Abstract
Honeybees have been shown to exhibit cognitive performances that were thought to be specific to some vertebrates. However, the molecular and cellular mechanisms of such cognitive abilities of the bees have not been understood. We have identified a novel gene, Mahya, expressed in the brain of the honeybee, Apis mellifera, and other Hymenoptera. Mahya orthologues are present in Deuterostomes but are absent or highly diverged in nematodes and, intriguingly, in two dipteran insects (fruit fly and mosquito) and Lepidoptera (silk moth). Mahya genes encode novel secretory proteins with a follistatin-like domain (Kazal-type serine/threonine protease inhibitor domain and EF-hand calcium-binding domain), two immunoglobulin domains, and a C-terminal novel domain. Honeybee Mahya is expressed in the mushroom bodies and antennal lobes of the brain. Zebra fish Mahya orthologues are expressed in the olfactory bulb, telencephalon, habenula, optic tectum, and cerebellum of the brain. Mouse Mahya orthologues are expressed in the olfactory bulb, hippocampus, and cerebellum of the brain. These results suggest that Mahya may be involved in learning and memory and in processing of sensory information in Hymenoptera and vertebrates. Furthermore, the limited existence of Mahya in the genomes of Hymenoptera and Deuterostomes supports the hypothesis that the genes typically represented by Mahya were lost or highly diverged during the evolution of the central nervous system of specific Bilaterian branches under the specific selection and subsequent adaptation associated with different ecologies and life histories.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The honeybee, Apis mellifera, belongs to Hymenoptera, one of four large holometabolous insect orders. Honeybees are highly social insects and have been used as a model system to study complex animal behavior in addition to learning and memory. Honeybees form colonies consisting of a single queen, hundreds of males, and thousands of female worker bees. In contrast to the queen and male bees, workers perform a wide variety of tasks to maintain the colony, including nursing and foraging for food. In comparison to performing tasks inside the dark hive, foraging involves extensive flight, learning and memorizing food locations, navigation using multiple cues (e.g., sun compass), and communication by a dance language (Fahrbach and Robinson 1995; Menzel and Muller 1996; Menzel and Giurfa 2001). These tasks may require better modules for sensory-input processing and enhanced learning and memory capabilities than other insects. The social behavior and cognitive ability of honeybees partially match those of higher vertebrates, a recognition that led to the launching of the honeybee genome project.
Cross-species comparison of genome sequences and EST data sets has given us enormous insight into the evolution of metazoan genomes. Regarding the origin and evolution of the central nervous system (CNS) of Protostomes and Deuterostomes, most of the nervous system-related genes of the planarian Dugesia japonica (Platyhelminthes, Tricladida) are shared with fruit fly, nematode, and human. This suggests at least the presence of a common ancestral CNS at the molecular level (Mineta et al. 2003). However, a few planarian genes are not conserved among the above animals, suggesting that gene loss occurred during the evolution of the Bilaterian CNS. This is also consistent with the high evolutionary rates of model invertebrates, fruit fly, and nematode compared to those of vertebrates (Mushegian et al. 1998; Zdobnov et al. 2002). Research on Acropora millepora (Anthozoan, Cnidaria) supports a view that a significant proportion of the genes present only in the vertebrates and absent in the model invertebrates is not a vertebrate-specific invention. Instead, these genes appear to have been lost in the specific metazoan lineages that branched from the common ancestors during evolution (Kortschak et al. 2003). The comparative analysis of honeybee EST data set with genome databases also demonstrates that over 100 honeybee cDNA sequences conserved with other organisms are absent in the fruit fly genome (Whitfield et al. 2002). Meanwhile, a number of studies examining genes involved in morphogenesis have revealed the spectacular conservation of the developmental programs between fruit fly and the vertebrates. These results thus demonstrate that the genes functioning for basic biological programs (e.g., the process of embryogenesis) were conserved across the metazoan species, but some genes were either generated or lost in the particular metazoan branches under positive and negative selection pressures.
In addition to two dipteran insects, Drosophila melanogaster and Anopheles gambiae, the genome projects of Apis mellifera, Bombyx mori (silk moth, Lepidoptera), and Tribolium castaneum (red flour beetle, Coleoptera) promise new insights into how the genomes of holometabolous insects evolved in comparison to other metazoans. While most of their genes are expected to be conserved, there will be some genes that are not shared among these five model insect species but are present in other metazoans. Characterization of these genes (their distribution across Bilateria and Cnidaria and their expression patterns) will give us a clearer picture of the ancient evolutionary states and the relationship between gene loss and evolutionary change.
Here, we report a novel gene, Mahya, that is conserved in Hymenoptera and Deuterostomes but is absent in fruit fly, mosquito, silk moth, and nematode. Mahya genes encode novel secretory proteins and are highly expressed in the specific brain regions of honeybee, zebra fish, and mouse where learning and memory and processing of sensory information take place. Thus, Mahya appears to be one of the CNS-related genes retained and evolved in specific Bilaterian branches during evolution.
