectodermal epithelia, CapI-Cdx brain expression persists (Fig. 2i,j, black arrows). At late larval and juvenile stages, brain expression decreases to low levels (Fig. 2k). CapI-Cdx brain expression overlaps both temporally and spatially with the expression of CapI-Gsx (compare Fig. 2b and g). CapI-Cdx is also prominently expressed in the forming gut, initially in ectodermal cells at the posterior pole of the embryo (Fig. 2h, black arrowhead) at approximately the same time anterior neuroectoderm expression appears (St. 4; Fig. 2h, black arrows). This is soon followed by ectodermal expression lateral to the stomodeum in the peristomal region (Fig. 2h) and will contribute to the anterior portion of the gut (Fig. 2h–j). Over time, the posterior expression in the presumptive hindgut shifts inward, becoming located beneath the surface epithelium (compare Fig. 2h and i). By late larval stages, CapI-Cdx expression decreases in all anterior and midbody regions, with the exception of weak ventral nerve cord expression (not shown). After metamorphosis in juvenile worms, CapI-Cdx is localized to the anterior digestive tract, ventral nerve cord, and is most prominent in the hindgut, which has expanded in area relative to late larval stages (Fig. 2k). CapI-Cdx is not expressed in the posterior growth zone or in mesoderm in juveniles (Fig. 2k,l). Juvenile expression in the hindgut persists and is detectable after generation of eight additional segments (Fig. 2l). Thus, in Capitella sp. I, CapI-Cdx is expressed in distinct tissues of all three germ layers (Fig. 3a) and spans the entire anterior–posterior axis of the body. The onset of CapI-Cdx in each distinct tissue precedes morphogenesis, consistent with a patterning role in the brain, segmental mesoderm, and the anterior and posterior regions of the gut.
ParaHox gene expression patterns in Capitella sp. I and comparison with expression in other animals. a Summary of ParaHox gene expression in Capitella sp. I. b Comparison of ParaHox gene activation order between chordates and Capitella sp. I. The temporal onset of ParaHox gene expression in Branchiostoma is reversed with respect to that observed in vertebrates. c Expression domains of ParaHox genes are conserved in tissues within and among diverse animal groups. The relative order of gene activation reflects the timing of tissue formation expressing each gene
Discussion
The data presented in this study represent the first report of expression for all three ParaHox genes in a single lophotrochozoan species during body plan formation. Our identification of all ParaHox gene orthologues in the polychaete Capitella sp. I supports the observation from a previous report in sipunculans that, unlike the ecdysozoans, lophotrochozoans have retained three ParaHox gene orthologues (Ferrier and Holland 2001). The presence of three ParaHox genes in lophotrochozoans is consistent with an ancestral bilaterian ParaHox cluster of three genes. Linkage data are not yet available for ParaHox genes from any lophotrochozoan, although the Capitella sp. I genome is currently being sequenced by the Joint Genome Institute (DOE).
Although the Capitella sp. I genome clearly has orthologues of all three ParaHox gene members, when considered as a group, their expression profiles do not completely fit predictions of spatial and temporal colinearity (Brooke et al. 1998; Holland 2001; Fig. 3a). The earliest Capitella sp. I ParaHox gene to be expressed is the ‘posterior’ gene Cdx, followed by the ‘anterior’ gene Gsx, and finally the ‘central’ gene Xlox (Fig. 3b). This temporal order does not correlate with the order of the ParaHox cluster seen in chordates. Spatially, CapI-Gsx is expressed anteriorly and CapI-Xlox is in the midbody; thus, these two genes exhibit spatial colinearity. In contrast, CapI-Cdx expression is not limited to the posterior; it extends to the length of the anterior–posterior axis during some stages of its expression, and it also overlaps with CapI-Gsx expression in both timing and position. Our observations that, as a group, spatial and temporal colinearity predictions from chordates are not perfectly met by the Capitella sp. I ParaHox genes can be interpreted in multiple ways. In one scenario, the bilaterian ancestor had a ParaHox cluster of three genes that exhibited spatial and temporal colinearity. According to this scenario, a genomic cluster organization may have been lost in Capitella sp. I, releasing the three ParaHox genes from a strictly coordinated spatiotemporal colinear expression pattern. This is the case for Ciona, in which ParaHox genes are not clustered and also do not exhibit either temporal or spatial colinearity (Ferrier and Holland 2002). Alternatively, in the last common ancestor of the deuterostomes and lophotrochozoans, ParaHox gene expression did not exhibit spatial and temporal colinearity, and possibly the ParaHox genes were not clustered in the genome. Additional data from other protostome taxa, especially regarding genomic organization, are necessary in providing more definitive evidence to distinguish between these alternatives.
