Abstract
Main conclusion
Young Seedling Stripe1 (YSS1) was characterized as an important regulator of plastid-encoded plastid RNA polymerase (PEP) activity essential for chloroplast development at rice seedling stage.
Chloroplast development is coordinately regulated by plastid- and nuclear-encoding genes. Although a few regulators have been reported to be involved in chloroplast development, new factors remain to be identified, given the complexity of this process. Here, we report the characterization of a temperature-sensitive young seedling stripe1 (yss1) rice mutant, which develops striated leaves at the seedling stage, particularly in leaf 3, but produces wild-type leaves in leaf 5 and onwards. The chlorotic leaves have decreased chlorophyll (Chls) accumulation and impaired chloroplast structure. Positional cloning combined with sequencing demonstrated that aberrant splicing of the 8th intron in YSS1 gene, due to a single nucleotide deletion around splicing donor site, leads to decreased expression of YSS1 and accumulation of an 8th intron-retained yss1 transcript. Furthermore, complementation test revealed that downregulation of YSS1 but not accumulation of yss1 transcript confers yss1 mutant phenotype. YSS1 encodes a chloroplast nucleoid-localized protein belonging to the DUF3727 superfamily. Expression analysis showed that YSS1 gene is more expressed in newly expanded leaves, and distinctly up-regulated as temperatures increase and by light stimulus. PEP- and nuclear-encoded phage-type RNA polymerase (NEP)-dependent genes are separately down-regulated and up-regulated in yss1 mutant, indicating that PEP activity may be impaired. Furthermore, levels of chloroplast proteins are mostly reduced in yss1 seedlings. Together, our findings identify YSS1 as a novel regulator of PEP activity essential for chloroplast development at rice seedling stage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plastid development from proplastids in shoot meristems to mature chloroplasts involves various metabolic processes during plant growth (Sakamoto et al. 2008). Chloroplasts are essential not only for photosynthesis in higher plants, but also for the synthesis and storage of other metabolites (Mullet 1993). Chloroplasts are semiautonomous organelles containing their own genomes originally derived through an endosymbiotic process from a photosynthetic cyanobacterium (Moreira et al. 2000; Sugimoto et al. 2004). Most of the genes are lost or transferred to nuclear genomes, but a few are retained to self-replication. The plastid genome in rice is 135 kb and contains 34 RNA-coding genes and nearly 120 protein-coding genes (Hiratsuka et al. 1989; Pfalz and Pfannschmidt 2013). Their functions are involved in transcription, translation, photosynthetic electron transfer, and photosynthetic metabolism (Sakamoto et al. 2008). Almost 3000 proteins function in the chloroplast and more than 95 % of them are encoded by nuclear genes and transported into chloroplast (Reumann et al. 2005). Chloroplast differentiation is controlled by both plastid and nuclear genes but is largely under nuclear control. Many of the chloroplast proteins required for various metabolic processes are encoded by nuclear genes and are needed for plastid gene expression (Sakamoto et al. 2008).
The transcriptional network in plastids of higher plants is mediated by two types of RNA polymerases: plastid-encoded plastid RNA polymerase (PEP) and nuclear-encoded phage-type RNA polymerase (NEP). The PEP complex consists of four core subunits (α, β, β′, and β″) and at least ten PEP-associated proteins (PAPs) (Tozawa et al. 2001; Steiner et al. 2011; Pfalz and Pfannschmidt 2013). In both Arabidopsis thaliana and rice, six σ recognition factors exist (Isono et al. 1997; Fujiwara et al. 2000; Kasai et al. 2004; Kubota et al. 2007). The core subunits of PEP complex are separately encoded by plastid genes rpoA, rpoB, rpoC1, and rpoC2 while the σ factors are encoded by nuclear genes required for transcription initiation and thus allow the nucleus to regulate PEP transcriptional activity (Allison 2000). The mutations of many PEP complex components (e.g., PAPs and AtSIG6) cause chlorotic phenotype accompanied by either decreased PEP activity or disrupted plastid transcription (Ishizaki et al. 2005; Pfalz and Pfannschmidt 2013). In contrast, the NEP complex is encoded by RPOTp or RPOTmp, knockout of which also leads to delayed chloroplast biogenesis and plant growth in Arabidopsis (Hricova et al. 2006; Kühn et al. 2009). PEP is responsible for the transcription of photosynthesis genes and plays an important role in chloroplast development (Hajdukiewicz et al. 1997). Decreased PEP activity may impair chloroplast development (Pfalz and Pfannschmidt 2013). However, the housekeeping genes and plastid ribosomal genes are transcribed by NEP (Hedtke et al. 1997). Protein–protein interactions between NEP and PEP complexes have not yet been identified, although they coincide in regulating plastid gene transcription temporally and spatially (Börner et al. 2015).