Materials and methods
Identification and isolation of Mahya orthologues from honeybee, ascidian, zebra fish, mouse, and human
We first identified the honeybee Mahya during a screen of genes differentially expressed between newly emerged bee and the forager brains (Tsuchimoto et al. 2004). The partial Mahya cDNA clone was used as a probe to screen the honeybee brain-specific cDNA library supplied by G. E. Robinson (University of Illinois). The 2.8 kb cDNA containing 3′ UTR was obtained, and the cDNA with 5′ end sequence was then isolated by a 5′ rapid amplification of cDNA end (RACE) method. The full-length Mahya cDNA (3,585 bp) was isolated by a reverse transcription (RT)–polymerase chain reaction (PCR) method with two primers (5′ CGCGGGGAGTACACCCCAAGCCATCAC 3′ and 5′ GGCCACCGATAGGGTAACGATTAGG 3′) and then sequenced (accession number: AB231585). To identify and clone zebra fish Mahya orthologues (drMahya-1 and drMahya-2), the zebra fish genome and EST database (http://www.sanger.ac.uk/Project3/D_rerio/) were searched by BLAST with the honeybee Mahya protein sequence as a query. We identified two different Mahya orthologues with the significant E values represented by one EST clone (accession number: BG739057) and several genome sequences. The primers for 5′ and 3′ RACE were then designed, and the cDNAs with 5′ and 3′ ends were isolated. The partial cDNAs filling the gaps between 5′ and 3′ RACE products were isolated by RT-PCR. All of these cDNAs were sequenced (accession number: AB231586 and AB231587). Human (accession numbers: NP064501 and AX135099) and mouse (accession numbers: NP848788 and NP796033) Mahya orthologues were identified through a nr database search by BLAST. The cDNA sequence of mMahya-2 (NP796033) contains one base insertion in the ORF that is corrected by assembling the two different reads of the same cDNA. The ascidian Mahya orthologue was identified by searching a Ciona intestinalis genome database (http://www.genome.jgi-psf.org/ciona4/ciona4.home.html) by BLAST with the mMahya-1 protein sequence as a query. The initial attempt was unsuccessful with the honeybee Mahya protein sequence as a query. Search for Mahya orthologues in other animal species was carried out by both BLASTP and TBLASTN analysis (with low complexity filter, expected value: 10) of their genome sequences using honeybee, ascidian, zebra fish, and mouse Mahya protein sequences as queries.
Analysis of the expression pattern of honeybee Mahya by Northern blot and RT-PCR
PolyA+RNA was isolated from the brains of 20 newly emerged bees, 7-day-old in-hive bees, and 28-day-old foragers. Two micrograms of polyA+RNA was electrophoresed with 1% agarose gel containing 2.2 M formaldehyde and then transferred to a nylon membrane. After the fixation of RNA, membranes were hybridized with 32P-labeled Mahya and EF-1alpha cDNAs in a buffer containing 0.5 M sodium phosphate (pH 7.2), 7% (w/v) SDS, and 1 mM EDTA (pH 7.0) at 68°C. The signal was detected and analyzed with a Bioimage analyzer BAS2000 (Fuji). The expression of Mahya in the brain, thorax, and abdomen of honeybee workers, and in the brains of males and queens, was analyzed by RT-PCR with polyA+RNA isolated from each sample and the following two primers: 5′ GTAATACCAACAGATAAAAATCCTG 3′ and 5′ ACAAATTGGATTTGAAGAATATGAAA 3′. The expression of beta actin was also analyzed as a control with the following two primers: 5′ AATGGCAACTGCTGCATCATCCTCAAGCTT 3′ and 5′ GAGATCCACATCTGTTGGAAGGTGGACA 3′. The amplified DNA fragments were sequenced to confirm their identity.
