Abstract
Floral organ identity B class genes are generally recognized as being required for development of petals and stamens in angiosperm flowers. Spinach flowers are distinguished in their complete absence of petals in both sexes, and the absence of a developed stamen whorl in female flowers. As such, we hypothesized that differential expression of B class floral identity genes is integral to the sexual dimorphism in spinach flowers. We isolated two spinach orthologs of Arabidopsis B class genes by 3′ and 5′ RACE. Homology assignments were tested by comparisons of percent amino acid identities, searches for diagnostic consensus amino acid residues, conserved motifs, and phylogenetic groupings. In situ hybridization studies demonstrate that both spinach B class genes are expressed throughout the male floral meristem in early stages, and continue to be expressed in sepal primordia in reduced amounts at later stages of development. They are also highly expressed in the third whorl primordia when they arise and continue to be expressed in these tissues through the development of mature anthers. In contrast, neither gene can be detected in any stage in female flowers by in situ analyses, although northern blot experiments indicate low levels of SpAP3 within the inflorescence. The early, strong expressions of both B class floral identity genes in male floral primordia and their absence in female flowers demonstrate that B class gene expression precedes the origination of third whorl primordia (stamen) in males and is associated with the establishment of sexual floral dimorphism as it initiates in the first (sepal) whorl. These observations suggest that regulation of B class floral identity genes has a role in the development of sexual dimorphism and dioecy in spinach rather than being a secondary result of organ abortion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Floral organ development is the best studied of the molecular genetic developmental pathways in plants. Characterization of the major genes and functional testing of their products as transcription factors have been established in the two model systems of Arabidopsis thaliana and Antirrhinum majus and is summarized in the ABC model (Coen et al. 1991; Coen and Meyerowitz 1991; Irish 1999; Jack 2004; Lohmann and Weigel 2002; Weigel and Meyerowitz 1994; Zik and Irish 2003a). However, the expansion of the ABC model to non-model species, as well as studies of variation within species, has been limited. First, comparative studies of the expression patterns of the floral homeotic genes in other species are still relatively few (Ainsworth et al. 1995; Angenent et al. 1993; Chuck et al. 1998; Kater et al. 2001; Kramer et al. 1998; Kramer and Irish 1999; Park et al. 2003; Riechmann and Meyerowitz 1998; Shu et al. 2000; Wang et al. 1999). Second, studies of intraspecific polymorphisms of homeotic genes and expression are almost non-existent (Cubas et al. 1999; Purugganan et al. 2000; Wang et al. 1999). At present, our understanding of the downstream regulatory effects of the master transcription factors is in its infancy. While interactions among some of the transcription factor genes are known (Busch et al. 1999; Doebley and Lukens 1998; Ito et al. 2004; Krizek and Meyerowitz 1996; Lohmann et al. 2001; Parcy et al. 1998; Samach et al. 1997; Wagner et al. 1999), potential downstream response genes are only beginning to be detected, even in Arabidopsis (Bey et al. 2004; Wellmer et al. 2004; Zik and Irish 2003b). Thus, how deviations from the Arabidopsis ABC model are related to morphological variation among floral structures in angiosperms, what proximal genes control morphological development in floral organs, and how environmental factors influence the developmental pathway remain largely unknown.
Plants with imperfect flowers provide a powerful tool for studying the floral developmental pathway as the developmental cascade found in hermaphroditic flowers must be altered to produce unisexual flowers. Two basic morphological pathways produce imperfect flowers (Heslop-Harrison 1964). In the first, all organ primordia are initiated and begin differentiation, only to degenerate or abort at a later stage. In the second, organ primordia for particular reproductive organs do not develop at all. In male flowers, carpel primordia do not form and in females stamen primordia are not initiated. Five species with imperfect flowers whose development have been studied on a molecular level, Zea mays (Ambrose et al. 2000; Delong et al. 1993; Munster et al. 2001), Rumex acetosa (Ainsworth et al. 1995), Silene latifolia (Hardenack et al. 1994; Janousek et al. 1996; Viskot et al. 1993), Cucumis sativus (Kater et al. 2001), and Asparagus officinalis (Park et al. 2003), all develop unisexual flowers following the first pathway.
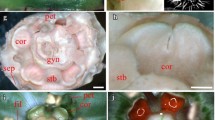
Cultivated spinach, Spinacia oleracea, follows the second developmental pathway (Pobursky 2000; Sherry et al. 1993), and thus provides a novel model for the analysis of floral development. In this dioecious species, both male and female flowers begin organ development with the growth of two opposite sepal primordia. The flowers then differentiate when the male sepal primordia are restricted in growth and two additional sepal primordia form in the space in the outer whorl between the initial sepal primordia. The two female sepal primordia extend laterally around the outer whorl and distally to overcanopy the central organs. Thus, sexual dimorphism is initiated before the appearance of reproductive organ primordia. Neither males nor females produce a second (petal) whorl in the perianth. The male flowers develop four stamens opposite the four sepals. The central space (fourth whorl) is flattened in the male flowers and no organs or primordia form (Fig. 1a). The female flowers do not establish organ primordia in the third whorl and only develop a single carpel in the central or fourth whorl. The initial fourth whorl primordia has a ring or girdling rim (Pobursky 2000; Sherry et al. 1993) similar to that found in Chenopodium album (Sattler 1973). This structure forms individual lobes that engulf the central region and fuse above it to form the pistil and stigmatic branches while the basal region forms the ovary wall. As the flower matures the central gynoecium protrudes from between the two sepals and a four-lobed stigma extends outward (Fig. 1b).