In addition to PEP and NEP, nucleoid proteins including some plastid transcriptionally active chromosomes (pTACs) may also participate in plastid transcription (Pfalz et al. 2006; Pfalz and Pfannschmidt 2013). Plastid DNAs are embedded in protein-DNA complex and form a large structure called nucleoid together with other components involved in gene expression and inheritance (Yagi et al. 2012). It is reported that about 127 candidates were proposed in maize plastid nucleoid (Majeran et al. 2012). Similar to nuclear, it is assumed that the nucleoid proteins are responsible for plastid transcription. However, the specific roles of nucleoid proteins are still vague. Recent proteomics studies identified several nucleoid proteins and genetic evidence showed that lacks of these proteins result in different degree of chlorotic leaf phenotypes and improper plastid development (Pfalz and Pfannschmidt 2013). Determination of plastid transcript in these nucleoid protein mutants revealed a Δ-rpo expression pattern, indicating that the proteins in nucleoid may be involved in plastid transcriptional regulation (Zhong et al. 2013). AtMSH1 is a nucleoid protein and lack of which exhibits a Δ-rpo expression pattern and results in undeveloped chloroplasts and a chlorotic phenotype (Xu et al. 2011). Similar observations are obtained in mutants of nucleoid proteins AtRugosa2 and AtPRIN2 (Quesada et al. 2011; Kindgren et al. 2012).
Chloroplast development is mostly blocked in rice Chl-deficient mutants. Stripe leaf mutant plants usually produce longitudinally green/white-striped leaves and abnormal chloroplasts at an early stage and develop normally during later growth stages. Previous studies showed that v1, v2, v3, st1, and ylc1 are temperature-sensitive mutants that generate severe leaf chlorosis at a constant 20 °C condition, but exhibit phenotypic changes towards normality as temperatures increase (Kusumi et al. 1997, 2011; Sugimoto et al. 2004, 2007; Yoo et al. 2009; Zhou et al. 2013). Many undifferentiated chloroplasts are present in leaf cells of v3 and st1 mutants under low temperatures (Yoo et al. 2009). Chloroplast development may also be affected in v1, v2, and ylc1 mutants grown at less than 20 °C, although data are not available. Accumulation of different gene transcripts might be closely related to the phenotype. It is reported that amylose content is regulated by alteration between pre-mRNA and mature mRNA of the Wx gene. Glutinous rice with no mature Wx mRNA shows extremely low amylose content. Conversely, mature Wx mRNA is abundant in non-glutinous cultivars that have higher amylose contents. Cultivars with intermediate levels of amylose produce both pre-mRNA and mature mRNA (Wang et al. 1995; Isshiki et al. 1998). However, as far as we know, such regulatory model for chloroplast development in rice remains unreported.
In this paper, we describe a temperature-sensitive seedling stripe mutant yss1 in rice. Under paddy field conditions, the striated phenotype in yss1 mutant is visible in leaf 2 and became evident in leaf 3, but slowly weakens until completely disappears in leaf 5 and thereafter. Decreased Chl accumulation and undifferentiated chloroplasts are displayed in the yss1 mutant in comparison to the wild type at 20 °C, but developed almost normal at 30 °C. A single base deletion in the YSS1 gene leads to an incomplete splicing of the 8th intron, thus producing two transcripts: the wild-type YSS1 transcript and an aberrant yss1 transcript. Expression analysis revealed that the expression levels of wild-type YSS1 transcripts are closely related to the yss1 phenotypic alternations. The YSS1 gene was isolated via the map-based cloning strategy; it encodes an unknown functional protein belonging to the DUF3727 superfamily. The YSS1 gene is more expressed in newly expanded leaves and up-regulated as temperatures increase and by light stimulus. The expression levels of genes required for PEP and NEP are differentially regulated and exhibited a Δ-rpo expression pattern in yss1 mutant. Furthermore, western blot analyses revealed that the contents of both nuclear-encoded and plastid-encoded chloroplast proteins are mostly lower in the yss1 mutant than in wild-type seedlings. These observations suggest that YSS1 mutation might affect chloroplast development through modulating PEP activity during the early leaf development in rice.