In situ hybridization of the brain sections of honeybee, zebra fish, and mouse
In situ hybridization of honeybee (foragers) brain sections with digoxigenin-labeled riboprobes was carried out as described in Funada et al. (2004). For the preparation of riboprobes, the partial honeybee Mahya cDNA (57-3580) with T7 and SP6 promoters was first PCR amplified, and then the sense and antisense riboprobes were made with either T7 or SP6 RNA polymerase and rNTPs containing digoxigenin-UTP. Hybridization was carried out overnight at 62°C. High-stringency posthybridization washes were performed at the same temperature. The brain sections were blocked and then incubated with 500-fold diluted alkaline phosphatase (AP)-conjugated antidigoxigenin antibody overnight at 4°C. The brain sections were washed, and then the AP substrate solution containing nitroblue terazolium and 5-bromo-4-chlor-3-indoly-phosphate was added. The color development was monitored under a microscope and carried out for the same periods on the samples hybridized with the sense and antisense riboprobes. In situ hybridization of zebra fish and mouse brain sections was carried out as described in Yoshimura et al. (2000). The antisense and sense 45 mer oligonucleotides were made against the following sequences—drMahya-1: 5′ GGTGTGTGGATCTGACGGACGCTTCCACCAGAACCACTGCGAGCT 3′; drMahya-2: 5′ CCTGTGTGCGGATCTGACGGGAAACTCTACCAGACCACTGTGAGC 3′; mMahya-1: 5′ GCGGAGCTGGTGCTGTCCACAGAGTGTCGGTGCTGAACCCCGGGT 3′; and mMahya-2: 5′ AGCTTGGACTATCTCTAGCACGCCAGGGGACCAGCCGACAGCCGG 3′. They were labeled with [33P] dATP and terminal deoxyribonucleotidyl transferase. The sagittal sections (20-μm thickness) of the brain were prepared with a Cryostat. Hybridization was carried out overnight at 42°C. Two high-stringency posthybridization washes were performed at 55°C. The sections were air-dried and exposed to a Biomax-MR film for 2 weeks. Following the exposure, each slide was dipped into type NTB2 autoradiography emulsion and developed 4 weeks later at 4°C. The sections were then observed with a microscope for the signal detection.
Analysis of Mahya in other bee species by genomic PCR
Mahya was originally identified in Apis mellifera, the only animal containing Mahya in the Protostomes. To obtain insight into a possible relationship between Mahya and a specific animal behavior, we further searched for Mahya in the genomes of various bee species that exhibit not only different morphology but also express a wide range of social behavior. Based on honeybee genome sequence, honeybee Mahya consists of 18 exons, and exons 11–17 encode a novel C-terminal half domain (Mahya domain). Five different sets of primers (28–30 mer length; T m at 49–53.6°C) corresponding to the 5′ and 3′ end sequences of each of exons 13–17 were used for genomic PCR. These exon sequences are absent in the genomes of fruit fly, mosquito, and nematode. Genomic DNA was isolated from bee samples, either fresh, frozen, or stored in absolute ethanol, by the methods as described in Ashburner (1989). PCR was carried out (annealing temperature at 45°C; number of cycles, 40), and PCR products were analyzed by 2% agarose gel electrophoresis. Any lack of amplification could be due to (1) large mismatches between the primer and genome sequences, (2) different exon–intron organization of Mahya between honeybee and the other bees (the insertion of large intron sequences in the particular exons), or (3) absence of particular exons in the genomes. The PCR-amplified bands of expected sizes were extracted from the gel and sequenced to confirm their identity as exons encoding the Mahya domain. The sequences were identical to those of Apis mellifera, except for base substitutions at several positions.
Results
Identification of Mahya from honeybee, ascidian, zebra fish, mouse, and human
The longest open-reading frame of the honeybee Mahya cDNA encodes a 101-kDa protein of 898 amino acids with the possible N-terminal signal sequence, Kazal-type serine/threonine protease inhibitor domain, EF-hand calcium-binding domain, two immunoglobulin domains, and the C-terminal novel domain. We also identified mouse (mMahya-1 and mMahya-2), human (hMahya-1 and hMahya-2), zebra fish (drMahya-1 and drMahya-2), and ascidian (ciMahya) orthologues of Mahya by BLAST search. All of the vertebrate Mahya and ciMahya proteins share the same domain structures as the honeybee Mahya (Fig. 1a). The N-terminal halves with known domains and the novel C-terminal halves of Mahya proteins are equally conserved across the species (Tables 1 and 2), indicating that the novel C-terminal domain is also important for the function of Mahya. We therefore name this domain as the Mahya domain. The phylogenetic relationship among Mahya proteins from different species (Fig. 1b) shows that the vertebrate Mahya-1 and Mahya-2 proteins cluster into the separate clades. Honeybee and ascidian Mahya are quite diverged from each other and from the vertebrate Mahya. The search for Mahya in the honeybee and ascidian genomes demonstrates that both species contain a single Mahya gene in the genome. Meanwhile, two different Mahya genes are present in mouse and human genomes. These results suggest that the ancestral Mahya gene was duplicated once during the evolution of the vertebrates.