Studies have demonstrated that sexual determination in spinach can be altered by environmental stress (Freeman and Vitale 1985; Vitale and Freeman 1985), hormonal treatment (Chailakhyan 1979; Pobursky 2000), and developmental state (Miglia and Freeman 1996). Additionally, ectopic fourth whorl organs in males can be found at low frequencies in cultivar America (Pobursky 2000). Thus, it appears that genes that can control the development of all sexual organs are present in both male and female spinach, and therefore, altered gene regulation rather than gene presence or absence appears to control sex-specific development in spinach. As the B class genes are responsible for petal and stamen identity in Arabidopsis and Antirrhinum, and as neither male nor female spinach flowers produce petals, we hypothesized that B class gene expression should be clearly different in the two flower morphs. Here we report on the isolation, characterization, and expression of spinach homologs to the Arabidopsis B class genes, APETALA3 and PISTILLATA. We demonstrate that the two genes are expressed at very early stages and have widely overlapping expression patterns in male spinach flowers, while neither is detectable by in situ hybridization in any stage of female flower development. We relate the regulatory interactions of B class genes with potential downstream targets suggested by studies in Arabidopsis to produce a model for the role of spinach B class genes in the development of sexual dimorphism in the first whorl.
Materials and methods
Spinach AP3 isolation and sequencing
DNA was extracted from bagged leaf spinach using a standard CTAB extraction protocol (Doyle and Doyle 1987). PCR primers were designed by comparison of A. thaliana AP3 (accession no. A42095, Jack et al. 1992, 1994), AG (acc. no. S10933, Yanofsky et al. 1990), A. majus DEF (acc. no. S12378, Sommer et al. 1990), PLE (acc. no. S52900, Bradley et al. 1993), and Brassica napus Bag1 (acc. no. M99415, Mandel et al. 1992) gene sequences. A degenerate forward primer, degMADSF, was based primarily on the Arabidopsis AP3 gene starting from the start codon (5′ ATG GCI AGR GGI AAR AT 3′). The reverse primer, AP3-131R, was derived from the Arabidopsis AP3 sequence. The 3′ end of the primer falls at position 131 of the coding sequence and is within the MADS box. Three of four amino acids coded at this 3′ end differ between AP3 and AG, thus giving specificity to the primer (5′ TGT TGG AGC TAG AGA ACA TGA TAA TCG 3′). A 25-μl PCR reaction using 3 mM MgCl2 was set up, and Taq polymerase (Gibco BRL, Gaithersburg, Md.) was added to the reaction after incubation at 94°C for 2 min (hot start). The reaction conditions were 40 cycles of 94°C at 40 s, 52°C at 40 s, and 72°C for 1 min. The reaction product was cloned into a TA cloning vector (Invitrogen, Carlsbad, Calif.) and sequenced on an ALF automated sequencing machine following the manufacturer’s suggested protocol (Pharmacia, Peapack, N.J.). Sequence comparisons were done using Sequencher 3.0 (Gene Codes, Ann Arbor, Mich.). Spinach AP3-specific primers, SPMADS38F (5′ ACA ATA CGA ATC GTC AAG T 3′) and SPMADS77F (5′ AAC GGT CTG TTC AAG AAG G 3′), were designed from the spinach sequence. The last five bases of SPMADS38F and the last seven bases of SPMADS77F are conserved in comparison with AP3 homologous sequences from Dianthus caryophyllus CMB2 (acc. no. L40405, Baudinette et al. 2000), S. latifolia SLM3 (acc. no. X80490, Hardenack et al. 1994), R. acetosa RaD1 (acc. no. X89113, Ainsworth et al. 1995), and A. thaliana AP3 (acc. no. A42095, Jack et al. 1992, 1994).
Individual plants of S. oleracea cv America were grown from seed in a 3:1 mixture of potting soil and vermiculite. The plants were grown under long-day light conditions (18 h light:6 h dark) at 20°C and were watered as needed. Total RNA was extracted either from anthers or complete bolted inflorescences by grinding the tissue in the presence of Trizol, extracting once with chloroform, and precipitating the RNA with isopropanol, following the supplier’s protocol (Gibco BRL, Gaithersburg, Md.). 3′ RACE was executed using 3′ RACE System for Rapid Amplification of cDNA Ends (Gibco BRL, Gaithersburg, Md.). Primary PCR was carried out in 50-μl reactions using the abridged universal adaptor primer (AUAP), the gene-specific SPMADS38F primer, Biolase (ISC Bioexpress, Kaysville, Utah), Biolase buffer (an NH4-based PCR buffer), and 3 mM MgCl2. The reactions were run in glass capillary tubes in an Idaho Technologies RapidCycler set at 94°C for 2 min followed by 35 cycles of 94°C for 12 s, 47°C for 12 s, and 72°C for 90 s. This was followed by a semi-nested secondary PCR using AUAP and SPMADS77F as primers under similar conditions for 30 cycles. PCR products were cloned into pCRII (Invitrogen, Carlsbad, Calif.) and sequenced using cycle-sequencing (Big Dye; PE Applied Biosystems, Foster City, Calif.). The precipitated sequencing reaction products were read on an ABI Prism Model 377, version 3.0.
Spinach PI isolation and sequencing
The spinach PI gene was isolated by 5′ and 3′ RACE as described above. Three nested forward primers were designed from the S. latifolia SLM2 sequence (acc. no. X80489, Hardenack et al. 1994) in comparison with sequences from A. majus GLO (acc. no. S28062, Tröbner et al. 1992), A. thaliana PI (acc. no. D30807, Goto and Meyerowitz 1994), and Petunia hybrida PMADS2 (acc. no. X69947, Kush et al. 1993). The primers were PI.2F (5′ TAA TGG GTA GAG GAA AAA T 3′), PI.59F (5′ CTT ACT CAA AGA GAA GAA ATG G 3′), and PI.100F (5′ GAG ATC ACT GTT CTT TGT GA 3′). PCR products were cloned using TOPO TA Cloning Kit with vector pCR 2.1 (Invitrogen, Carlsbad, Calif.) and sequenced using ABI Prism Model 377, version 3.0. From these sequences, nested primers for 5′ RACE were designed. PI primers used were PI.542R (5′ AAA CCC GTA AGG AAG GTA 3′), PI.308R (5′TCC TCT CCA TTC AAG TGC 3′), and PI.216R (5′ TAG CAT CCC ACA ACC TCT TAC C 3′). 5′ RACE products were cloned and sequenced as above.