Materials and methods
Plant materials and treatments
The seedling stripe mutant yss1 was obtained from a 60Co-irradiated population of indica rice cultivar II-32B. Plants were grown in a paddy field during the rice growing season or in a growth chamber. For temperature treatments, seedlings were grown in a growth chamber with 12 h of light/12 h of darkness at a constant temperature of 20 °C (C20), 23 °C (C23), 30 °C (C30), and 12 h of light at 30 °C/12 h of darkness at 20 °C (L30/D20). For light treatments, seedlings were cultivated in a growth chamber with 15 h of light at 30 °C/9 h of darkness at 25 °C and 10 h of light at 30 °C/14 h of darkness at 25 °C. For nutrition treatments, seedlings were grown in hydroponic solutions (1 mM NH4NO3, 0.32 mM NaH2PO4·2H2O, 1.5 mM KCl, 1.7 mM MgCl2·6H2O, 0.67 mM CaCl2, 0.4 mM Na2SiO3·9H2O, 9 μM MnCl2·4H2O, 0.6 μM Na2MoO4·2H2O, 0.14 μM ZnSO4·7H2O, 0.16 μM CuSO4·5H2O, and 44.8 μM FeSO4·7H2O, pH 5.6). Crosses between yss1 and rice varieties II-32B and 02428 were made for genetic analysis and mapping. The phenotype of yss1 mutant was identified from the L2 to L5 stage. The varieties II-32B and 02428 were obtained from the Chinese National Key Facility for Crop Gene Resources and Genetic Improvement.
Determination of chlorophyll content
Fresh leaves of wild type and yss1 mutant from different developmental stages (L3–L5) or from temperature treatments were collected to determinate Chl contents as described previously (Zhou et al. 2013). The leaf samples were marinated in 95 % ethanol for 48 h in darkness, and then the absorbance of supernatants was measured with a DU 800 UV/Vis Spectrophotometer (Beckman Coulter).
Confocal and transmission electron microscopy (TEM) analyses
To investigate the number of Chl-containing cells, leaf samples of wild type and the yss1 mutant at the L3 stage under different temperature conditions (C20, C23, C30, and L30/D20) were collected and observed with a confocal laser scanning microscope (Carl Zeiss LSM700). For TEM, transverse sections of wild-type and the yss1 mutant leaves from the L3 stage at C20, C23, C30, and L30/D20 were fixed in 2.5 % glutaraldehyde in a phosphate buffer and further fixed in 1 % OsO4 overnight at 4 °C. After staining with uranyl acetate, tissues were further dehydrated through an ethanol series, and then embedded in Spurr’s medium prior to ultrathin sectioning. Sections were air-dried, stained again, and viewed with a Hitachi H-7650 transmission electron microscope. For measuring the percentages of normal and undifferentiated chloroplasts in mesophyll cells, 50 cells were examined for each determination.
Mapping of YSS1 and complementation test
For linkage analysis, 94 typical individuals showing the mutant phenotype were generated from the 02428/yss1 F2 population. A further 1223 F2 mutant plants were used for fine mapping. Insertion/deletion (InDel) markers were developed with Primer Premier 5.0 based on differences in the entire genomic sequence between japonica rice Nipponbare and indica cultivar 93-11. Primer pair K83 (F: 5′-ATTTCAGTGGCTGGATTGC-3′, R: 5′-AACATCACAGTGCTCCATTCTAT-3′) was designed to distinguish YSS1 transcript from yss1 transcript in the yss1 mutant. In a search for candidate genes, both full-length cDNA and genomic DNA of wild type and the yss1 mutant were amplified and sequenced.