Domain structure of Mahya and the phylogenetic relationship among Mahya proteins from different species. The schematic representation of Mahya is shown in a. Mahya contains the N-terminal signal sequence (solid oval), Kazal-type serine/threonine protease inhibitor domain (Kazal), EF-hand calcium-binding domain (EFh), two immunoglobulin domains found in cell adhesion molecule (IgCAM), and C-terminal novel domain (Mahya domain). Mahya proteins from all species examined share the same domain structure. The phylogenetic relationship among Mahya orthologues is shown in b. The vertebrate Mahya-1 and Mahya-2 proteins cluster into the separate clades. Honeybee and ascidian Mahya proteins are diverged from each other and from the vertebrate Mahya proteins. The tree was constructed by the neighbor-joining method. The scale bar indicates percent divergence or distance between the sequences. Branch lengths indicate phylogenetic divergence. Numbers at the nodes of the tree are bootstrap values
Mahya is absent in the genomes of fruit fly, mosquito, silk moth, and nematode
When we searched the genome databases for Mahya orthologues by BLASTP and TBLASTN, we were able to find them in Strongylocentrotus purpuratus (sea urchin), Takifugu rubripes and Tetraodon nigroviridis (teleost fish), Xenopus tropicalis (frog), Gallus gallus (chicken), Rattus norvegicus (rat), and Pan troglodytes (chimpanzee) genomes in addition to ascidian, zebra fish, mouse, and human. In contrast, we could not find the Mahya orthologues with the significant E values in the genomes of Drosophila melanogaster, Anopheles gambiae, Bombyx mori, and Caenorhabditis elegans. The exons encoding the Kazal-type serine/threonine protease inhibitor domain, EF-hand calcium-binding domain, and immunoglobulin domain are, of course, present in various genes of the above animals. However, the genome DNA sequences capable of encoding the Mahya domain cannot be identified by TBLASTN search with any Mahya proteins as a query. Honeybee belongs to Hymenoptera, and both fruit fly and mosquito are diptera. All are holometabolous insect orders with a similarly structured CNS. Honeybee and fruit fly are thought to have separated 280 million years ago (Carpenter and Burnham 1985). These results therefore suggest that the last common ancestors of Bilateria had the ancestral gene of Mahya, but it was lost or highly diverged in most of Protostome branches during evolution.
Expression profile of Mahya in honeybee
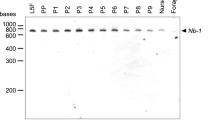
In contrast to EF-1alpha mRNA, Mahya mRNA level is higher in the brains of 7-day-old in-hive bees and 28-day-old foragers than newly emerged bees (Fig. 2a). Thus, Mahya is one of the up-regulated genes in the brains of honeybee workers by age or task. Mahya mRNA is detected in the brain but not in the thorax and abdomen of workers (Fig. 2b), suggesting that Mahya is expressed at high level in the brain compared to thoracic and abdominal ganglia. No caste-specific expression of Mahya is observed (Fig. 2b). By in situ hybridization, we found that Mahya is predominantly expressed in the antennal lobes and mushroom bodies, which are the centers of learning and memory in insect brains (Fig. 3). In the antennal lobes, Mahya mRNA is detected in the lateral and medial cells surrounding the antennal lobes (Fig. 3c). They make synaptic contacts with the glomerular neuropils, where the sensory input from antennae is integrated. Mahya is also expressed at high level in the intrinsic Kenyon cells within the calyces of mushroom bodies and the neurons at the inner border of the calyces in the posterior protocerebrum (Fig. 3a). It is likely that these neurons are a different population of Kenyon cells. The level of Mahya mRNA is higher in the small cell-body Kenyon cells located in the center of each of the calyces of mushroom bodies than in the large cell-body Kenyon cells (Fig. 3b) located within the calyces surrounding the central smaller Kenyon cells. Most of the small Kenyon cells project to the basal ring of the mushroom body, and the large Kenyon cells project to the lip and collar region of the mushroom body.
Expression profile of honeybee Mahya. Mahya and EF-1alpha mRNA levels are analyzed in the brains of newly emerged bees (day 0), 7-day-old in-hive bees, and 28-day-old foragers by Northern blot (a). The expression of Mahya and beta-actin mRNAs in the abdomens (a), brains (b), and thoraxes (t) of workers and the brains of workers (W), males (M), and queens (Q) was analyzed by RT-PCR. These bands are not detected without reverse transcription. The numbers at the right side are molecular weight markers (b)
Expression of honeybee Mahya in the brain. The expression areas of Mahya in the brain were analyzed by in situ hybridization of the antisense and sense riboprobes to the cryosections of honeybee brain. Frontal sections of the entire brain area (a), mushroom bodies (b), and antennal lobes (c) displaying specific expression of Mahya are shown. The black arrows indicate the Kenyon cells in the calyces of mushroom bodies, the white arrow indicates the neurons at the inner border of the calyces, and the black arrowheads point to the lateral neurons surrounding the antennal lobes in (a). The black arrows in (b) show the small cell-body Kenyon cells expressing the high level of Mahya mRNA. The above signals are absent in the section hybridized with the sense probes (d)
Expression areas of drMahya-1 and drMahya-2 in the zebra fish brain
A high level of drMahya-1 mRNA is detected in the dorsal telencephalon and ventral corpus cerebelli of zebra fish brain (Fig. 4a,b,d). Meanwhile, the expression of drMahya-2 mRNA is very high in the habenula and moderately high in the olfactory bulb and optic tectum (Fig. 4f,g,i).