Sequence alignment and analysis
Sequences were imported into Sequencher 3.0 (Gene Codes, Ann Arbor, Mich.) and multiply aligned. Alignments were manually adjusted. Amino acid sequences were projected using the universal code. Amino acid sequences of the spinach AP3 and PI proteins were first aligned with SLM3 (accession number X80490), SLM2 (acc. no. X80489), AP3 (acc. no. D21125), PI (acc. no. D30807), GLO (acc. no. X68831), and DEFA (acc. no. X52023) using the nucleotide sequence as a guide. Following this initial alignment, the spinach sequences were aligned with additional proteins according to the alignment in Kramer et al. (1998). The initial alignments based on nucleotide sequences and those of Kramer et al. (1998) differed substantially at the carboxyl end. As such, the sequences were truncated to position 201 of Appendix 2 in Kramer et al. (1998) approximately 20 positions before the PI motif regions. Phylogenetic analysis of the amino acid sequences were completed using PAUP 3.0 (Swofford 1993) and Protdist and Neighbor programs of PHYLIP 3.573c (Felsenstein 1993). The parsimony searches used ten replicates with the TBR and MULPARS options in effect. Strength of parsimony branching inferences were tested in 1,000 bootstrap resamplings using the same options.
Northern blot
Total RNA was extracted from floral, and vegetative tissues by grinding the tissue in the presence of Trizol, extracting once with chloroform, and precipitating the RNA with isopropanol, following the supplier’s protocol (Gibco BRL, Gaithersburg, Md.). The RNA was size fractionated by electrophoresis and transferred to a nylon membrane using the Northen Max-Gly protocols (Ambion, Austin, Tex.). Truncated clones of SpAP3 and SpPI, which included only the C regions of each gene, were subcloned into pBluescript SK. Antisense RNA probes were generated for both SpAP3 and SpPI by transcription using the T7 and T3 promoters of pBluescript, respectively, and labeled by incorporation of digoxigenin-11- UTP. A portion of the spinach G6pdh gene was cloned using information from the published sequence AJ000182. An antisense probe of G6pdh was used as a positive control. The probes were hybridized with ULTRAhyb hybridization buffer following the manufacturer’s protocol (Ambion, Austin, Tex.) and detected by conjugation with anti-digoxigenin-AP and exposure to CDP-star following standard protocols.
In situ hybridization
In situ hybridizations were carried out following a modification of the mRNAlocator-Hyb system (Ambion, Austin, Tex.). Male and female inflorescences were dissected from cv America plants and incubated in fixative at 2–8°C for 8–12 h. They were then dehydrated in an ethanol-xylene series and imbedded in paraffin. The paraffin block was cooled on ice during sectioning. Sections were floated in nuclease-free water and positioned on Fisherbrand Superfrost/Plus microscope slides. Slides were incubated at 60°C overnight. The sections were deparaffinized and rehydrated in a reverse xylene-ethanol series. Tissues were treated with proteinase K for 20–30 min. Initial hybridizations using biotin-labeled probes resulted in non-specific background staining. Use of digoxygenin-labeled probes produced highly defined and sex-specific hybridization as detected by dark blue NBT staining with low background. SpAP3 expression was first tested using a probe containing the SpAP3 I, K, and C regions. Non-specific hybridization occurred throughout the floral tissues which may reflect cross hybridization to paralogous genes. A truncated probe containing just the unique C region (from coding position 432–834) generated tissue-specific hybridization patterns. Based on these results, SpPI expression was detected using an SpPI C region (from position 526 to the poly-A site) probe only. Sections were hybridized with digoxigenin RNA probes at 50–55°C. Hybridization was detected by incubation with anti-digoxigenin-AP followed by an NBT/BCIP solution. Slides were subsequently dehydrated, cleared, mounted, and stored at −20°C under desiccant.
Results
Spinach AP3 and PI isolation and sequence analysis
Utilizing a 3′ RACE, we isolated a 930-bp fragment, which was cloned and sequenced. The majority of clones gave identical sequences (shown in the Electronic Supplementary Material Fig. 1) in comparison with the S. latifolia AP3 ortholog SLM3 (X80490, Hardenack et al. 1994). A second cloned sequence was identical except for a 14-bp addition (AATGCTTGCTGGGT) at the 3′ UTR before the poly-A tail. This sequence has been deposited in GenBank as accession AY604514.
A second spinach B class sequence was differentially amplified by using PI consensus primers. The spinach PI- sequence was isolated in two overlapping fragments from independent 3′ and 5′ RACE reactions. The nucleotide sequence was aligned with Arabidopsis PI and Silene SLM2 (Electronic Supplementary Material Fig. 2). The spinach PI sequence has been deposited in GenBank as accession AY604515.
