Abstract
Main Conclusion
A wheat RPP13-like isoform interacting with WPP1 contributes to quantitative and/or basal resistance to powdery mildew (Blumeria graminis f. sp. tritici) by restricting the development of Bgt conidia.
Abstract
Plant disease resistance (R) genes confer an ability to resist infection by pathogens expressing specific avirulence genes. Recognition of Peronospora parasitica 13-like (RPP13-like) genes belong to the nucleotide-binding site and leucine-rich repeat (NBS-LRR) superfamily and play important roles in resistance to various plant diseases. Previously, we detected a TaRPP13-like gene located on chromosome 3D (TaRPP13L1-3D) in the TaSpl1 resided region, which is strongly induced by the cell death phenotype (Zhang et al. 2021). Here, we investigated the expression and functional role of TaRPP13L1-3D in wheat responding to fungal stress. TaRPP13L1-3D encoded a typical NB-ARC structure characterized by Rx-N and P-loop NTPase domains. TaRPP13L1-3D transcripts were strongly upregulated in wheat by powdery mildew (Blumeria graminis f. sp. tritici; Bgt) and stripe rust (Puccinia striiformis f. sp. tritici; Pst) infection although opposing expression patterns were observed in response to wheat-Bgt in incompatible and compatible backgrounds. Overexpression of TaRPP13L1-3D enhanced disease resistance to Bgt, accompanied by upregulation of the defense-related marker genes encoding phytoalexin-deficient4 (PAD4), thaumatin-like protein (TLP) and chitinase 8-like protein (Chi8L), while silencing of TaRPP13L1-3D disrupted the resistance to Bgt infection. Subcellular localization studies showed that TaRPP13L1-3D is located in both the plasma membrane and nucleus, while yeast-two-hybrid (Y2H) assays indicated that TaRPP13L1-3D interacts with WPP domain-containing protein 1 (TaWPP1). This indicates that TaRPP13L1-3D shuttles between the nucleus and cytoplasm membrane via a mechanism that is mediated by the RanGAP-WPP complex in nuclear pores. This insight into TaRPP13L1-3D will be useful in dissecting the mechanism of fungal resistance in wheat, and understanding the interaction between R gene expression and pathogen defense.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat powdery mildew (caused by Blumeria graminis f. sp. tritici; Bgt) and stripe rust (Puccinia striiformis f. sp. tritici; Pst) are important fungal diseases of wheat leading to significant yield losses. Deployment of disease resistance genes were thought of as one of the most economical and effective methods to control fungal disease outbreaks (Wellings 2011). To date, over 85 powdery mildew resistance (R) genes (including alleles) and 83 stripe rust R genes have been documented in cultivated wheat and related species (Ma et al. 2020; Lu et al. 2020a; Jia et al. 2020; Morales et al. 2021; Pang et al. 2021). However, the emergence of resistant cultivars with a limited number of resistance genes (even as few as 1 or 2) as well as mutations of increases the frequency of virulence genes, commonly resulting in decreased specific resistance function (Zhao et al. 2013, 2016; Zhang et al. 2016). Pyramiding of genes is reported to be an effective method for improving R gene utilization efficiency (Krattinger and Keller 2016). Therefore, identification of resistance-related genes and their effective introduction into cultivated wheat has become an important focus of plant breeding to improve resistance to stripe rust and powdery mildew.
Plant disease resistance gene products recognize avirulence (Avr) gene products, triggering a chain of signal transduction events that culminate in the activation of defense responses (Adachi et al. 2019; Wang et al. 2020a). To date, 300 R genes have been identified in crops, of which, about 40 have been shown to participate in the responses of wheat to powdery mildew (Kourelis and van der Hoorn 2018; Lu et al. 2020a; Liu et al. 2020; Hewitt et al. 2021) and stripe rust (Fu et al. 2009; Liu et al. 2014; Wang et al. 2019). Most R proteins have leucine-rich repeat (NB-LRR) domains, but show structural and functional diversity (Baggs et al. 2017). According to sequence structure, these proteins are classified as serine/threonine kinases, transmembrane receptor proteins, ATP enzymes, and nucleotide-binding site (NBS)-LRR resistance proteins. Based on their N-terminal characteristics, NBS-LRR proteins are divided into Toll/interleukin-1 receptor (TIRs) or Coiled-coil (CC) domain subfamilies (Eitas and Dangl 2010; Sekhwal et al. 2015). RPP13 is a single dominant resistance gene that confers specific recognition of Peronospora parasitica (downy mildew) effector ATR13 (Hall et al. 2009) and elicits an interaction phenotype of flecking or extensive pitting necrosis (Bittner-Eddy et al. 1999). The RPP13 protein has a typical CNL domain, which induces disease resistance. TaRPP13-3 was reported to function in promoting disease resistance to wheat powdery mildew (Liu et al. 2020). A single dominant resistance gene, TaSpl1, has been shown to simulate an interaction phenotype of flecking necrosis in the absence of inhibitors (Zhang et al. 2021). Referencing the genome of Chinese Spring, the candidate gene TaSpl1 was predicted to be located in the physical region from 1,337,605 to 15,925,318 bp in chromosome 3D. Among these intra-segment genes, TraesCS3D02G032100 was identified as a differentially expressed gene and annotated as RPP13-like protein 1, with 98% identity. Since programmed cell death (PCD) is typical symbol of plant resistance to stripe rust and powdery mildew, we investigated the function of RPP13L in the responses of wheat to Bgt and Pst pathogens.