Expression of zebra fish Mahya orthologues in the brain. The expression areas of drMahya-1 (a–e) and drMahya-2 (f–j) in the brain were analyzed by in situ hybridization of the antisense and sense oligonucleotide probes to the cryosections of zebra fish brain. The sagittal sections of the entire brain areas hybridized with the antisense probes are shown (a, f). The high level of drMahya-1 mRNA is detected in the dorsal telencephalon (the white arrowhead in b) and the ventral corpus cerebelli (the white arrowhead in d). These signals are absent in the sections hybridized with the sense probe (c, e). The high level of drMahya-2 mRNA is detected in the habenula (the white arrowhead in g). The intermediate level of the expression is also observed in the olfactory bulb (the white arrow in g) and the optic tectum (the white arrowhead in i). These signals are absent in the sections hybridized with the sense probe (h, j)
Expression areas of mMahya-1 and mMahya-2 in the mouse brain
The level of mMahya-1 mRNA is particularly high in the olfactory bulb, hippocampus, and cerebellum of mouse brain (Fig. 5a). The detailed analysis at high magnification demonstrates that mMahya-1 mRNA is expressed in the granular layer of the cerebellum (Fig. 5c), the hippocampal CA3 region (Fig. 5f), and the glomerular layer and mitral cell layer of the olfactory bulb (Fig. 5i). Meanwhile, the expression level of mMahya-2 is lower than that of mMahya-1, although it is expressed in the hippocampus (Fig. 5k) and weakly in the cerebellum and olfactory bulb (Fig. 5m,s). The observation at high magnification indicates that mMahya-2 mRNA is expressed in the CA3 pyramidal cells and dentate gyrus granular cells of hippocampus (Fig. 5p).
Expression of mouse Mahya orthologues in the brain. The expression areas of mMahya-1 (a–j) and mMahya-2 (k–t) in the brain were analyzed by in situ hybridization of the antisense and sense oligonucleotide probes to the cryosections of mouse brain. The sagittal sections of the entire brain areas hybridized with the antisense probes are shown (a, k). The Nissl staining patterns (b, e, h, l, o, r) and autoradiographs are shown. The high level of mMahya-1 mRNA is detected in the granular layer of the cerebellum (c), hippocampal CA3 region (f), and the glomerular and mitral cell layers of the olfactory bulb (i). These signals are absent in the sections hybridized with the sense probe (d, g, j). The high level of mMahya-2 mRNA is detected in the hippocampal CA3 region and dentate gyrus granular cell layer (p). The low level of the expression is also observed in the granular layer of the cerebellum (m) and the mitral cell layer of the olfactory bulb (s). These signals are absent in the sections hybridized with the sense probe (n, q, t)
Analysis of the existence of Mahya in the genomes of bee species of Apoidea
All five exons encoding the Mahya domain are present in both Apis mellifera and A. cerana (Fig. 6a), suggesting that A. cerana contains Mahya. Three stingless bees, Nannotrigona perilampoides, Trigona guianae, and Tetragonula fuscobalteata, are positive for all of the five exons except exon 14 in Nannotrigona perilampoides (Fig. 6b). These results demonstrate that the highly social stingless bees contain Mahya. The five exons are also present in four bumblebee species, Bombus diversus, B. griseocollis, B. bimaculatus, and B. transversalis (Fig. 6c). The bumblebees with intermediate sociality have Mahya. The mostly solitary orchid bee, Eulaema meriana, is positive for all five exons, although the efficiency of PCR amplification is low, probably due to sequence mismatches (Fig. 6b). Nevertheless, this solitary bee contains Mahya. The same results were obtained with the communal carpenter bees, Xylocopa virginica and X. appendiculata (Fig. 6d), demonstrating that they have Mahya. We then analyzed the existence of Mahya in the genomes of bees belonging to the broader Apoidea. Andrena tsukubana is positive for all of the five exons, Megachile tsurugensis is positive for exons 13, 15, and 17, Lasioglossum occidens is positive for only exon 17, and Colletes patellatus is positive only for exon 15 (Fig. 6e). Thus, Andrena tsukubana and Megachile tsurugensis are likely to contain Mahya. Lasioglossum occidens and Colletes patellatus also may have Mahya since none of the Mahya domain-encoding exons is present in the genomes of fruit fly, mosquito, silk moth, and nematode. All of PCR products were sequenced to confirm that they correspond to exons encoding the Mahya domain.