To infer homology of these sequences to published B class genes, we first compared nucleotide and translated amino acid sequences to known genes for general similarities and presence of diagnostic motifs. The SpAP3 (spinach APETALA3) sequence was aligned with A. thaliana APETALA3 (D21125, Thomas et al. 1992), and S. latifolia SLM3 (Hardenack et al. 1994). The percent amino acid identities between the spinach and Arabidopsis, spinach and Silene, and Arabidopsis and Silene are 76%, 82%, and 82% within the MADS domain, 53%, 53%, and 42% within the I region, and 44%, 65%, and 52% within the K box. Theißen et al. (1996) identified residues that are particularly characteristic of the different paralogs in the MADS gene family (marked in red in Electronic Supplementary Material Fig. 1). Specifically, the SpAP3 protein has a conserved phenylalanine at position 29 and a conserved methionine at position 47. It differs from the consensus residues at position 36 where it has a serine instead of a threonine (as does Antirrhinum DEF) and at position 42 where it has a threonine instead of a lysine (as does Silene SLM3; Electronic Supplementary Material Fig. 1).
Kramer et al. (1998, 2000) identified two conserved C terminal regions in AP3 homologs. The first region is reminiscent of a PI motif and is found in nearly all angiosperms listed. The second is an AP3-specific motif that occurs either as a euAP3 (derived) or paleoAP3 (ancestral) sequence. The spinach AP3 C terminal sequence is shown in Fig. 2a in comparison with that of S. latifolia. The PI Motif LALRLQPC is conserved between S. latifolia and spinach. The remaining QPNLH and the subsequent EuAP3 motif are absent in spinach and are replaced by eight residues, LMLVQDHV, before terminating. On the nucleotide level, this abrupt divergence in sequence is due to a 10-bp deletion that results in a frameshift (Fig. 2b). A second 6-bp deletion in the spinach relative to the Silene sequence is further downstream and occurs in the intervening, variable region between the PI motif and the euAP3 motif. If the spinach sequence were to be translated in the original reading frame, an euAP3 motif (GS) CVTTYTLL would be present, indicating that the euAP3 motif was conserved on the nucleotide level following the 6-bp deletion. This conservation of a translated sequence suggests that the upstream 10-bp deletion is the more recent of the two deletions, and that the spinach sequence shares remnants of the expected motif consistent with the inferred homology.
Comparisons of euAP3 motifs and PI motifs between SpAP3 and SLM3. a Comparison of the amino acid sequences at the carboxyl ends of SLM3 and SpAP3. The euAP3-motif and PI-motif regions are boxed for comparison. The black triangles under the SLM3 sequence indicate the positions of the SpAP3 deletions on the nucleotide level. b Aligned nucleotide sequences of the ends of the coding regions of both SLM3 and SpAP3. Gaps are introduced into the SpAP3 sequence to improve sequence similarity and are marked with a colon. The SpAP3 sequence is then translated with the gaps to demonstrate the conserved coding sequence of this putative carboxyl end in comparison with SLM3. The first gap is a 10-bp deletion whereas the second gap is an in-frame 6-bp deletion
The SpPI (spinach PISTILLATA) translated amino acid sequences demonstrate a high degree of identity in the MADS box with the spinach sequence identity ranging from 88% compared to the Silene MADS box and 74% compared to the Arabidopsis sequence. The PI I region is more highly conserved than the comparable AP3 I region with 76% identity between spinach and Silene, and 59% identity between spinach and Arabidopsis, whereas the K region is comparable at 64% and 50% identities between spinach and Silene and spinach and Arabidopsis. Also, following the Theißen et al. (1996) prediction, the translated spinach sequence has the conserved PI serine residue at position 14 (marked in red in Electronic Supplementary Material Fig. 2).
The carboxyl terminus of the translated protein is highly variable and requires the placement of indels in the cDNA sequence to align the sequences. However, as opposed to the AP3 sequences, all indels are in frame. The first 13 amino acids following the K region are identical in Silene and spinach, but the sequences are then marked by extensive variation including indels until reaching the PI motif. The PI motif (Kramer et al. 1998; Kramer and Irish 2000) is present in the spinach sequence (PYGFRGQPNQ QG) two residues before the carboxyl end of the protein, but differs from the consensus by a deletion of a PNL motif (dotted underline in Electronic Supplementary Material Fig. 2). Similar internal deletions are reported in other species by Kramer et al. (2000). Therefore, both general sequence similarity and presence of diagnostic motifs support the homology of the two isolated spinach sequences to previously characterized B class genes.
Phylogenetic analysis of spinach B class genes
To test for orthology of the spinach sequences, we carried out phylogenetic analyses to assess systematically consistent grouping of the spinach sequences to those of other species. Parsimony analysis produced two equally parsimonious trees (Fig. 3a). The two maximum parsimony trees support the separation of AP3/DEF orthologs and PI/GLO orthologs into two distinct clades. They differ only in the placement of the AP3 root, resulting in the placement of the PhAP3 sequence as either a sister taxon to the CpAP3/DeAP3 clade or to the remaining AP3 orthologs. The consensus tree of 1,000 bootstrap resamplings using parsimony shows 100% support for the separate AP3 and PI clades. Furthermore, a number of other hierarchical groupings are also strongly supported. Of particular interest in this study, SLM2 and SpPI form a clade with 90% bootstrap support, and SLM3 and SpAP3 form a clade with 95% bootstrap support which would be predicted given accepted phylogenetic relationships of the species. The Neighbor-joining tree does not differ significantly from that of the parsimony analysis and again shows the placement of SpAP3 with SLM3 in the AP3 lineage, and SpPI with SLM2 in the PI lineage (Fig. 3b). Thus, phylogenetic analyses support the assignment of SpAP3 and SpPI as AP3 and PI orthologs.