Materials and methods
Plant materials and pathogen stress treatment
For Bgt tests, a pair of near isogenic lines (NILs), designated N9134R2 (resistant) and N9134S2 (susceptible), were developed from the Shaanyou 225/7 × N9134 backcross (PmAS846) (Guo et al. 2021). For Pst tests, a pair of recombinant inbred lines (RILs), designated PD34R and PD34S, were developed from Shaanmai 159 × Pindong 34 (Yr61) (Zhou et al. 2014). The Bgt isolate E09 and the Pst isolate CYR34 were maintained on Shaanyou 225 (SY225) and Mingxian 169 (MX169) wheat plants, respectively. The contrasting NIL (N9134R2/S2) and RIL (PD34R/S) plants were cultivated in soil in a growth chamber at 18 ˚C with a 16-h light/8-h dark photoperiod. At the three-leaf stage, seedlings were inoculated with Bgt or Pst conidia from pre-inoculated SY225 or MX169 seedlings. The Bgt-E09 inoculated and mock-inoculated N9134R2/S2 leaves were harvested at 0, 6, 12, 24, 36, 48, 72, and 96 h post-inoculation (hpi), and immediately frozen in liquid nitrogen for storage at − 80 ˚C. Similarly, the Pst-PD34R/S leaves were harvested at 0, 24, 48, 96 and 192 hpi. The second and third leaves from three plants were collected and mixed as one sample, and three biological replicates were included for all analyses.
Wheat RPP13L1-3D homolog cloning and sequence analysis
Wheat RPP13L1-3D was amplified by PCR with specific primers cRPP13L1-3D-F/R (Table S1) covering the whole open reading frame and using cDNA from leaves of N9134R2 at 2 days post-inoculation with avirulent Bgt race E09 as the template. The PCR products were purified from agarose gel and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA) according to the manufacturer’s protocol. The nucleotide sequences of the positive clones were determined by AuGCT DNA-SYN Biotechnology Co. Ltd (Yangling, China). The protein sequence encoded by TaRPP13L1 was predicted using InterProScan (http://www.ebi.ac.uk/) and the deduced amino acid sequence was aligned with other RPP13 homologue genes using the MegAlign program in Bioedit. A phylogenetic neighbor-joining tree was generated with MEGA software (Kumar et al. 2018).
Real-time quantitative PCR analysis
TaRPP13L1-3D expression profiles in infected wheat leaves of NILs/RILs were determined by real-time quantitative PCR (Q-PCR) analysis of cDNA samples using SYBR Green. Q-PCR was performed on the QuantStudio 7 Flex Real-Time PCR System (Life Technologies Corporation). Sequence specific primers qRPP13L1-3D-S/AS, qGAPDH-S/AS and qActin-S/AS (Table S1), designed by the Primer 5 program, were used to quantify the accumulation of TaRPP13L1-3D transcripts and to normalize the amounts of cDNA in samples, respectively. The amplification was conducted in a 20-μL volume according to the SYBR Premix Ex Taq manual (Takara, Dalian, China) with the following conditions: 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s and 63 °C for 34 s. For each sample, reactions were carried out in triplicate and three non-template negative controls were included. Products were analyzed by melting curves obtained at the end of process to confirm amplification of a single product. The standard 2−ΔΔCT method was employed to quantify the relative gene expression. Mean values and standard errors were calculated with Microsoft Excel software. Data were analyzed by Student’s t-test with the SPSS 16.0 program to assess the significance of any differences between the control and treated samples or between time-points. The threshold for statistical significance was set at P < 0.05.
Vectors construction, subcellular localization and overexpression assay
The overexpression plasmid was based on the PYJ::GFP vector driven by the Cauliflower mosaic virus (CaMV) 35S promoter (35Spro). To construct PYJ:TaRPP13L1-3D:GFP, the TaRPP13L1-3D open reading frame (ORF) sequences were ligated into the SpeI-cut plasmid using ClonExpressII One Step Cloning Kit (Vazyme Biotech, Nanjing, China). The recombinant plasmid was transformed into Agrobacterium tumefaciens EHA105 using the freeze–thaw method. A single colony was inoculated into 50 mL liquid LB medium (50 mg/L rifampicin and 50 mg/L kanamycin) and cultured at 28 °C. The bacterial culture was then centrifuged for 10 min. For overexpression, the pellet was resuspended in suspension buffer (10 mM MgCl2, 10 mM MES, and 150 μm acetosyringone) to obtain an OD600 value of 0.8–1.0 according to a previously described protocol (Sparkes et al. 2006). Bacteria suspended in infiltration media were injected into wheat leaves at the two-leaf stage with a syringe; leaves were injected with A. tumefaciens carrying the PYJ::GFP empty vector as a control. After 36 h, the wheat leaves were inoculated with powdery mildew E09. Tests were replicated three times in 12 plants. The efficiency TaRPP13 overexpression was evaluated by quantification of transcript levels in leaves collected at 12, 48 hpi with E09 infection. The injected leaves were also collected at 24, 48 and 72 hpi with powdery mildew fungus, and spore development was observed and photographed under a microscope after Coomassie brilliant blue R-250 staining. The mycelium length was measured by ImageJ (Barry et al. 2009). For determination of subcellular localization, bacteria cultured in infiltration media were injected into 4-week-old tobacco (Nicotiana benthamiana) leaves and cultured for approximately 48 h in a growth chamber under normal growth conditions. The transformed epidermal cells were also stained for 3 min with FM4-64 dye (Invitrogen) to mark the plasma membrane. To stain the nucleus, the treated onion epidermis was immersed in 4',6-diamidino-2-phenylindole (DAPI) working solution at room temperature for 5–20 min (depending on the staining results of the experimental materials). After 20 min, the localization of cells was observed using a fluorescence confocal microscope at a detection wavelength 488 nm.
Gene silencing induced by tobacco-transcribed BSMV-RNA in wheat
The barley stripe mosaic virus (BSMV) RNA-induced gene silencing (VIGS) system was utilized to silence TaRPP13L1-3D. Briefly, a 178-bp fragment of the TaRPP13L1 homolog gene isolated by RT-PCR was inserted into SapI-digested pCB301-BSMV-γ by homologous recombination (Clontech). The resulting plasmid, designated γRPP13L, was sequenced by AuGCT DNA-SYN Biotechnology Co. Ltd. After the pCB301-BSMV-α, -β and -γRPP13L constructs were transformed into A. tumefaciens EHA105, the purified Agrobacterium cultures α, β, and γRPP13L were mixed and transformed into Nicotiana benthamiana to transcribe RNA in vivo. The empty γ0 vector was used as a negative control, while a construct encoding a 214-bp fragment of the wheat phytoene desaturase (PDS) gene, designated γPDS, was also generated. After 5 days, the second leaves of N9134R2/S2 seedlings (2.5-leaf stage) were inoculated with the phosphate diluted tissue abrasive fluid BSMV-RPP13L, BSMV-blank vectors, tap water (referred to as mock-BSMV inoculation) and BSMV-PDS. The seedlings were then incubated at 22 °C under 80% relative humidity for 12 h in the dark and then at 18 °C–22 °C in a growth cabinet (RLD-1000D-4DW, Ningbo, China). After approximately 7 days (when the PDS silencing phenotype appeared), the three types of infected seedlings and the corresponding control plants (SY225 for powdery mildew) were challenged with Bgt race E09, and maintained at 18 °C–22 °C until the susceptible variety SY225 expressed powdery mildew symptoms. Tests were replicated three times. For monitoring of silencing, the efficiency of TaRPP13L1-3D silencing was evaluated by quantification of transcript levels in leaves collected at 0, 12, 48 hpi with E09.