The existence of Mahya in the genomes of various bee species of Apoidea. The presence of exon sequences encoding the Mahya domain (exons 13–17) in the genomes of different bee species was analyzed by genomic PCR. The results of honeybees (a, Apis mellifera and A. cerana), orchid bee (b, Eulaema meriana), stingless bees (b, Nannotrigona perilampoides, Trigona guianae, and Tetragonula fuscobalteata), bumblebees (c, Bombus diversus, B. griseocollis, B. bimaculatus, and B. transversalis), and carpenter bees (d, Xylocopa virginica and X. appendiculata) are shown. All of the above bees belong to Apidae. The results of four different Apoidea bee species other than Apidae (e, Andrena tsukubana, Lasioglossum occidens, Megachile tsurugensis, and Colletes patellatus) are also shown. Three bands indicated by the white asterisks in X. appendiculata and Andrena tsukubana are nonspecifically amplified products that are not related to the Mahya domain encoding exons. The numbers at the left sides of the panels are molecular weight markers. All of PCR products were sequenced to confirm their identity as exons encoding the Mahya domain
Discussion
Functional implication of Mahya genes
Based on the amino acid sequences of Mahya proteins from all species examined, they appear to be secretory proteins with several function-known domains (Fig. 1a). In fact, a fraction of mMahya-1 protein can be detected in the culture medium when it is expressed in HEK293 cells (not shown). The N-terminal domain structure of Mahya with a Kazal-type serine/threonine protease inhibitor domain and EF-hand calcium-binding domain is also found in follistatin and follistatin-like proteins that bind the TGF-beta family to inhibit their activities (Balemans and Hul 2002). The TGF-beta family exerts its diverse functions by binding its specific receptors to activate the cell-signaling pathways mediated by the Smad family (Shi and Massague 2003). Several TGF-beta proteins are expressed in many brain areas including the hippocampus and are known to stimulate dendrite outgrowth and neuronal differentiation (Ishihara et al. 1994). In addition to the above domains, Mahya also contains two immunoglobulin domains that bind a wide variety of proteins. These results suggest that Mahya may act as a scaffold protein linking the TGF-beta family and other unknown proteins to modulate their functions in the brain. The C-terminal novel Mahya domain has a weak homology to a beta chain of quinohemoprotein amine dehydrogenase that forms a seven-bladed beta-propeller and is a part of the enzyme active site (Datta et al. 2001). The Mahya domain may fold into a similar structure.
According to the expression patterns of Mahya genes in honeybee, zebra fish, mouse, and human, Mahya appears to have major functions in the brain compared to other nervous systems and tissues (Fig. 2). The antennal lobes of the honeybee brain and the olfactory bulb of the vertebrate brain, where Mahya is expressed, share the same functions: the integration and processing of olfactory information by the projection of olfactory neurons. Honeybee and fruit fly have structurally similar antennal lobes, but the expression of Mahya in honeybee and not fruit fly neurons (Fig. 3c) indicates the mechanistic differences of the integration and processing of olfactory cues. Honeybee Mahya is also expressed in the Kenyon cells of the mushroom bodies (Fig. 3b). The dendrites of the Kenyon cells arborize calyces, where the olfactory and visual inputs from projection neurons of the antennal and optic lobes are integrated. The axons of Kenyon cells bifurcate into two lobes, where the efferent connections with other protocerebral neurons are made. Mushroom bodies are thus thought to be higher-order neuronal structures engaged in multisensory integration, learning, and memory in insects. The basic structures of the mushroom bodies are also similar between honeybee and fruit fly, suggesting that Mahya is related to the functional but not gross structural differences between honeybee and fruit fly Kenyon cells.
The optic tectum of teleost fish where Mahya is expressed (Fig. 4i) is the integration area of visual and other sensory inputs. Because the optic tectum also projects to many other brain regions, it is the brain area of connecting various sensory input and motor output (Yoshimoto and Ito 1993). The habenula, another Mahya-expressing brain area (Fig. 4g), was shown to integrate photic inputs from the pineal as well as other brain regions and also project to many other brain areas (Yanez and Anadon 1996). However, its precise functions have not been identified yet. The dorsal region of the teleost fish telencephalon also expresses Mahya (Fig. 4b) and functions in the processing of multiple sensory inputs (olfactory, auditory, mechanosensory, and visual; Prechtl et al. 1998). The recent study with goldfish (Portavella et al. 2004) also demonstrates that the medial and lateral telencephalic regions are involved in the emotional memory system and the spatial and relational memory system, respectively. Thus, the medial and lateral telencephalic regions appear to correspond to the mammalian amygdala and hippocampus, respectively. The mammalian hippocampus is involved in spatial learning, relational memory, and processing of temporal attributes of events and situations. Both mMahya-1 and mMahya-2 are expressed in the CA3 region and the dentate gyrus of the hippocampus (Fig. 5). The vertebrate cerebellum, where drMahya-1 and mMahya-1 are highly expressed, functions in motor coordination and motor learning and memory. Moreover, the cerebellum may be involved in emotional and fear memory (Yoshida et al. 2004; Sacchetti et al. 2002). The expression of human Mahya (hMahya-1 and hMahya-2) mRNAs is also highest in the brain compared to the other tissues. In the human brain, both hMahya-1 and hMahya-2 mRNAs are detected in most of areas; however, hMahya-1 mRNA is relatively high in the cerebellum and amygdala, and hMahya-2 mRNA is high in the amygdala and thalamus (Kikuno et al. 2004; http://www.kazusa.or.jp/huge/gfpage/KIAA1263/ for hMahya-1 and http://www.kazusa.or.jp/huge/gfpage/KIAA1061/ for hMahya-2). The thalamus integrates all sensory information except olfactory and transmits these information back and forth between the cortex. Thus, Mahya genes are expressed in the functionally equivalent brain areas of the honeybee, zebra fish, mouse, and human, suggesting that the basic function of Mahya should be conserved in the brains of honeybee and vertebrates.