For the complementation test, a 960-bp wild-type YSS1 coding region was amplified (primer pair YSS1GBD, Suppl. Table S1) from II-32B and inserted downstream of the UBIQUITIN promoter in the binary vector pCUbi1390 to generate the transformation plasmid pUbi::YSS1. Due to difficulties in obtaining callus of the yss1 mutant (an indica variety), the recombinant plasmid pUbi::YSS1 was introduced into individuals with the homozygous yss1 allele in a japonica genetic background selected from one 02428/yss1 F2:3 line by Agrobacterium-mediated transformation as described previously (Hiei et al. 1994). To determine whether the abnormal yss1 transcript is responsible for the mutant phenotype, the full-length yss1 cDNA sequence from yss1 mutant was also amplified (primer pair YSS1GBD, Suppl. Table S1) and cloned into the binary vector pCUbi1390 to generate the transformation plasmid pUbi::yss1, which was then transformed into rice cultivar Kitaake as described above. The expression levels of total YSS1 transcripts and abnormal yss1 transcript in transgenic plants were separately detected by real-time RT-PCR primers YSS1 and yss1 (Suppl. Table S1).
Quantitative real-time RT-PCR analysis
Total RNA was extracted using an RNA Prep Pure Plant kit (Tiangen Co., Beijing, China) and reverse-transcribed using a SuperScript II kit (TaKaRa) following the manufacturer’s instructions. Real-time RT-PCR was performed using a SYBR® Premix Ex Taq™ kit (TaKaRa) on an ABI prism 7500 Real-Time PCR System. Primers for real-time RT-PCR designed using GenScript (https://www.genscript.com/ssl-bin/app/primer) are listed in Suppl. Table S1. The rice ubiquitin gene (LOC_Os03g13170) was used as the internal control (primer pair Ubq, Suppl. Table S1). The 2−ΔΔCT method was used to analyze relative gene expression (Livak and Schmittgen 2001).
Phylogenetic and sequence analysis
Sequence information of YSS1 gene was found with the NCBI accession number: NM_001060901.1. Homologous sequences of YSS1 protein were identified using NCBI (http://www.ncbi.nlm.nih.gov/) and sequences alignment was performed with BioEdit software. A maximum parsimony (MP) tree was generated using PAPU4.0B software with 1000 bootstrap replications.
Subcellular localization
Subcellular localizations of YSS1 protein and its chloroplast-targeting signal were predicted using ChloroP (Emanuelsson et al. 1999) and TargetP (Emanuelsson et al. 2000), respectively. The coding sequences of YSS1 and yss1 were amplified and cloned to the N-terminus of green fluorescent protein (GFP) in the transient expression vector pA7-GFP to form YSS1-GFP and yss1-GFP fusion plasmids (primer pairs shown in Suppl. Table S1). Both constructs were separately transformed or co-transformed with chloroplast nucleoid marker (PEND-CFP) into rice protoplasts and incubated in darkness at 28 or 23 °C for 16 h before examination (Terasawa and Sato 2005; Chen et al. 2006). The full-length cDNA of YSS1 was also inserted into the binary vector 1305-GFP (Ren et al. 2014; primer pairs YSS1-1305GFP are shown in Suppl. Table S1), and then introduced into rice cultivar Kitaake by Agrobacterium-mediated transformation. Fluorescence of GFP was observed using a confocal laser scanning microscope (Carl Zeiss LSM700).
Protein extraction and Western blot analysis
Fresh rice leaves were weighed and ground with liquid nitrogen. Total plant proteins were extracted with moderate NB1 buffer (50 mM Tris-Mes, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT and protease inhibitor cocktail CompleteMini tablets, pH 8.0), followed by denaturation at 95 °C for 5 min. The extracts were resolved in 10 or 15 % SDS–PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (0.45 μm, Millipore). The proteins were detected with relevant antibodies (Huada Co., Beijing, China) using an ECL Plus Western Blotting Detection Kit (Thermo). The software “Quantity One” was used to quantify the intensity of chloroplast proteins.