Phylogenetic analysis of amino acid sequences of AP3 and PI homologs. a One of two maximum parsimony trees. The trees were generated using heuristic search settings with random taxon additions in PAUP. Optimal trees were search using TBR branch-swapping using the MULPARS option. The shortest trees required 1,013 steps. b Neighbor-joining tree. Distances were generated using the Dayhoff PAM matrix. The tree was generated using the Neighbor program of PHYLIP 3.5
Northern hybridization of SpAP3 and SpPI
Northern hybridization analysis was carried out to determine whether our sequences hybridized to unique or multiple mRNA molecules and to determine in which organs the genes are expressed. Antisense probes of the 3′ ends of both genes were used to challenge two independent total RNA blots The blots included extracts from male inflorescences, female inflorescences, stem, and leaf tissues. Both probes generated a single band in the male inflorescence RNA (Fig. 4). In the case of SpAP3, a weak signal was detected in the female inflorescence RNA, but not in the stem or leaf extracts. SpPI was not detected in any additional tissue extracts. Positive controls using a spinach G6pdh antisense probe indicate hybridization in all RNA extracts. These results indicate that each probe hybridizes to a unique transcript and that the gene expression patterns are broadly floral and gender specific.
Northern blot hybridizations of SpAP3 and SpPI. Hybridizations using SpPI and SpAP3 3′-end probes. The first lane is male inflorescence RNA; the second lane is female inflorescence RNA; the third lane is stem RNA; the fourth lane is leaf RNA. The control hybridization using a spinach G6pdh probe of the filter is shown below
In situ hybridization analyses of SpAP3 and SpPI
Fine scale spatial and temporal expression patterns were determined by in situ hybridization of SpAP3 and SpPI on thin sections of male and female inflorescences. SpAP3 and SpPI demonstrated similar expression patterns. SpAP3 was detected through all stages of male flower development. At the earliest developmental stage when no organ primordia can be distinguished, SpAP3 was detected throughout the floral meristem (Fig. 5a). As the sepal primordia form, SpAP3 expression could be seen in the sepal primordia, mainly in the central cells. Once the stamen primordia formed, SpAP3 expression became more concentrated in the developing stamen (Fig. 5e). Expression in the sepals was reduced at this stage, though was still detectable in cells in the central layer. As the locules differentiate within the developing anthers and the microsporangia form by meiosis, SpAP3 transcripts were detected in the tapetum of the locule walls as well as in the microsporangia (Fig. 5I). There was no detectable signal in the male flowers at any stage using the sense probe of the SpAP3 C region (Fig. 5m).
In situ hybridization of spinach SpAP3 and SpPI in male and female flowers. Male flowers are in a, e, I, m of the first column , c, g, k, o of the third column , and q and r. Female flowers are in b, f, j, n of the second column , d, h, l, p of the fourth column , and s and t. Stages 1 and 2 male flowers are shown in a and c. Stages 4 and 5 are shown in e, g, m, o, q, and r. Transverse sections of stage 6 anthers showing locules and microsporangia are seen in I and k. Female stages 1 and 2 flowers are seen in b and d. Stages 3 and 4 are seen in f, h, j, l, n, and p. f, l Transverse sections through female flowers. SpAP3 antisense hybridization is shown in a, b, e, f, j, and l. m, n Male and female negative controls using an SpAP3 sense probes. SpPI antisense hybridization is shown in c, d, g, h, k, and l. o, p Male and female SpPI negative controls using an SpPI sense probe. q, s Spinach G6pdh positive controls using an antisense probe for males and females, respectively. s, t G6pdh negative controls. Additional time for NBT signal development was given to the female samples in order to detect faint signal should it be present
SpPI is also expressed throughout all stages of male flower development. SpPI transcripts were first detected in the earliest stages (before the establishment of stamen primordia) of the male flower (Fig. 5c). SpPI was expressed at high levels in the stamen primordia as they developed (Fig. 5g). At this stage, SpPI transcripts were also detected in the sepals, though at lower levels. As the stamen developed further, SpPI became more strongly expressed in the anthers, eventually with stronger signals within the locule (Fig. 5k). As with SpAP3, there is no detectable signal at any stage using the sense strand probe (Fig. 5o).
Figure 5b, f and j shows sections of female flowers in progressive stages of development. There was no detectable level of SpAP3 hybridization in any female section. These results are in contrast to the northern blot in which a faint SpAP3 signal was detected. The lack of detection in the in situ hybridizations may be the result of low and/or transient signal per cell. Figure 5d, h, and l shows sections of female flowers in comparable developmental stages to the males hybridized with SpPI and stained following the conditions used on male tissue. As with SpAP3, there is no detectable level of SpPI expression in any of the female sections. Negative controls of sense probes for SpAP3 and SpPI are shown in Fig. 5n and p, respectively. In positive controls using a spinach G6pdh probe, hybridization was detected throughout the female inflorescence using anti-sense probes and no hybridization was detected using sense probes (Fig. 5q–t). This indicates that the lack of B class signal in female tissue is a gene-specific effect and not a result of the inability to hybridize to female tissue preparations.
In summary, the in situ hybridization experiments demonstrate that B class genes are exclusively expressed together in developing male flowers in spinach and not in female flowers. This gender-specific pattern differs from the Arabidopsis expression pattern in its early expression throughout the male floral primordium prior to the appearance of sepal primordia and its persistent, although declining, expression in the sepals after the stamen primordia form.
Discussion
Our results indicate that spinach flower sexual differentiation is initiated at a very early stage and is accompanied by early dimorphism. The prediction that spinach homologs of the Arabidopsis B class genes, AP3 and PI, would be differentially expressed in males versus females has been verified by in situ hybridization. In addition to the expected expression patterns, however, further deviations from the Arabidopsis model indicate that spinach B class gene expression is temporally correlated with morphological differentiation in terms of organ number and placement outside of their expected effects in whorls two and three.