Mahya appears to be present in diverse bees classified as Apoidea (Fig. 6). The possibility that the Mahya genes we detected in the various bees by genomic PCR are pseudogenes could not be completely ruled out at this stage (it is, of course, necessary to isolate and sequence the full-length cDNAs). Nevertheless, we think this is unlikely because both Drosophila melanogaster and Anopheles gambiae genomes were shown to have small numbers of pseudogenes (176 and 166, respectively; Zdobnov et al. 2002) compared to mammals (more than 10,000). Also, the Mahya domain encoding exon sequence is absent in these model insects and silk moth genome. The bees apparently containing Mahya include both solitary and highly social bees. However, these bee species construct nests and bring food back to their nest for storage and for raising progeny. These results thus suggest that Mahya is not related to the high degree of sociality (colony maintenance by multiple individuals, caste differentiation, and generation overlap) but may be involved in the cognitive ability of bees associated with accurate spatial and olfactory recognition and memory (Giurfa et al. 2001).
What is the function of Mahya in sea urchin and sea squirt with relatively simple nervous systems, which do not exhibit complex behavior? If Mahya genes are expressed in the nervous systems of above animals, they would be involved in the primary processing of chemosensory information and not high order of learning and memory processes. Thus, the basic functions of Mahya conserved between Deuterostomes and Hymenoptera should be related with the mechanistic aspects of sensory information processing, which are different from those in fruit fly, mosquito, silk moth, and nematode. However, the possibility that Mahya genes are expressed in tissues other than nervous systems of sea urchin and sea squirt cannot be ruled out at this point.
The genome of Hymenoptera
It was reported that approximately 10% of the annotated genes in Drosophila melanogaster and Anopheles gambiae show better sequence matches with noninsect genes (Zdobnov et al. 2002). Although the percentage of this category of genes will likely be reduced after the completion of the Apis mellifera, Bombyx mori, and Tribolium castaneum genome projects, each insect must contain genes with specific functions shared between noninsect animal species. The existence of these genes reflects the selection and subsequent adaptation associated with different ecologies and life histories. In the case of Apis mellifera, we could predict the genes related with the high degree of sociality and cognitive ability. Furthermore, we can also expect honeybee-specific genes involved in polyphenism (caste differentiation) and in sex determination via haplodiploidy.
Gene loss and divergence in the evolution of metazoan genomes
Distribution of Mahya across Bilaterian species and phylogenetic relationship among Mahya proteins from different species suggest that the last common ancestors of Bilateria (Urbilateria) had the ancestral Mahya genes. It also indicates that Urbilateria was genetically quite complex with diverse sets of genes. We propose that the ancestral gene of Mahya was lost or became highly diverged in many metazoan species other than Hymenoptera and Deuterostomes. Even if highly diverged Mahya is present in many metazoan genomes, the ancestral function of Mahya would be more conserved between Hymenoptera and Deuterostomes. In terms of searching for the true ancestor of Mahya, it will be interesting to see if Mahya is present in the genomes of Cnidaria. This type of gene loss is now recognized as one of the scenarios leading to metazoan diversity (Kortschak et al. 2003). Even between two dipteran insects, fruit fly and mosquito, that diverged 250 million years ago, only mosquito contains genes homologous to human leukotriene B4 12-hydroxy dehydrogenase and calcineurin-binding intracellular regulatory protein (Zdobnov et al. 2002). Since fruit fly and honeybee diverged about 280 million years ago (Carpenter and Burnham 1985), we could also expect honeybee-specific gene loss and retention. Cross-species genome sequence comparison and gene functional analysis will undoubtedly give insights into how gene emergence, divergence, and loss have led the spectacular diversity of metazoans.