Results
yss1 is a young seedling stripe mutant
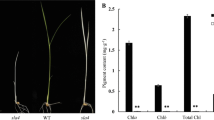
The yss1 mutant was isolated from a 60Co-irradiated population of indica variety II-32B. Under paddy conditions, the mutant seedlings exhibited an obvious striated leaf phenotype compared to the green leaves of wild-type plants (Fig. 1a). To characterize the yss1 mutant in detail, we performed time-course analysis from the L3–L5 stage. Notably, irrespective the developmental stage, the striated leaf phenotype occurred in leaf 2 to leaf 4, with the significant difference detectable in leaf 3. Interestingly, the phenotypic defects in leaf color completely disappeared in leaf 5 and onwards (Fig. 1b). Furthermore, examination of the Chl contents showed that compared with the wild type, the defective yss1 leaves had reduced both Chl a and Chl b levels, and that reduction degree was positively related to the leaf color defects. The chlorotic leaves (i.e., leaf 2 to leaf 4) did not become green and the Chl contents also failed to restore to normal levels as the plants growth (Fig. 1b, c). Together, these data suggested that yss1 is a young seedling stripe mutant.
Phenotypic analysis of the yss1 mutant. a Phenotype of wild type and the yss1 mutant at the seedling stage grown in field. b Detailed observations of leaf color appearance of the yss1 mutant at the L3, L4, and L5 stages grown in field, respectively. The numbers indicate different leaves. c Determination of both Chl a and Chl b contents in different leaves corresponding to b (L3-2, L3-3, L4-2, etc. indicate different leaves; for example, L3-2 represents leaf 2 of the L3 stage). Values are mean ± SD from three independent repeats
To verify whether the yss1 phenotype was temperature-dependent, we planted wild-type and yss1 mutant seedlings in a growth chamber with 12 h of light/12 h of darkness under different temperature conditions, and examined the expressivity of the yss1 phenotype and Chl contents. As shown in Fig. 2a, under constant 20 °C (C20) condition, the yss1 mutant exhibited extreme chlorosis and contained only traces of Chl a and b, compared with the moderate chlorosis phenotype of the wild-type plants. As temperatures increased, the mutant plants showed lighter chlorotic symptoms and higher accumulations of Chls at constant 23 °C (C23) condition (Fig. 2b). Under optimum 12 h of light at 30 °C/12 h of darkness at 20 °C (L30/D20) condition for rice growth, yss1 mutant plants displayed mild striping and accumulated more Chls than at either C23 or C20 conditions (Fig. 2c). Notably, the yss1 mutant produced light chlorosis only at the tip of leaf 3 and had much higher Chl concentrations at constant 30 °C (C30) condition (Fig. 2d), indicating that high temperature could weaken the yss1 phenotype. These observations suggested that the mutation in yss1 causes a temperature-sensitive chlorotic defect in rice seedlings.
Phenotypic characteristics and Chls levels of wild type and yss1 mutant under different temperature and photoperiod conditions. Leaf 3 of wild-type and the yss1 mutant seedlings grown in a growth chamber with 12 h of light/12 h of darkness under C20 (a), C23 (b), L30/D20 (c) and C30 (d) and under short day (SD) and long day (LD) (e) was used for Chl measurement, respectively. Values are mean ± SD of three replicates. FW fresh weight, C20 constant 20 °C. C23 constant 23 °C, L30/D20 12 h of light at 30 °C/12 h of darkness at 20 °C, C30 constant 30 °C. LD represents 15 h of light at 30 °C/9 h of darkness at 25 °C. SD represents 10 h of light at 30 °C/14 h of darkness at 25 °C
Next, we tested whether the yss1 phenotype was also affected by other external conditions. The yss1 chlorotic phenotype appeared more severe under short day than under long day (Fig. 2e). However, the observed phenotypic defects of yss1 mutant plants grown under various nutrient-deficient mediums showed no significant difference from those grown under normal condition, except for slightly delayed growth such as in N and K deficiencies (Suppl. Fig. S1a). These data collectively suggested that the phenotypic defects of the yss1 mutant are more sensitive to photoperiod than nutrient deficiencies.
The yss1 mutant impairs chloroplast development
To investigate the effect of the yss1 mutation on chloroplast development, we compared the number of Chl-containing cells and the ultrastructure of chloroplasts in yss1 mutant and wild-type plants under different temperature conditions. Confocal microscopic observations demonstrated that under L30/D20, green leaves in the wild-type plants had almost normal numbers of Chl-containing cells except for stomatal and motor cells (Suppl. Fig. S2a). In contrast, yss1 mutant leaves had less numbers of Chl-containing cells at C20 (Suppl. Fig. S2b), and the number of Chl-containing cells increased with the increase of temperature (Suppl. Fig. S2b–d).