Yeast two-hybrid assays
Protein–protein interactions among TaRPP13L1, RanGAP2, and WPPs were evaluated using the MatchMaker yeast-two-hybrid (Y2H) system (Clontech Inc.) as previously described (Guo et al. 2021). The full-length TaRPP13L1-3D open reading frame (ORF) was amplified by PCR with specific primers with homologous arm designed by Primer5 and ligated into PGBKT7 plasmid predigested with EcoRI and BamHI (Table S1). To evaluate transcriptional activity, tenfold serial dilutions of the transformants were prepared, and 3-μL aliquots of each dilution were used to inoculate SD/ − Trp/ − His/ − Ade medium and SD/ − Trp/ − Leu medium containing X-α-Gal (Clontech). The inoculated media were incubated for 4 d at 30 °C. The coding sequences were subcloned into the pGBKT7 (DNA-binding domain, BD) and pGADT7 (activation domain, AD) vectors.
Statistical analysis
Indexes were measured and analyzed with at least three biological replicates, and statistical analyses were performed with three or more technical replicates. Statistical analysis and plotting were carried out using Microsoft Excel 2010 and GraphPad Prism 8.0.2, and statistical comparisons were performed using one-way ANOVA in SPSS 16.0. Mapping data were presented as mean value ± SD; P < 0.05 was set as the threshold for statistical significance.
Results
Isolation of wheat RPP13 homolog and characterization of the encoding proteins
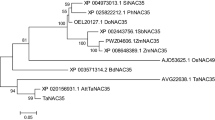
TraesCS3D02G032100 is an upregulated gene identified in the cDNA library of spontaneous cell death germplasm. In this study, one cDNA clone was isolated by RT-PCR from the total RNA of N9134R2 leaves. The deduced amino acid sequence was highly homologous to the RPP13-like 1 protein from Aegilops tauschii (XP_020190966.1), hypothetical protein CFC21 from Triticum aestivum (KAF7027114.1), E3 ubiquitin-protein ligase SINA-like 11 (KAE8768389.1) and putative disease resistance protein RGA4 (KAE8804069.1) from Hordeum vulgare, and uncharacterized protein (XP_037409381.1) and RGA2-like (XP_037472798.1) from Triticum dicoccoides. This cDNA, designated TaRPP13L1 homolog, contains an ORF of 1,431 nucleotides encoding a polypeptide of 487 amino acids, with an estimated molecular weight of 55,387.92 Da and a PI of 9.04. The CD Search (Lu et al. 2020b) and InterProScan (Blum et al. 2021) results showed that TaRPP13L1-3D consists of an Rx-N domain and a typical NB-ARC structure domain characterized by the P-loop NTPase motif (Fig. 1a). However, alignment indicated that TaRPP13L1-3D is a partial coding sequence of the Rx protein due to the loss of the LRR domain (Fig. 1b). Subsequently, a neighbor-joining phylogenetic tree was generated with TaRPP13L1 and RPP protein sequences from 25 other plant species using MEGA X (Kumar et al. 2018). Among eight sub-groups, TaRPP13L1 was closely related to Aegilops tauschii RPP13-like 1 and Salix suchowensis RPP13 proteins (Fig. S1).
The predicted protein sequence of TaRPP13L1-3D and its functional domain. a Characterization of amino acid sequences and conserved motif. b The functional domain predicted by CD Search and InterPro Scan. The full-length sequence is divided into two domains corresponding to the amino terminal CC and NB-ARC domains. The overall motif sequence in the NBS domain includes the hhGRExE, P-loop, Walker B and RNBS-B motifs, which are underlined. Three NLS consensus motifs are indicated by dashed underlining
Expression of TaRPP13L1-3D in wheat-Bgt and wheat-Pst interactions
The expression profiles of TaRPP13L1-3D in the leaves of the N9134R2 and PD34R (resistant), N9134S2 and PD34S (susceptible) wheat genotypes infected with Bgt-E09 and Pst-CYR34 are shown in Fig. 2. Following inoculation with Bgt, TaRPP13L1-3D transcripts expression was upregulated and reached the maximum level in N9134R2 (Fig. 2a), while the transcript level was repressed at 12 hpi in N9134S2 (Fig. 2b). Subsequently, the accumulation of TaRPP13L1-3D transcripts was decreased rapidly to 48 hpi in N9134R2, while the accumulation of transcripts peaked at 48 hpi in N9134S2. These observations indicated that TaRPP13L1 is involved in plant-pathogen interactions. Notably, opposing patterns of TaRPP13L1 transcript expression were detected in wheat-Bgt incompatible and compatible backgrounds. Interestingly, the TaRPP13L1 expression profiles were similar in resistant and susceptible (PD34R/S) wheat lines after infection with Pst CYR34. TaRPP13L1 expression was consistently induced after Pst infection, with maximum relative levels (stress vs. mock) detected at 48 hpi in both the resistant and susceptible lines (Fig. 2c and d), indicating that the expression of the TaRPP13 homolog is also associated with stress induced by stripe rust infection. The accumulation of Pst-induced TaRPP13L1 transcripts in the susceptible genotype (PD34S) was consistently higher than that in the resistant genotype (PD34R), while the relative ratio (PD34S vs. PD34R) reached 7.8 at 48 hpi. This hinted that high expression of TaRPP3L1 homolog may be associated with basic resistance to stripe rust, especially when the specific resistance is disabled.