References
Ashburner M (1989) Drosophila a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Balemans W, Hul WV (2002) Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol 250:231–250
Carpenter FM, Burnham L (1985) The geological record of insects. Annu Rev Earth Planet Sci 13:297–314
Datta S, Mori Y, Takagi K, Kawaguchi K, Chen ZW, Okajima T, Kuroda S, Ikeda T, Kano K, Tanizawa K, Mathews FS (2001) Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking. Proc Natl Acad Sci U S A 98:14268–14273
Fahrbach SE, Robinson GE (1995) Behavioral development in the honey bee: toward the study of learning under natural conditions. Learn Mem 2:199–224
Funada M, Yasuo S, Yoshimura T, Ebihara S, Sasagawa H, Kitagawa Y, Kadowaki T (2004) Characterization of the two distinct subtypes of metabotropic glutamate receptor from honeybee, Apis mellifera. Neurosci Lett 359:190–194
Giurfa M, Zhang S, Jenett A, Menzel R, Srinivasan MV (2001) The concepts of ‘sameness’ and ‘difference’ in an insect. Nature 410:930–933
Ishihara A, Saito H, Abe K (1994) Transforming growth factor-beta 1 and -beta 2 promote neurite sprouting and elongation of cultured rat hippocampal neurons. Brain Res 639:21–25
Kikuno R, Nagase T, Nakayama M, Koga H, Okazaki N, Nakajima D, Ohara O (2004) HUGE: a database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE. Nucleic Acids Res 32(database issue):D502–D504
Kortschak RD, Samuel G, Saint R, Miller DJ (2003) EST analysis of the Cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol 13:2190–2195
Menzel R, Giurfa M (2001) Cognitive architecture of a mini-brain: the honeybee. Trends Cogn Sci 5:62–71
Menzel R, Muller U (1996) Learning and memory in honeybees: from behavior to neural substrates. Annu Rev Neurosci 19:379–404
Mineta K, Nakazawa M, Cebria F, Ikeo K, Agata K, Gojobori T (2003) Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc Natl Acad Sci U S A 100:7666–7671
Mushegian AR, Garey JR, Martin J, Liu LX (1998) Large-scale taxonomic profiling of eukaryotic model organisms: a comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res 8:590–598
Portavella M, Torres B, Salas C (2004) Avoidance response in gold fish: emotional and temporal involvement of medial and lateral telencephalic pallium. J Neurosci 24:2335–2342
Prechtl JC, von der Emde G, Wolfart J, Karamursel S, Akoev GN, Andrianov YN, Bullock TH (1998) Sensory processing in the pallium of a mormyrid fish. J Neurosci 18:7381–7393
Sacchetti B, Baldi E, Lorenzini CA, Bucherelli C (2002) Cerebellar role in fear-conditioning consolidation. Proc Natl Acad Sci U S A 99:8406–8411
Shi Y, Massague J (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113:685–700
Tsuchimoto M, Aoki M, Takada M, Kanou Y, Sasagawa H, Kitagawa Y, Kadowaki T (2004) The changes of gene expression in honeybee (Apis mellifera) brains associated with ages. Zoolog Sci 21:23–28
Whitfield CW, Brand MR, Bonaldo MF, Kumar CG, Liu L, Pardinas JR, Robertson HM, Soares MB, Robinson GE (2002) Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res 12:555–566
Yanez J, Anadon R (1996) Afferent and efferent connections of the habenula in the rainbow trout (Oncorhynchus mykiss): an indocarbocyanine dye (Dil) study. J Comp Neurol 372:529–543
Yoshida M, Okamura I, Uematsu K (2004) Involvement of the cerebellum in classical fear conditioning in gold fish. Behav Brain Res 153:143–148
Yoshimoto M, Ito H (1993) Cytoarchitecture, fiber connections, and ultrastructure of the nucleus pretectalis superficialis pars magnocellularis (PSm) in carp. J Comp Neurol 336:433–446
Yoshimura T, Suzuki Y, Makino E, Suzuki T, Kuroiwa A, Matsuda Y, Namikawa T, Ebihara S (2000) Molecular analysis of avian circadian clock genes. Brain Res Mol Brain Res 78:207–215
Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, Mueller H-M, Dimopoulos G, Law JH, Wells MA, Birney E, Charlab R, Halpern AL, Kokoza E, Kraft CL, Lai Z, Lewis S, Louis C, Barillas-Mury C, Nusskern D, Rubin GM, Salzberg SL, Sutton GG, Topalis P, Wides R, Wincker P, Yandell M, Collins FH, Ribeiro J, Gelbart WM, Kafatos FC, Bork P (2002) Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298:149–159
Acknowledgements
We thank M. Mizunami for helpful discussion. This study was supported by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to T.K.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Desplan
Rights and permissions
About this article
Cite this article
Tsuchimoto, M., Yasuo, S., Funada, M. et al. Conservation of novel Mahya genes shows the existence of neural functions common between Hymenoptera and Deuterostome. Dev Genes Evol 215, 564–574 (2005). https://doi.org/10.1007/s00427-005-0021-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-005-0021-z