Based on ParaHox developmental expression domains in chordates, a model was proposed implicating ParaHox genes in patterning the intestinal tract (Holland 2001). According to this scenario, the Eubilaterian displayed colinearity of the ParaHox genes along the digestive tract, with Gsx anterior, Xlox in the middle, and Cdx posterior. Of the Capitella sp. I ParaHox genes, CapI-Xlox expression in the larval midgut fits this model, although CapI-Gsx and CapI-Cdx do not. The model further suggests that Gsx was expressed in the region of the Eubilaterian mouth (Holland 2001). Lack of Gsx expression in the anterior gut of deuterostomes was explained as a loss of the primary mouth and evolution of a new secondary mouth in the deuterostome clade (Holland 2001). If this were the case, protostomes should maintain Gsx expression in anterior gut structures. Our results do not support such a model since CapI-Gsx expression is limited to a restricted region of the forming brain. Additionally, CapI-Cdx is expressed in both anterior and posterior gut regions.
Comparative analyses across the animal kingdom show conservation of ParaHox gene expression domains in distinct tissues (Fig. 3c). Gsx is expressed in the cerebral vesicle in Branchiostoma (Brooke et al. 1998), in primitive neuroepithelial cells of the forming forebrain and neural tube in mice (Hsieh-Li et al. 1995), and in the anterior neuroectoderm during brain formation in Capitella sp. I. Drosophila Gsx (ind) is expressed prominently in a column of neuroblasts along the length of the ventral nerve cord and also in a subset of brain neuroblasts, including a few of the protocerebrum (Urbach and Technau 2003; Weiss et al. 1998). Such expression patterns are consistent with a conserved function in brain development. CapI-Xlox midgut expression is similar to that in leeches (Wedeen and Shankland 1997; Wysocka-Diller et al. 1995), the only other protostome pattern reported for Xlox. It is likely that the highly conserved midgut expression of Xlox found in a number of deuterostomes (Brooke et al. 1998; Hwang et al. 2003; Ohlsson et al. 1993; Wright et al. 1989) and protostomes (Wedeen and Shankland 1997; Wysocka-Diller et al. 1995) may reflect an ancestral function in midgut epithelia formation and maturation.
CapI-Cdx expression in the brain and anterior digestive tract is somewhat distinctive from other organisms. However, the expression of CapI-Cdx in multiple germ layers in the posterior part of the body is reminiscent of Cdx expression patterns in chordate tissues, including neural tube, mesoderm and posterior regions of the gut and endoderm of vertebrates (Duprey et al. 1988; Gamer and Wright 1993; Gont et al. 1993; Marom et al. 1997; Pillemer et al. 1998; Reece-Hoyes et al. 2002; Subramanian et al. 1995), and posterior neural tube and gut in Branchiostoma (Brooke et al. 1998). Posterior Cdx expression in the presomitic mesoderm in vertebrates and the posterior growth zone of short-germ arthropods has previously been noted (Wu and Lengyel 1998), and an ancestral role has been proposed for Cdx in posterior axis elongation and segmentation (Copf et al. 2004). The expression of Cdx in mesoteloblastic stem cells of Capitella sp. I is consistent with this scenario. It is noteworthy that in the mollusc, P. vulgata, Cdx is also expressed in mesoteloblasts, although P. vulgata is unsegmented and lacks terminal growth. CapI-Cdx is expressed in mesoteloblasts that generate mesoderm of larval segments, but not in the posterior growth zone contributing to new juvenile and adult tissues (Fig. 2k,l). This suggests that CapI-Cdx may have a function associated with the formation of larval but not adult segments and is consistent with a mechanistic distinction between larval and adult segment formation as has been previously suggested for serpulid and nereid polychaetes (Iwanoff 1928). In contrast, in a recent report for Cdx expression in another polychaete, Platynereis dumerulii, Cdx is expressed in the posterior growth zone of juveniles and in regenerating animals (de Rosa et al. 2005). Conserved Cdx expression in the posterior growth zone in representatives from all three bilaterian superclades supports an ancestral role in axis elongation.