Patterns of TaRPP13L1 gene expression induced in wheat by powdery mildew and stripe rust pathogen. a and b Gene expression levels were assessed in Bgt-infected N9134R2 (resistant) and N9134S2 (susceptible) lines by qRT-PCR at 6, 12, 24, 36, 48, 72, and 96 hpi. c and d Gene expression levels were assessed in Pst-infected RILs PD34R/S at 24, 48, 96, and 192 hpi. Data were normalized to the GAPDH expression level. Data represent the mean ± standard deviation of three independent replicates. Statistical comparisons (one-way ANOVA) are presented for each variable (*P < 0.05, **P < 0.01). The mock column indicates the relative expression of TaRPP13L1 in mock inoculation plants at the same time point
Overexpression and silencing of TaRPP13L1 substantiated its positive role in the response of wheat to Bgt
To explore its role in the response of wheat to Bgt, we overexpressed TaRPP13L1-3D in 2-week-old N9134S2 leaves. At 7 days after inoculation with Bgt E09, 80% (10/12) of N9134S2 plants pre-treated with PYJ-TaRPP13L1-3D showed relieved symptoms of infection, in contrast to plants pre-treated with PYJ0 and mock infection, in which dense fungal spores were observed on the leaves (Fig. 3a). Overexpression of TaRPP13L1-3D in Bgt-inoculated plants at 12 and 48 hpi was confirmed by Q-PCR (19.9-and 3.8-fold increase vs. mock) (Fig. 3b). Histological evaluation showed that the Bgt hyphal length in N9134S2 leaves pre-inoculated with PYJ0 and mock control was significantly longer and more pronounced at 48 and 72 hpi than that in TaRPP13L1-3D overexpressing wheat leaves (Fig. 3c and Fig. S2). These results indicated that upregulation of TaRPP13L1-3D in wheat enhances disease resistance to Bgt. To gain insights into the molecular mechanism underlying the TaRPP13L1 phenotypes, we examined the transcript levels of several defense-related genes in plants overexpressing TaRPP13L1 at different time-points after Bgt infection. It has been shown that fungal resistance is accompanied by accumulation of salicylic acid (SA) (Shi et al. 2013). Therefore, we selected several widely used marker genes related to the SA pathway, including phytoalexin deficient4 (PAD4), thaumatin-like protein (TLP) and chitinase (Chi8L), all of which are upregulated when defense responses are activated (Bernacki et al. 2019; Zhang et al. 2021). The qRT-PCR analysis showed that all these defense-related genes were upregulated more strongly in plants overexpressing TaRPP13L1. The expression levels of Chi8L (TraesCS5B02G403700) and TLP (TraesCS7B02G483400) were 5.3-fold and 3.1-fold higher, respectively, in TaRPP13L1-overexpressing plants than those in GFP0 injected plants at 12 hpi, while the levels of PAD4 (TraesCS4D02G096400) were 4.6-fold and 7.9-fold higher at 12 and 48 hpi, respectively. These results indicated that TaRPP13L1 is upregulated along with defense-related genes resulting in enhanced resistance.
Effect of TaRPP13L1-3D overexpression on the response of N9134S2 leaves to Bgt E09 stress. a Images of infection symptoms 10 days after inoculation with Bgt E09. The reconstructed vectors, PYJ:TaRPP13L1-3D, and PYJ0 were applied to the leaves before inoculation with Bgt pathogen. The mock group was treated with buffer in the same way. b Relative expression level of TaRPP13L1 and defense-related marker genes. Data represent the mean ± standard deviation of three independent replicates (Student’s t-test, **P < 0.01). c Bgt hyphal length after infection. Data represent the mean ± standard deviation (*P< 0.05)
The silenced phenotype appeared in leaves 7 days after inoculation with BSMV-PDS (γPDS) (Fig. 4a), thus confirming that the BSMV silencing system was effective. For BSMV-RPP13L treated N9134R2, we noted a slight, but visible fading phenotype change in leaves after inoculation with Bgt E09 (Fig. 4b), while a denser network of powdery mildew conidia was observed in BSMV-RPP13L treated N9134S2 plants compared with that in mock and BSMV-blank treated plants (Fig. 4c). The fading phenotype in BSMV-RPP13L treated N9134R2 is similar to the hypersensitive resistance response (HR) after fungal spore puncture the ‘skin’ of wheat leaves tissue. The qRT-PCR analysis showed that TaRPP13L1-3D expression was decreased by 63.4%-85% (Fig. 4d, e). while expression of the marker gene, Chi8L, was also decreased by 80–95% in both the N9134R2 and N9134S2 genotypes. Strikingly, the expression of TLP and PAD4 was strongly induced in RPP13L-silenced N9134R2 plants compared with the mock after inoculation with Bgt E09 (Fig. 4d). Considering the immune resistance of N9134R2 to Bgt E09 infection, the reduced resistance level in BSMV-RPP13L plants indicated that silencing TaRPP13L1 affected resistance to Bgt. However, it is not enough altering the phenotype of resistance to powdery mildew in incompatible genetic background (Fig. 4b). Thus, our observation that knockdown of TaRPP13L1 increases susceptibility indicated that TaRPP13L1 plays a role in quantitative and/or basal resistance to Bgt E09.