Spinach B class genes follow the Arabidopsis/Antirrhinum expression patterns in the overlapping expression of SpAP3 and SpPI, and their dominant concentration in the third whorl by stage 4 and following stages. The absence of detectable expression in female flowers that do not have second or third whorl primordia is consistent with this pattern. It should be noted that the homologous genes SLM3 and SLM2 of S. latifolia are initially expressed in petals and stamens of both male and female flowers (Hardenack et al. 1994). Similar to its spinach ortholog in males, SLM3 appears to have a more diffuse expression pattern in the initial floral meristem but then becomes concentrated in the petal and sepal primordia. This is consistent with B class gene expression being spatially fixed in second and third whorls in higher eudicots, regardless of the ultimate fate of the organs. However, in contrast, the spinach expression patterns differ in the very early expression of both genes in stage 1, and the expression in sepals beginning in stage 2 in males and persisting at lower intensities even after the stamen primordia form and develop. In the case of the first whorl primordia, flowers of both sexes physically retain the organs. Thus, in spinach, there is a novel region of expression that is sex-specific and independent of the presence/absence of the organs involved.
In Arabidopsis, primordia are initiated and subsequently differentiate; i.e. organ identity is separate from and occurs subsequent to organ primordia initiation. Many molecular studies have demonstrated that the organ fate of floral organ primordia is determined by organ identity genes (Bowman et al. 1989, 1991, 1993; Bowman and Smyth 1998; Chuang et al. 1999; Hill and Lord 1989; Irish and Sussex 1990; Jack et al. 1992; Komaki et al. 1988; Kunst et al. 1989; Weigel and Meyerowitz 1994). Therefore, the order and placement of organ primordia are considered to occur independently of the function of floral identity genes. Genes that control the number and placement of organ primordia primarily do so by altering cell division and differentiation in meristem growth (Chuang et al. 1999; Clark et al. 1993; Fletcher 2001; Running et al. 1998; Running and Meyerowitz 1996). Larger meristems, particularly larger areas in the circumference where cell differentiation begins, often results in the development of additional primordia and ultimately additional organs.
The development of spinach sexual dimorphism may deviate from this strict model of distinction between organ identity genes and organ number genes. The sex of a spinach flower can be accurately predicted by sepal number, which is determined before either stamen or carpel primordia form. In contrast to the studies of mutants in Arabidopsis in which organ number is modified by an increase in meristem size or rate of cell differentiation, the increased number of sepals in males appears to be a result of the restriction of the lateral field of sepal growth around the first whorl found in females. The additional male sepal primordia then develop in the spaces in the circumference of the first whorl. Several genes that establish organ boundaries in Arabidopsis, Petunia, and Pisum belong to the NAC gene family (Aida et al. 1997, 1999; Liu et al. 1999) which differs from gene families or systems that control meristem growth and differentiation. Similarly, an unrelated gene SUPERMAN controls organ number in both Arabidopsis and Petunia by regulating cell division in regions around primordia and between whorls (Bowman et al. 1992; Nakagawa et al. 2004; Sakai et al. 1995; Schultz et al. 1991). It is noteworthy that both NAP, an Arabidopsis NAC family gene, and SUPERMAN expressions have been reported to be positively regulated by AP3 and PI (Sablowski and Meyerowitz 1998; Sakai et al. 2000). Thus, B class gene expression can potentially affect organ number through the regulation of such genes. In Arabidopsis, this control is not exclusive, as NAP is expressed in tissues in the absence of B class expression (Sablowski and Meyerowitz 1998). Our observations show that spinach floral sexual dimorphism of non-reproductive organs is tightly associated with sex-specific B class gene expression patterns. This suggests that, in spinach, the regulation of organ identity genes may be associated with developmental features in addition to organ identity. The involvement of B class organ identity genes on organ number through regulation of downstream organ boundary genes would be a likely mechanism.
The gender-specific B class expression patterns in spinach differ in significant ways from those found in other species with unisexual flowers that have been previously studied (reviews in Ainsworth et al. 1998; Grant et al. 1994; Juarez and Banks 1998; Tanurdzic and Banks 2004). In Z. mays and C. sativus, hormone levels, especially of gibberellic acid, play a role in determination of the ultimate sex of the flower (Delong et al. 1993; Hansen 1976; Kahana et al. 1999; Perl-Treves 1999; Yin and Quinn 1995). In comparison, regulation of floral organ identity genes has not been causally related to sex determination. In R. acetosa, S. latifolia, A. officinalis, and C. sativus, B class genes are initially expressed in primordia of the second and third whorls following the Arabidopsis pattern. The expression becomes non-detectable upon degeneration of the respective organs (Ainsworth et al. 1995; Hardenack et al. 1994; Kater et al. 2001; Park et al. 2003). Similarly, in Z. mays, the B class homolog SILKY1 is initially identically expressed in both male and female flowers (Ambrose et al. 2000). There is evidence in Silene that differential methylation is correlated with sexual phenotypes and methylation inhibition results in the production of hermaphroditic plants with male karyotypes (Janousek et al. 1996; Viskot et al. 1993). Yet, in general, these studies suggest that sex determination is not a result of initial regulation of floral identity genes in species that develop imperfect flowers through abortion of existing or developing organs. The unique pattern of early suppression of B class expression in spinach female floral primordia may, therefore, reflect the unique development of unisexual flowers without organ abortion.