We further observed the ultrastructure of chloroplasts using TEM. Under C30, similar to the wild type, most of chloroplasts in yss1 mutant exhibited well-developed lamellar structures, which were equipped with normally stacked grana and thylakoid membranes (Fig. 3a, b). Under L30/D20, chlorotic leaf cells in the yss1 mutant contained more undifferentiated chloroplasts (Fig. 3c). Under both C23 and C20, most of chloroplasts were undifferentiated in the chlorotic leaves of yss1 mutant and abnormalities were greater at C20 than at C23 (Fig. 3d, e). The percentages of normal (NC) versus undifferentiated chloroplasts (UC) also increased with temperature (Fig. 3f). These data revealed that the yss1 mutation disrupts the chloroplast differentiation especially at low temperature conditions.
Ultrastructures of chloroplasts in the leaf 3 of wild type (a) and yss1 mutant (b-e) at the L3 stage under various temperature conditions. Electron micrographs showing ultrastructures of chloroplasts from the leaf 3 of wild-type (a) and yss1 mutant (b–e) seedlings grown in a growth chamber with 12 h of light/12 h of darkness under C30 (a, b), L30/D20 (c), C23 (d) and C20 (e). The right micrographs in a–e represent magnified chloroplasts as indicated by arrowheads in the left micrographs. C30 constant 30 °C, L30/D20 12 h of light at 30 °C/12 h of darkness at 20 °C, C23 constant 23 °C, C20 constant 20 °C, TM thylakoid membrane, N nucleus, M mitochondrion, NC normal chloroplast, UC undifferentiated chloroplast. Bars 2 μm (left); 0.5 μm (right). f Percentage of NC and UC in mesophyll cells of wild type and yss1 mutant at different temperatures; 50 cells were examined for each determination. The data are from two independent repeats
Cloning of the YSS1 gene
Genetic analyses of crosses yss1/II-32B, II-32B/yss1, and yss1/02428 showed that the yss1 phenotype was controlled by a single nuclear recessive gene (Suppl. Table S2). Cross 02428/yss1 was used to generate an F2 mapping population; 94 F2 individuals with typical striated leaves were used to map the yss1 locus to a 2.7-cM (centimorgan) segment distal to the simple sequence repeat (SSR) marker RM127 at the end of chromosome 4L (Fig. 4a). We further narrowed the yss1 locus to a 16.4-kb region between the insertion/deletion (InDel) markers JW12-16 and JW12-18 on bacterial artificial chromosome (BAC) clone OSJNBb0020J19 based on 1223 F 2 homozygous yss1 individuals (Fig. 4b). There are four open reading frames (ORFs) in the segment and the genes were predicted using FGENESH 2.2 (http://www.softberry.com). Comparison with the wild-type genomic sequences revealed that the third ORF (expression protein designated LOC_Os04g59570) in the yss1 mapping interval carried a single base ‘A’ deletion neighboring the splicing donor site of the 8th intron (Fig. 4c, d). cDNA sequencing further showed that the single base deletion in yss1 mutant caused an incomplete splicing of the 8th intron (83 bp in length), thus producing two types of transcripts. One is the wild-type YSS1 transcript; the other is an 8th intron-retained yss1 transcript (Fig. 4e). If translated, the later would encode a truncated protein, named yss1, which is missing the C-terminal 36 amino acid residues, but contains an extra 4 amino acid residues resulting from the frame-shift translation. To more clearly distinguish the aberrant yss1 transcript from the wild-type YSS1 transcript in yss1 mutant, primer pairs K83-F/R amplifying a smaller DNA fragment was used (YSS1 607 bp vs yss1 690 bp; Fig. 4e). YSS1 with ten exons and nine introns was identified as gene encoding a polypeptide of 319 amino acid residues belonging to the DUF3727 superfamily. Blast analysis and sequence alignment showed only one copy of the YSS1 gene in rice and no paralogs. The YSS1 protein had high similarity to DUF3727 superfamily proteins in other species in a region of nearly 200 amino acids in the C-terminus (Suppl. Fig. S3). Phylogenetic analysis demonstrated that YSS1 orthologs broadly exist in many photosynthetic organisms and evolve from cyanobacteria, suggesting that YSS1 is probably transferred from prokaryotic to eukaryotic genomes (Suppl. Fig. S4).