Effect of TaRPP13L1-3D silencing on the response of N9134R2/S2 leaves to Bgt E09 stress. a Images showing the appearance of the silenced phenotype 7 days after inoculation with BSMV-PDS (γPDS). b and c The effect of the reconstructed vectors BSMV-RPP (γRPP13L) on inoculated leaves of N9134R2 (b) and N9134S2 (c) leaves after Bgt infection; the mock and BSMV-Blank (γ0) groups were treated with buffer in the same way. d and e Quantitative RT-PCR analysis of the expression levels of TaRPP13L1-3D and defense-related marker genes in the TaRPP13L1-3D-silenced N9134R2 (d) and N9134S2 (e) plant leaves compared with the Bgt-infected mock plants. Gene expression was normalized against Actin. Data represent the mean ± standard deviation of three independent replicates. *P< 0.05 or **P< 0.01, γ0 versus γRPP13L at the same time-point
TaRPP13L1 functioned in the nucleus and plasma membrane
Based on the presence of specific subcellular targeting motifs in the amino acid sequence, TaRPP13L1 was assumed to be localized in the cytoplasmic, mitochondrial and nuclear fractions (https://www.genscript.com/psort.html). To investigate the subcellular localization of TaRPP13L1, GFP fusion constructs (GFP was fused to the C-terminus) were agro-infiltrated into N. benthamiana leaves and GFP fluorescence was examined in non-plasmolyzed epidermal cells of the abaxial side of the leaf. The TaRPP13L1 GFP fusion constructs were not only expressed in the nucleus, but also co-expressed with the membrane-selective fluorescent dye FM4-64 in the plasma membrane at 2 dpi (Fig. 5). The control GFP0 was detected throughout the cytoplasm and nucleus. To evaluate TaRPP13L1 localization in the nucleus, TaRPP13L1-GFP fusion constructs were expressed in the onion epidermal cells. We observed that the GFP signal in the nucleus overlapped with that of the nuclei-selective fluorescent dye 4',6-diamidino-2-phenylindole (DAPI) (Fig. S3). This confirmed that TaRPP13L1 is localized in both the plasma membrane and nucleus. Intriguingly, particular GFP fluorescence signals were also commonly detected in the RPP13L1-overexpressing epidermal cells, while GFP aggregation signals migrate slowly from the plasma membrane to the nucleus (Supplemental Video 1).
Transient expression and localization of TaRPP13L1 fusion proteins in Nicotiana benthamiana non-plasmolyzed epidermal cells. The fusion vector pYJ:TaRPP13L1:GFP and the pYJ::GFP control (Ck, marker with GFP0) vectors were transformed into tobacco epidermal cells by Agrobacterium. The subcellular distribution of GFP in the epidermal cells was revealed by fluorescence scanning microscopy. Dark field images show green and red fluorescence (GFP and FM4-64 line) and bright field images show cell morphology. Merged images are shown for localization. Scale bars: 80 μm
TaRPP13L1 interacted with the RanGAP2-WPP complex
The migration of TaRPP13L1-3D to the nucleus from the plasma membrane indicated its ability to traverse nuclear envelope (NE) channels. In eukaryotes, nuclear pore complexes (NPC), also known as SUN–KASH (INM Sad1/UNC-84 and Klarsicht/ANC-1/Syne-1 homology) NE bridges, are responsible for connecting the nucleoplasm and cytoplasm (Zhou et al. 2015; Yang et al. 2017). Arabidopsis thaliana WPP domain-interacting proteins (AtWIPs) were identified as plant specific KASH proteins, while Ran GTPase activating protein (RanGAP) is a member of the WPP domain-containing protein family, which is associated with the NE by protein–protein interactions (Zhou et al. 2012; Graumann and Evans 2017). More importantly, RanGAP2 was identified as an Rx-interacting protein (Sacco et al. 2007). Therefore, we test the interaction between RPP13L1 with RanGAP2L and WPP1 to further clarify the mechanism by which RPP13L1 is transferred from the cytoplasm into the nucleus. After the expression of the proteins in the Y2H system was confirmed on SD medium lacking Trp and Leu (double dropout, DDO), our results showed that the yeast strains with BD-TaRPP13L1 and AD-WPP1 (TraesCS2B02G210200) grew successfully on SD/ − Trp/ − Leu/ − His medium (Triple dropout, TDO), indicating that TaRPP13L1 interacts directly with TaWPP1 (Fig. 6). However, no interaction between TaRPP13L1 and RanGAP2L (TraesCS3B02G433100) was detected using the Y2H system. This provided evidence that TaRPP13L1 maybe cross the NE by RanGAP-mediated nucleocytoplasmic transport (i.e., via the SUN–KASH NE bridge).
The interaction between TaRPP13L1-3D, WPP1 and RanGAP2 proteins in a yeast two-hybrid system. TaRPP13L1 was cloned into pGBKT7 and the corresponding proteins were cloned into the pGADT7. Y2H cells harboring the indicated plasmid combinations were grown on either the nonselective (SD-L-W) or selective (SD-L-W–H) medium containing 20 μg mL−1 X-α-gal. The interaction between SV40 large T-antigen (T) and murine p53 (53), T-AD + 53-BD, was used as the positive control, while the interaction between T-antigen and human lamin C(Lam), T-AD + Lam-BD, was used as the negative control
Discussion
Previously, we detected that a differentially expressed gene was annotated as RPP13-like protein in the location region of the spotted necrosis gene TaSpl1. Notably, RPP13 and several homologue were proposed to play an essential role in regulation of responses to a variety of external stimuli, including biotic and abiotic stresses (Peter et al. 2000; Ramachandran et al. 2017; Liu et al. 2020; Yang et al. 2021). To understand its’ potential role in wheat responding to fungal stress, here we tested the expression of TaRPP13L1-3D gene in wheat under Pst and Bgt infection. Furthermore, we substantiated the positive role of TaRPP13L1-3D in wheat-powdery mildew interaction through transient overexpression and gene silencing. This provides useful information to further explore the activity and mechanism of TaRPP13L1 in defense against biotic stress.
Plant NBS-LRR proteins contain many highly conserved functional motifs. According to the classifying rule based on the presence of an aspartic acid residue in TIRs and a tryptophan in non-TIRs (Miller et al. 2008), TaRPP13L1-3D is categorized as a C-terminal truncated CNL protein. The potato resistance protein RX confers resistance against potato virus X. Previous studies have suggested that the Rx-NB domain (Rairdan et al. 2008) initiates defense signaling and causes cell death due to the interaction of the Rx protein with RanGAP2 via the CC domain surrounding the EDVID motif (Bendahmane A et al. 2002; Hao et al. 2013; Bernacki et al. 2019). Compared to RX, we found that the TaRPP13L1 protein contained differences in the Rx-CC domain, and the EDVID motif was absent. We did not detect any interaction between TaRPP13L1 and RanGAP2L in this study using the Y2H system, indicating that the amino acid side chains in Rx-N may be an important influence on the formation of the RanGAP2-WPP complex. These observations also further support the notion that R proteins have diverse functions in CC/NB-LRR interactions. This evidence is consistent with previous studies of functional motifs and/or amino acid residues implicated in the transfer of proteins across nuclear membranes and their functions in plant hypersensitive resistance (HR) responses (Xu et al. 2007; Gu et al. 2016; Yang et al. 2017).