The ParaHox genes appear to have taken an evolutionary path distinct from that of its evolutionary sister, the Hox cluster. Unlike Hox genes, which have expanded in gene number and remained a distinct cluster with complex patterns of colinear expression, ParaHox genes may have undergone gene loss. Additionally, in species such as Capitella sp. I, the ParaHox genes may have been released from potential constraints of coordinated gene regulation, allowing for co-option of novel developmental roles (e.g., CapI-Cdx expression in the brain). Our results support the hypothesis that the ancestral role for the proto-Hox/ParaHox cluster was in the formation of specific tissue types along the anterior–posterior axis. It seems that a conserved feature of the temporal order of ParaHox gene activation reflects the relative species-specific order of tissue and organ formation, rather than a strict ordered position along the chromosome.
References
Brooke NM, Garcia-Fernandez J, Holland PWH (1998) The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392:920–922
Chawengsaksophak K, Beck F (1996) Chromosomal location of cdx2, a murine homologue of the Drosophila gene caudal, to mouse chromosome 5. Genomics 34:270–271
Copf T, Schroder R, Averof M (2004) Ancestral role of caudal genes in axis elongation and segmentation. PNAS 101:17711–17715
de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, Carroll S, Balavoine G (1999) Hox genes in brachiopods and priapulids and protostome evolution. Nature 399:772–776
de Rosa R, Prud’homme B, Balavoine G (2005) Caudal and even-skipped in the annelid Platynereis dumerilii and ancestry of posterior growth. Evol Dev 7:574–587
Duprey P, Chowdhury K, Dressler G, Balling R, Simon D, Guenet J, Gruss P (1988) A mouse gene homologous to the Drosophila gene caudal is expressed in epithelial cells from the embryonic intestine. Genes Dev 12A:1647–1654
Eisig H (1899) Zur Entwicklungsgeschichte der Capitelliden. Mitt Zool Stn Neapel 13:1–292
Ferrier DEK, Holland PWH (2001) Sipunculan ParaHox genes. Evol Dev 3:263–270
Ferrier DEK, Holland PWH (2002) Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol Phylogenet Evol 24:412–417
Fiedorek F, Kay E (1995) Mapping of the insulin promoter factor 1 gene (IPF1) to distal mouse chromosome 5. Genomics 28:581–584
Gamer L, Wright C (1993) Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech Dev 43:71–81
Garcia-Fernandez J (2005) Hox, ParaHox, ProtoHox: facts and guesses. Heredity 94:145–152
Gont L, Steinbeisser H, Blumberg B, de Robertis E (1993) Tail formation as a continuation of gastrulation: the multiple cell populations of the Xenopus tailbud derive from the late blastopore lip. Development 119:991–1004
Holland PWH (2001) Beyond the Hox: how widespread is homeobox gene clustering? J Anat 199:13–23
Hsieh-Li HM, Witte DP, Szucsk JC, Weinstein M, Li H, Potter S (1995) Gsh-2, a murine homeobox gene expressed in the developing brain. Mech Dev 50:177–186
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Hwang S, Wu J, Chen C, Hui C, Chen C (2003) Novel pattern of AtXlox gene expression in starfish Archaster typicus embryos. Dev Growth Differ 45:85–93
Inoue H, Riggs A, Tanizawa Y, Ueda K, Kuwano A, Liu L, Donis-Keller H, Permutt M (1996) Isolation, characterization, and chromosomal mapping of the human insulin promoter factor 1 (IPF-1) gene. Diabetes 45:789–794
Iwanoff PP (1928) Die Entwicklung der Larvalsegmente bei den Anneliden. Z Morphol Okol Tiere 10:62–161