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Aida M, Ishida T, Tasaka M (1999) Shoot apical meristem and cotyledon formation during arabidopsis embryogenesis: interaction among the cup-shaped cotyledon and shoot meristemless genes. Development 126:1563–1570
Ainsworth C, Crossley S, Buchanan Wollaston V, Thangavelu M, Parker J (1995) Male and female flowers of the dioecious plant sorrel show different patterns of MADS box gene expression. Plant Cell 7:1583–1598
Ainsworth C, Parker J, Buchanan Wollaston V (1998) Sex determination in plants. Curr Top Dev Biol 38:167–223
Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky M, Schmidt RJ (2000) Molecular and genetic analysis of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5:569–579
Angenent GC, Franken J, Busscher M, Colombo L, van Tunen AJ (1993) Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J 4:101–112
Baudinette SC, Stevenson TW, Savin KW (2000) Isolation and characterisation of the carnation floral-specific MADS box gene, cmb2. Plant Sci 155:123–131
Bey M, Stuber K, Fellenberg K, Schwarz-Sommer Z, Sommer H, Saedler H, Zachgo S (2004) Characterization of antirrhinum petal development and identification of target genes of the class B MADS box gene deficiens. Plant Cell 16(12):3197–3215
Bowman JL, Smyth DR (1998) Patterns of petal and stamen reduction in Australian species of Lepidium l. (Brassicaceae). Int J Plant Sci 159:65–74
Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1:37–52
Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112:1–20
Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM (1992) Superman, a regulator of floral homeotic genes in Arabidopsis. Development 114:599–615
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by apetala1 and interacting genes. Development 119:721–743
Bradley D, Carpenter R, Sommer H, Hartley N, Coen E (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of antirrhinum. Cell 72:85–95
Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285:585–587
Chailakhyan MK (1979) Genetic and hormonal regulation of growth, flowering, and sex expression in plants. Am J Bot 66:717–736
Chuang CF, Running MP, Williams RW, Meyerowitz EM (1999) The perianthia gene encodes a bzip protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev 13:334–344
Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the apetala2-like gene indeterminate spikelet1. Genes Dev 12:1145–1154
Clark SE, Running MP, Meyerowitz EM (1993) Clavata1, a regulator of meristem and flower development in Arabidopsis. Development 119:397–418
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
Coen ES, Doyle S, Romero JM, Elliot R, Murphy G, Carpenter R (1991) Homeotic genes controlling flower development in Antirrhinum. Development Suppl 1:149–155
Cubas P, Vincent C, Coen E (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401:157–160
Delong A, Calderon-Urrea A, Dellaporta S (1993) Sex-determination gene tasselseed2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74:757–768
Doebley J, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10:1075–1082
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11
Felsenstein J (1993) Phylip (phylogeny inference package). Department of Genetics, University of Washington, Seattle, Wash.
Fletcher JC (2001) The ultrapetala gene controls shoot and floral meristem size in Arabidopsis. Development 128:1323–1333
Freeman DC, Vitale JJ (1985) The influence of environment on the sex ratio and fitness of spinach. Bot Gazette 146:137–142
Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene pistillata. Genes Dev 8:1548–1560
Grant S, Houben A, Vyskot B, Siroky J, Pan W-H, Macas J, Saedler H (1994) Genetics of sex determination in flowering plants. Dev Genet 15:214–230
Hansen DJ (1976) Gibberellic acid-controlled sex expression of corn tassels. Crop Sci 16:371–374
Hardenack S, Ye D, Saedler H, Grant S (1994) Comparison of MADS box gene expression in developing male and female flowers of the dioecious plant white campion. Plant Cell 6:1775–1787
Heslop-Harrison J (1964) Sex expression in flowering plants. In: Brookhaven National Laboratory (eds) Brookhaven symposia in biology. Brookhaven National Laboratory, Upton, N.Y., pp 109–125
Hill JP, Lord EM (1989) Floral development in Arabidopsis thaliana: comparison of the wild type and the homeotic pistillata mutant. Can J Bot 67:2922–2936
Irish VF (1999) Patterning the flower. Dev Biol 209:211–220
Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2:741–753
Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM (2004) The homeotic protein agamous controls microsporogenesis by regulation of sporocyteless. Nature 430:356–360
Jack T (2004) Molecular and genetic mechanisms of floral control. Plant Cell 16:S1–S17
Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene apetala3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68:683–697
Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene apetala3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76:703–716
Janousek B, Siroky J, Vyskot B (1996) Epigenetic control of sexual phenotype in a dioecious plant, Melandrium album. Mol Gen Genet 250:483–490
Juarez C, Banks JA (1998) Sex determination in plants. Curr Opin Plant Biol 1:68–72
Kahana A, Silberstein L, Kessler N, Goldstein RS, Perl-Treves R (1999) Expression of acc oxidase genes differs among sex genotypes and sex phases in cucumber. Plant Mol Biol 41:517–528
Kater M, Franken J, Carney K, Colombo L, Angenent G (2001) Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 13:481–493
Komaki MK, Okada K, Nishino E, Shimura Y (1988) Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development 104:195–203
Kramer EM, Irish VF (1999) Evolution of genetic mechanisms controlling petal development. Nature 399:144–148
Kramer EM, Irish VF (2000) Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. Int J Plant Sci 161:S29-S40
Kramer EM, Dorit RL, Irish VF (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the apetala3 and pistillata MADS-box gene lineages. Genetics 149:765–783
Krizek BA, Meyerowitz EM (1996) Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc Natl Acad Sci USA 93:4063–4070
Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW (1989) Ap2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1:1195–1208
Kush A, Brunelle A, Shevell D, Chua NH (1993) The cDNA sequence of two MADS box proteins in petunia. Plant Physiol 102:1051–1052