Recently, we found that TaYRG1, a typical CC-NB-ARC-LRR protein, triggered cell death though splice transcript loss of the CC domain, while the artificial mutant with the NB domain also caused moderate cell death following transient expression in tobacco (Wang 2021, dissertation). In this study, RPP13L1 overexpression characterized by the CC-NB structure (C-terminal truncation) failed to trigger cell death in Nicotiana benthamiana. Intriguingly, both the CC domain and CC-NB fragment of MLA10 were shown to trigger cell death in N. benthamiana conversely, while the NB domain did not (Bai et al. 2012). Additionally, NB-ARC regulated the defense response to rust infection though fusing with NPR1 post-inoculation (Bailey et al. 2018; Wang et al. 2020b). This evidence of structural and functional diversity indicates that the domains of NB-LRR resistance proteins play versatile roles in hypersensitive resistance (HR) response to pathogen stress (Baggs et al. 2017). This functional diversity was also supported by two recent studies in which TaRPP13-3 and ZmRPP13-LK3 were shown to play an important role in plant resistances to pathogen infections and heat stress, respectively (Liu et al. 2020; Yang et al. 2021). In the present study, we provide another example of the quantitative contribution of TaRPP13L1 not only to resistance to Bgt stress, but also to the response to Pst Stress, which further enriches our knowledge of the functions of RPP13 homologs in plants. The results of the current study suggest two possible mechanisms that responsible for the functions of RPP13-like genes; one is that TaRPP13L1 directly modulates signals that trigger a defense response in both resistant and susceptible plants due to it being able to transfer between the nucleus and plasma membrane, while the second is that differential sensitivity to pathogen stress or modulation of the defense pathway interacts with other proteins and/or complexes in different genetic background. Thus, our analysis indicates that TaRPP13L1 may be an important component of the plant basal defense pathway against Bgt colonization, although the biological functions remain to be fully elucidated.
During the co-evolution of plants and pathogens, resistance is usually overcome by the emergence of new races of the pathogen carrying virulence alleles that are compatible with the resistance alleles of the plant (Huang et al. 2016; Zhao et al. 2016). The evidence presented here indicates that TaRPP13L may be a positive indicator that links plant-Bgt innate immune defense capacity in hexaploid wheat, and/or a negative indictor in plant-Pst interaction. However, further research is required to determine the value of TaRPP13L as a marker that can be used in genetic analyses of immune resistance. Although it is unclear why Bgt and Pst infection induce different pattern and/or levels of TaRPP13L1-3D expression in wheat leaves, and why higher responding levels are expressed in the susceptible line after infection by Pst, our results raise the possibility that the functions of TaRPP13L1 in the responses of plants to Pst CYR34 challenge are controlled by the level of expression. This also supported the possibility that a single gene can be linked to susceptibility to one fungal species and related to resistance in another (Howard et al. 2010). In addition, the opposing patterns of TaRPP13L1-3D expression in the resistance and susceptible backgrounds after Bgt attack should be noted. This demonstrated that the potential of a single gene to play various role in the response not only to different pathogen, but also to different in genetic backgrounds, and substantiated our previously results obtained by comparing transcriptome expression data (Zhang et al. 2014; Guo et al. 2021).
In conclusion, these insights into TaRPP13L1 function will be useful in clarifying the mechanism of stripe rust and powdery mildew resistance in wheat, and enriching our knowledge of the interaction between R genes and pathogen defense responses.
Author contribution statments
H.Z. and W.J. conceived and designed the study. X.Z. and H.Z. analyzed data and wrote the article. X.Z., M.W., H.G. and L.Z. performed the research. X.Z., X.Q. and G.W. contributed by collecting the samples and visualization. T.L. and Y.W. contributed to the development of material. All authors read and approved the final manuscript.
Data availability statements
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- Bgt :
-
Blumeria graminis F. sp. tritici
- BSMV:
-
Barley stripe mosaic virus
- Hpi:
-
Hours post-inoculation
- Pst :
-
Puccinia striiformis F. sp. tritici
- R gene/genes:
-
Resistance gene/genes
- RPP13:
-
Recognition of Peronospora parasitica 13
- TLP :
-
Thaumatin-like protein gene
- WPP1:
-
WPP domain-containing proteins 1
References
Adachi H, Derevnina L, Kamoun S (2019) NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr Opin Plant Biol 50:121–131. https://doi.org/10.1016/j.pbi.2019.04.007
Baggs E, Dagdas G, Krasileva KV (2017) NLR diversity, helpers and integrated domains: making sense of the NLR IDentity. Curr Opin Plant Biol 38:59–67. https://doi.org/10.1016/j.pbi.2017.04.012
Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FL, Schulze-Lefert P, Shen QH (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. Plos Pathog 8(6):e1002752. https://doi.org/10.1371/journal.ppat.1002752
Bailey PC, Schudoma C, Jackson W, Baggs E, Dagdas G, Haerty W, Moscou M, Krasileva KV (2018) Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol 19(1):23. https://doi.org/10.1186/s13059-018-1392-6