Positional cloning of the YSS1 gene. a The yss1 locus was mapped to a 2.7-cM region containing six bacterial artificial chromosome (BAC) clones at the terminal region of chromosome 4L. b Fine mapping of YSS1. The yss1 locus was narrowed to a 16.4-kb interval between InDel markers JW12-16 and JW12-18 on the BAC clone OSJNBb0020J19 using 1223 F 2 homozygous mutant plants. c Four open reading frames (ORFs) were predicted in the region. The third ORF has a deletion of base ‘A’ approaching the splicing donor site of the 8th intron. Gray boxes indicate 5′ and 3′ UTR. Black boxes indicate exons and lines between them indicate introns. ATG and TGA represent the start and stop codons, respectively. d Diagrams depicting the partial exon/intron structure of YSS1 and yss1 transcripts. The intron sequences are in lowercase and italic. “gt”, “ag”, “-”, and “TAA” are in red and indicate splicing donor site, splicing acceptor site, the deleted “A” base, and newly formed stop codon, respectively. e PCR amplification using primer pairs K83-F/R as indicated in c. Note that two bands were amplified in yss1 mutant (up yss1 transcript; down YSS1 transcript) but only one in wild type
Decreased accumulation of YSS1 transcripts causes the yss1 phenotype
To determine whether downregulation of YSS1 transcript or increased accumulation of yss1 transcript caused the yss1 phenotypes, we performed a set of stable transgenic experiments. Firstly, we transformed a 960-bp wild-type YSS1 coding sequence under the control of the UBIQUITIN promoter into the yss1 homozygous mutant. Four green and two chlorotic plants derived from T1 transgenic progenies were selected for further real-time RT-PCR analysis using primer pairs YSS1-F/R common for both YSS1 and yss1 transcripts as well as primer pairs yss1-F/R specific for yss1 transcript. As shown in Fig. 5a, the wild-type plant accumulated high level of YSS1 transcript, while the yss1 mutant expressed significantly lower levels of both YSS1 and yss1 transcripts, indicating primer pairs used here could distinguish and quantify both transcripts efficiently. Further analyses showed that YSS1 transcript was differently overexpressed in four green plants but not in two chlorotic plants (Fig. 5a). And the four YSS1-overexpressed transgenic plants showed a complete rescue of the mutant phenotypes, including leaf color appearance, Chls accumulation and chloroplast structure (Fig. 5b–d). Next, to investigate the possibility that the yss1 had modified inhibitory effect on wild-type YSS1, we overexpressed the yss1 transcript driven by an UBIQUITIN promoter in rice cultivar Kitaake. The transgenic plants failed to phenocopy the yss1 mutant defects, although they show distinctly increased accumulations of the yss1 transcript (Fig. 5e, f). This result was consistent with our genetic analysis demonstrating that yss1 phenotype was controlled by a single nuclear recessive gene. Together, these data collectively suggested that LOC_Os04g59570 indeed corresponds to the YSS1 gene, and that it is the declined expression of YSS1 transcript but not the accumulation of yss1 transcript confers the yss1 mutant phenotype.