Liu C-M, Johnson S, di Gregorio S, Wang T (1999) Single cotyledon (sic) mutants of pea and their significance in understanding plant embryo development. Dev Genet 25:11–22
Lohmann JU, Weigel D (2002) Building beauty: the genetic control of floral patterning. Dev Cell 2:135–142
Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105:793–803
Mandel MA, Bowman JL, Kempin SA, Ma H, Meyerowitz EM, Yanofsky MF (1992) Manipulation of flower structure in transgenic tobacco. Cell 71:133–143
Miglia K, Freeman DC (1996) The effect of delayed pollination on stigma length, sex expression, and progeny sex ration of spinach: a test of Fisher’s prediction. Am J Bot 83:326–332
Munster T, Wingen LU, Faigl W, Werth S, Saedler H, Theissen G (2001) Characterization of three globosa-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene 262:1–13
Nakagawa H, Ferrario S, Angenent GC, Kobayashi A, Takatsuji H (2004) The petunia ortholog of Arabidopsis superman plays a distinct role in floral organ morphogenesis. Plant Cell 16:920–932
Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395:561–566
Park JH, Ishikawa Y, Yoshida R, Kanno A, Kameya T (2003) Expression of aodef, a B-functional MADS-box gene, in stamens and inner tepals of the dioecious species Asparagus officinalis l. Plant Mol Biol 51:867–875
Perl-Treves R (1999) Male to female conversion along the cucumber shoot: approaches to studying sex genes and floral development in Cucumis sativus. In: Ainsworth C (ed) Sex determination in plants. Bios, Oxford, pp 189–216
Pobursky KJ (2000) Early flower development and the influence of gibberellic acid on sex expression in Spinacia oleracea. MSc, Wayne State University, USA
Purugganan MD, Boyles AL, Suddith JI (2000) Variation and selection at the cauliflower floral homeotic gene accompanying the evolution of domesticated Brassica oleracea. Genetics 155:855–862
Riechmann JL, Meyerowitz EM (1998) The ap2/erebp family of plant transcription factors. Biol Chem 379:633–646
Running MP, Meyerowitz EM (1996) Mutations in the perianthia gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122:1261–1269
Running MP, Fletcher JC, Meyerowitz EM (1998) The wiggum gene is required for proper regulation of floral meristem size in Arabidopsis. Development 125:2545–2553
Sablowski RWM, Meyerowitz EM (1998) A homolog of no apical meristem is an immediate target of the floral homeotic genes apetala3/pistillata. Cell 92:93–103
Sakai H, Medrano LJ, Meyerowitz EM (1995) Role of superman in maintaining Arabidopsis floral whorl boundaries. Nature 378:199–202
Sakai H, Krizek BA, Jacobsen SE, Meyerowitz EM (2000) Regulation of sup expression identifies multiple regulators involved in Arabidopsis floral meristem development. Plant Cell 12:1607–1618
Samach A, Kohalmi SE, Motte P, Datla R, Haughn G (1997) Divergence of function and regulation of class B floral organ identity genes. Plant Cell 9:559–570
Sattler R (1973) Organogenesis of flowers. University of Toronto Press, Toronto
Schultz EA, Pickett FB, Haughn GW (1991) The flo10 gene product regulates the expression domain of homeotic genes ap3 and pi in Arabidopsis flowers. Plant Cell 3:1221–1237
Sherry RA, Eckard KJ, Lord EM (1993) Flower development in dioecious Spinacia oleracea (Chenopodiaceae). Am J Bot 80:283–291
Shu GP, Amaral W, Hileman LC, Baum DA (2000) Leafy and the evolution of rosette flowering in violet cress (Jonopsidium acaule, Brassicaceae). Am J Bot 87:634–641
Sommer H, Beltran J, Huijser P, Pape H, Lonnig W, Saedler H, Schwarz-Sommer Z (1990) Deficiens, a homeotic gene involved in the control of flower morphogenesis. EMBO J 9:605–613
Swofford DL (1993) PAUP: phylogenetic analysis using parsimony. Illinois Natural History Survey, Champaign, Ill.
Tanurdzic M, Banks JA (2004) Sex-determining mechanisms in land plants. Plant Cell 16:S61–S71
Theißen G, Kim JT, Saedler H (1996) Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in morphological evolution of eukaryotes. J Mol Evol 43:484–516
Thomas J, Laura BL, Meyerowitz EM (1992) The homeotic gene apetala3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 683–697
Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lonnig WE, Saedler H, Sommer H, Schwarz-Sommer Z (1992) Globosa: a homeotic gene which interacts with deficiens in the control of antirrhinum floral organogenesis. EMBO J 11:4693–4704
Viskot B, Araya A, Veuskens J, Negrutiu I, Mouras A (1993) DNA methylation of sex chromosomes in a dioecious plant, Melandrium album. Mol Gen Genet 239:219–224
Vitale JJ, Freeman DC (1985) Secondary sex characteristics in Spinacia oleraceae l.: quantitative evidence for the existence of at least three sexual morphs. Am J Bot 72:1061–1066
Wagner D, Sablowski RWM, Meyerowitz EM (1999) Transcriptional activation of apetala1 by leafy. Science 285:582–584
Wang R-L, Stec A, Hey J, Lukens L, Doebley J (1999) The limits of selection during maize domestication. Nature 398:236–239
Weigel D, Meyerowitz EM (1994) The abcs of floral homeotic genes. Cell 78:203–209
Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16:1314–1326
Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346:35–39
Yin TJ, Quinn JA (1995) Tests of a mechanistic model of one hormone regulating both sexes in Cucumis sativus (Cucurbitaceae). Am J Bot 82:1537–1546
Zik M, Irish VF (2003a) Flower development: initiation, differentiation, and diversification. Annu Rev Cell Dev Biol 19:119–140
Zik M, Irish VF (2003b) Global identification of target genes regulated by apetala3 and pistillata floral homeotic gene action. Plant Cell 15:207–222
Acknowledgements
We are indebted to D. Carl Freeman for his introduction to and insight into the sexual dimorphism and lability of spinach. We thank John M. Lopes, Aleksandar Popadic, Markus Friedrich, and two anonymous reviewers for their insightful comments on a previous version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by G. Jürgens
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Pfent, C., Pobursky, K.J., Sather, D.N. et al. Characterization of SpAPETALA3 and SpPISTILLATA, B class floral identity genes in Spinacia oleracea, and their relationship to sexual dimorphism. Dev Genes Evol 215, 132–142 (2005). https://doi.org/10.1007/s00427-004-0459-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-004-0459-4