Barry DJ, Chan C, Williams GA (2009) Morphological quantification of filamentous fungal development using membrane immobilization and automatic image analysis. J Ind Microbiol Biotechnol 36(6):787–800. https://doi.org/10.1007/s10295-009-0552-9
Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site–leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32(2):195–204
Bernacki MJ, Czarnocka W, Rusaczonek A, Witon D, Keska S, Czyz J, Szechynska-Hebda M, Karpinski S (2019) LSD1-, EDS1- and PAD4-dependent conditional correlation among salicylic acid, hydrogen peroxide, water use efficiency and seed yield in Arabidopsis thaliana. Physiol Plant 165(2):369–382. https://doi.org/10.1111/ppl.12863
Bittner-Eddy P, Can C, Gunn N, Pinel M, Tor M, Crute I, Holub EB, Beynon J (1999) Genetic and physical mapping of the RPP13 locus, in Arabidopsis, responsible for specific recognition of several Peronospora parasitica (downy mildew) isolates. Mol Plant Microbe in 12(9):792–802. https://doi.org/10.1094/Mpmi.1999.12.9.792
Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49(D1):D344–D354. https://doi.org/10.1093/nar/gkaa977
Eitas TK, Dangl JL (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13(4):472–477. https://doi.org/10.1016/j.pbi.2010.04.007
Fu DL, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen XM, Sela HA, Fahima T, Dubcovsky J (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323(5919):1357–1360. https://doi.org/10.1126/science.1166289
Graumann K, Evans DE (2017) The nuclear envelope - Structure and protein interactions. Annu Plant Rev 46:19–56. https://doi.org/10.1002/9781119312994.apr0498
Gu Y, Zebell SG, Liang Z, Wang S, Kang BH, Dong X (2016) Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell 166(6):1526–1538. https://doi.org/10.1016/j.cell.2016.07.042
Guo H, Zhang H, Wang G, Wang C, Wang Y, Liu X, Ji W (2021) Identification and expression analysis of heat-shock proteins in wheat infected with powdery mildew and stripe rust. Plant Genome-Us 14(2):e20092. https://doi.org/10.1002/tpg2.20092
Hall SA, Allen RL, Baumber RE, Baxter LA, Fisher K, Bittner-Eddy PD, Rose LE, Holub EB, Beynon JL (2009) Maintenance of genetic variation in plants and pathogens involves complex networks of gene-for-gene interactions. Mol Plant Pathol 10(4):449–457. https://doi.org/10.1111/j.1364-3703.2009.00544.x
Hao W, Collier SM, Moffett P, Chai JJ (2013) Structural basis for the interaction between the Potato Virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). J Biol Chem 288(50):35868–35876. https://doi.org/10.1074/jbc.M113.517417
Hewitt T, Muller MC, Molnar I, Mascher M, Holusova K, Simkova H, Kunz L, Zhang J, Li J, Bhatt D, Sharma R, Schudel S, Yu G, Steuernagel B, Periyannan S, Wulff B, Ayliffe M, McIntosh R, Keller B, Lagudah E, Zhang P (2021) A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol 229(5):2812–2826. https://doi.org/10.1111/nph.17075
Howard AF, Koenraadt CJ, Farenhorst M, Knols BG, Takken W (2010) Pyrethroid resistance in Anopheles gambiae leads to increased susceptibility to the entomopathogenic fungi Metarhizium anisopliae and Beauveria bassiana. Malar J 9:168. https://doi.org/10.1186/1475-2875-9-168
Huang L, Raats D, Sela H, Klymiuk V, Lidzbarsky G, Feng L, Krugman T, Fahima T (2016) Evolution and adaptation of wild emmer wheat populations to biotic and abiotic stresses. Annu Rev Phytopathol 54:279–301. https://doi.org/10.1146/annurev-phyto-080614-120254
Jia M, Xu H, Liu C, Mao R, Li H, Liu J, Du W, Wang W, Zhang X, Han R, Wang X, Wu L, Liang X, Song J, He H, Ma P (2020) Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front Genet 11:241. https://doi.org/10.3389/fgene.2020.00241
Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30(2):285–299. https://doi.org/10.1105/tpc.17.00579
Krattinger SG, Keller B (2016) Molecular genetics and evolution of disease resistance in cereals. New Phytol 212(2):320–332. https://doi.org/10.1111/nph.14097
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Liu W, Frick M, Huel R, Nykiforuk CL, Wang XM, Gaudet DA, Eudes F, Conner RL, Kuzyk A, Chen Q, Kang ZS, Laroche A (2014) The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS-LRR sequence in wheat. Mol Plant 7(12):1740–1755. https://doi.org/10.1093/mp/ssu112
Liu X, Zhang C, Zhang L, Huang J, Dang C, Xie C, Wang Z (2020) TaRPP13-3, a CC-NBS-LRR-like gene located on chr 7D, promotes disease resistance to wheat powdery mildew in Brock. J Phytopathol 168(11–12):688–699. https://doi.org/10.1111/jph.12949
Lu P, Guo L, Wang Z, Li B, Li J, Li Y, Qiu D, Shi W, Yang L, Wang N, Guo G, Xie J, Wu Q, Chen Y, Li M, Zhang H, Dong L, Zhang P, Zhu K, Yu D, Zhang Y, Deal KR, Huo N, Liu C, Luo MC, Dvorak J, Gu YQ, Li H, Liu Z (2020a) A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat Commun 11(1):680. https://doi.org/10.1038/s41467-020-14294-0
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A (2020b) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48(D1):D265–D268. https://doi.org/10.1093/nar/gkz991
Ma PT, Han GH, Zheng Q, Liu SY, Han FP, Wang J, Luo QL, An DG (2020) Development of novel wheat-rye chromosome 4R translocations and assignment of their powdery mildew resistance. Plant Dis 104(1):260–268. https://doi.org/10.1094/Pdis-01-19-0160-Re
Miller RN, Bertioli DJ, Baurens FC, Santos CM, Alves PC, Martins NF, Togawa RC, Souza MT Jr, Pappas GJ Jr (2008) Analysis of non-TIR NBS-LRR resistance gene analogs in Musa acuminata Colla: isolation, RFLP marker development, and physical mapping. BMC Plant Biol 8:15. https://doi.org/10.1186/1471-2229-8-15
Morales L, Michel S, Ametz C, Dallinger HG, Loschenberger F, Neumayer A, Zimmerl S, Buerstmayr H (2021) Genomic signatures of selection for resistance to stripe rust in Austrian winter wheat. Theor Appl Genet 134:3111–3121. https://doi.org/10.1007/s00122-021-03882-3