Kourakis MJ, Martindale MQ (2000) Combined-method phylogenetic analysis of Hox and ParaHox genes of the Metazoa. J Exp Zool 288:175–191
Le Gouar M, Lartillot N, Adoutte A, Vervoort M (2003) The expression of a caudal homologue in a mollusc, Patella vulgata. Gene Expr Patterns 3:35–37
Macdonald P, Struhl G (1986) A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature 324:537–545
Marom K, Shapira E, Fainsod A (1997) The chicken caudal genes establish an anterior–posterior gradient by partially overlapping temporal and spatial patterns of expression. Mech Dev 64:41–52
Matsuo K, Yoshida H, Shimizu T (2005) Differential expression of caudal and dorsal genes in the teloblast lineages of the oligochaete annelid Tubifex tubifex. Dev Genes Evol 215:238–247
McGinnis W, Krumlauf R (1992) Homeobox genes and axial patterning. Cell 68:283–302
Ohlsson H, Karlsson K, Edlund T (1993) IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J 12:4251–4259
Pillemer G, Epstein M, Blumberg B, Yisraeli J, De Robertis E, Steinbeisser H, Fainsod A (1998) Nested expression and sequential downregulation of the Xenopus caudal genes along the anterior–posterior axis. Mech Dev 71:193–196
Pollard S, Holland PWH (2000) Evidence for 14 homeobox clusters in human genome ancestry. Curr Biol 10:1059–1062
Reece-Hoyes J, Keenan I, Isaacs H (2002) Cloning and expression of the Cdx family from the frog Xenopus tropicalis. Dev Dyn 223:134–140
Ruvkun G, Hobert O (1998) The taxonomy of developmental control in Caenorhabditis elegans. Science 282:2033–2041
Seaver EC, Kaneshige LM (in press) Expression of ‘segmentation’ genes during larval and juvenile development in the polychaetes Capitella sp. I and H. elegans. Dev Biol
Seaver EC, Thamm K, Hill SD (2005) Growth patterns during segmentation in the two polychaete annelids, Capitella sp. I and Hydroides elegans: comparisons at distinct life history stages. Evol Dev 7:312–326
Subramanian V, Meyer B, Gruss P (1995) Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell 83:641–653
Swofford DL (2002) PAUP* 4.0: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, MA
Urbach R, Technau G (2003) Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development 130:3621–3637
Wedeen CJ, Shankland M (1997) Mesoderm is required for the formation of a segmented endodermal cell layer in the leech Helobdella. Dev Biol 191:202–214
Weiss JB, Von Ohlen T, Mellerick DM, Dressler G, Doe CQ, Scott MP (1998) Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev 12:3591–3602
Wright C, Schnegelsberg P, De Roberts EM (1989) XlHbox 8: a novel Xenopus homeo protein restricted to a narrow band of endoderm. Development 105:787–794
Wu LH, Lengyel JA (1998) Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development 125:2433–2442
Wysocka-Diller JW, Aisemberg GO, Macagno ER (1995) A novel homeobox cluster expressed in repeated structures of the midgut. Dev Biol 171:439–447
Acknowledgements
This work was supported by NSF (IBN00-94925). We are grateful to members of Kewalo Marine Lab for discussions and comments on the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.A. Weisblat
Rights and permissions
About this article
Cite this article
Fröbius, A.C., Seaver, E.C. ParaHox gene expression in the polychaete annelid Capitella sp. I. Dev Genes Evol 216, 81–88 (2006). https://doi.org/10.1007/s00427-005-0049-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-005-0049-0