Pang Y, Wu Y, Liu C, Li W, St Amand P, Bernardo A, Wang D, Dong L, Yuan X, Zhang H, Zhao M, Li L, Wang L, He F, Liang Y, Yan Q, Lu Y, Su Y, Jiang H, Wu J, Li A, Kong L, Bai G, Liu S (2021) High-resolution genome-wide association study and genomic prediction for disease resistance and cold tolerance in wheat. Theor Appl Genet 134:2857–2873. https://doi.org/10.1007/s00122-021-03863-6
Bittner-Eddy PD, Ian RC, Eric B, Beynon HJL (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J 21(2):177–188
Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P (2008) The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20(3):739–751. https://doi.org/10.1105/tpc.107.056036
Ramachandran SR, Yin C, Kud J, Tanaka K, Mahoney AK, Xiao F, Hulbert SH (2017) Effectors from wheat rust fungi suppress multiple plant defense responses. Phytopathology 107(1):75–83. https://doi.org/10.1094/PHYTO-02-16-0083-R
Sacco MA, Mansoor S, Moffett P (2007) A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J 52(1):82–93. https://doi.org/10.1111/j.1365-313X.2007.03213.x
Sekhwal MK, Li P, Lam I, Wang X, Cloutier S, You FM (2015) Disease resistance gene analogs (RGAs) in plants. Int J Mol Sci 16(8):19248–19290. https://doi.org/10.3390/ijms160819248
Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D (2013) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25(3):1143–1157. https://doi.org/10.1105/tpc.112.107904
Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1(4):2019–2025. https://doi.org/10.1038/nprot.2006.286
Wang S, Li QP, Wang J, Yan Y, Zhang GL, Yan Y, Zhang H, Wu J, Chen F, Wang X, Kang Z, Dubcovsky J, Gou JY (2019) YR36/WKS1-mediated phosphorylation of PsbO, an extrinsic member of photosystem II, inhibits photosynthesis and confers stripe rust resistance in wheat. Mol Plant 12(12):1639–1650. https://doi.org/10.1016/j.molp.2019.10.005
Wang W, Feng B, Zhou JM, Tang D (2020a) Plant immune signaling: advancing on two frontiers. J Integr Plant Biol 62(1):2–24. https://doi.org/10.1111/jipb.12898
Wang X, Zhang H, Nyamesorto B, Luo Y, Mu X, Wang F, Kang Z, Lagudah E, Huang L (2020b) A new mode of NPR1 action via an NB-ARC-NPR1 fusion protein negatively regulates the defence response in wheat to stem rust pathogen. New Phytol 228(3):959–972. https://doi.org/10.1111/nph.16748
Wellings CR (2011) Global status of stripe rust: a review of historical and current threats. Euphytica 179(1):129–141. https://doi.org/10.1007/s10681-011-0360-y
Xu XM, Meulia T, Meier I (2007) Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol 17(13):1157–1163. https://doi.org/10.1016/j.cub.2007.05.076
Yang Y, Wang W, Chu Z, Zhu JK, Zhang H (2017) Roles of nuclear pores and nucleo-cytoplasmic trafficking in plant stress responses. Front Plant Sci 8:574. https://doi.org/10.3389/fpls.2017.00574
Yang H, Zhao Y, Chen N, Liu Y, Yang S, Du H, Wang W, Wu J, Tai F, Chen F, Hu X (2021) A new adenylyl cyclase, putative disease-resistance RPP13-like protein 3, participates in abscisic acid-mediated resistance to heat stress in maize. J Exp Bot 72(2):283–301. https://doi.org/10.1093/jxb/eraa431
Zhang H, Yang Y, Wang C, Liu M, Li H, Fu Y, Wang Y, Nie Y, Liu X, Ji W (2014) Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics 15:898. https://doi.org/10.1186/1471-2164-15-898
Zhang H, Xu X, Wang M, Wang H, Deng P, Zhang Y, Wang Y, Wang C, Wang Y, Ji W (2021) A dominant spotted leaf gene TaSpl1 activates endocytosis and defense-related genes causing cell death in the absence of dominant inhibitors. Plant Sci 310:110982. https://doi.org/10.1016/j.plantsci.2021.110982
Zhang H, Zhang L, Wang C, Wang Y, Zhou X, Lv S, Liu X, Kang Z, Ji W (2016) Molecular mapping and marker development for the Triticum dicoccoides-derived stripe rust resistance gene YrSM139–1B in bread wheat cv. Shaanmai 139. Theor Appl Genet 129(2):369–376. https://doi.org/10.1007/s00122-015-2633-7
Zhao J, Wang M, Chen X, Kang Z (2016) Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Annu Rev Phytopathol 54:207–228. https://doi.org/10.1146/annurev-phyto-080615-095851
Zhao Z-H, Huang J, Lu M, Wang X-M, Wu L-F, Wu X-F, Zhao X, Li H-J (2013) Virulence and genetic diversity of Blumeria graminis f.sp.tritici collected from Shandong and Hebei Provinces. Acta Agron Sin 39(8):1377–1385. https://doi.org/10.3724/sp.J.1006.2013.01377
Zhou X, Graumann K, Evans DE, Meier I (2012) Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol 196(2):203–211. https://doi.org/10.1083/jcb.201108098
Zhou XL, Han DJ, Chen XM, Gou HL, Guo SJ, Rong L, Wang QL, Huang LL, Kang ZS (2014) Characterization and molecular mapping of stripe rust resistance gene Yr61 in winter wheat cultivar Pindong 34. Theor Appl Genet 127(11):2349–2358. https://doi.org/10.1007/s00122-014-2381-0
Zhou X, Graumann K, Meier I (2015) The plant nuclear envelope as a multifunctional platform LINCed by SUN and KASH. J Exp Bot 66(6):1649–1659. https://doi.org/10.1093/jxb/erv082
Acknowledgements
This work was supported financially by the National Natural Science Foundation of China (31971741) and the Shaanxi Innovation Team Project (2018TD-004). The PYJ::GFP vector was kindly gifted by Dr. Jiang. We also thank Dr. Karen Bysouth for editing the English text of a draft manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 (MP4 88300 KB)
Rights and permissions
About this article
Cite this article
Zhang, X., Wang, G., Qu, X. et al. A truncated CC-NB-ARC gene TaRPP13L1-3D positively regulates powdery mildew resistance in wheat via the RanGAP-WPP complex-mediated nucleocytoplasmic shuttle. Planta 255, 60 (2022). https://doi.org/10.1007/s00425-022-03843-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03843-0