YSS1 complementation test. a Real-time RT-PCR analysis showing the expression levels of both YSS1 and yss1 transcripts in four green and two chlorotic T1 transgenic progenies grown in a greenhouse. ‘YSS1’ and ‘yss1’ indicate normal and abnormal transcripts, respectively. The ubiquitin gene was used as an internal control. Data are mean ± SD of three repeats. b Phenotype of the yss1 mutant and four complemented transgenic plants at the L3 stage. c Chls levels of wild type, yss1 and four complemented plants at the L3 stage, respectively. d TEM analysis of chloroplast ultrastructure in wild type, yss1 mutant, and a representative complemented plant C9. Positive plants C1, C5, C9, and C10 were selected from the T1 transgenic progenies C1–C10. e Expression levels of YSS1 and yss1 in Kitaake and 3 overexpression lines of yss1 transcript (OE1–OE3) using real-time RT-PCR. OE overexpression. f Phenotypic characteristics of the leaf 4 at the L4 stage in 3 yss1-overexpressed lines (OE1–OE3)
Subcellular localization of YSS1
ChloroP (Emanuelsson et al. 1999) and TargetP (Emanuelsson et al. 2000) prediction analyses showed that YSS1 possessed a chloroplast-targeting signal (29 amino acid residues in length and probably 3.1 kDa in size) in the N-terminus, and likely localized to the chloroplasts. To determine the actual localization of YSS1, we constructed the plasmid that fused YSS1 to the N-terminus of GFP and transiently expressed them in rice protoplasts. Confocal microscopic observations demonstrated that YSS1-GFP displayed a spotty localization seemingly associated with chloroplasts (labeled by Chl autofluorescence; Fig. 6a). To further verify the subcellular localization of YSS1 in vivo, we constructed the P35S:YSS1-GFP binary vector and transformed it into the rice japonica cultivar Kitaake. Positive T1 transgenic seeds were grown for 5 days, and used to observe GFP signals. Fluorescence microscopy observation revealed that YSS1-GFP exhibited a similar localization pattern in the transgenic leaf sheath and roots to that observed in the rice protoplasts (Fig. 6b; Suppl. Fig. S5). In addition, further fluorescence microscopy observation demonstrated that the lack of C-terminal 36 amino acid residues in yss1 mutant did not interfere with the localization pattern of yss1-GFP fusion proteins (Fig. 6c). It is reported that GFP-tagged plastid binding protein PEND also appeared in small dot-like structures throughout the chloroplasts and can be used as an fluorescent marker protein characteristic for plastid nucleoids (Terasawa and Sato 2005). To determine the nature of YSS1-GFP-labeled punctate compartments, we coexpressed YSS1-GFP and PEND-CFP in rice protoplasts. As shown in Fig. 6d, YSS1-GFP colocalized with PEND-CFP, suggesting that YSS1 may be a chloroplast nucleoid-association protein.
Subcellular location of YSS1 and yss1 proteins. GFP signals of YSS1-GFP fusion protein were located in the chloroplasts by transient expression analyses in rice protoplasts (a). b Localization of YSS1 protein in leaf sheath cells from 5-day-old YSS1-GFP transgenic plants. c yss1-GFP protein was also localized in the chloroplasts of rice protoplasts. d Colocalization of YSS1 (GFP) and PEND (CFP) within chloroplast nucleoids. GFP, GFP signals of YSS1 (a–b, d) and yss1 (c); Auto chlorophyll autofluorescence; Bright bright field; CFP CFP signals of chloroplast nucleoid marker PEND; Merged merged images. e Western blot analysis with transformed cells expressing YSS1-GFP and yss1-GFP in rice protoplasts, respectively. The diagram below indicates the calculated band sizes of YSS1-GFP and yss1-GFP fusion proteins with or without processing. TPL transit peptide-like sequence, MP mature protein
Next, we transformed the fusion plasmids YSS1-GFP and yss1-GFP into rice protoplasts, respectively. Western blot analyses showed that two bands were separately produced in transformed protoplasts expressing either YSS1-GFP or yss1-GFP, indicating that both YSS1 and yss1 proteins could be posttranslationally processed in chloroplasts and accumulated stably. In addition, it is worth noting that the detected proteins had a larger molecular weight than the calculated size, most likely resulting from the potential occurrence of modifications such as glycosylation or/and phosphorylation (Fig. 6e). These results further supported the notion that YSS1 may be targeted to the chloroplasts by the predicted 29 amino acid transit peptides.
Expression pattern of YSS1 and its response to external conditions
To investigate the expression profile of YSS1, we analyzed YSS1 expression in various tissues by real-time RT-PCR. YSS1 was constitutively expressed in culms, flag leaves, young panicles and leaf sheaths at the booting stage, as well as young leaves and roots at the seedling stage. The strongest expression was detected in young leaves (Fig. 7a). In addition, we further determined the expression of YSS1 in different sections of seedlings at the L3 stage. The wild-type YSS1 plants showed significantly higher expression in leaf 3 than in other tissues. In contrast, YSS1 transcripts (including wild-type YSS1 transcript and the aberrant yss1 transcript) were also most abundant in leaf 3 of the yss1 mutant, but the overall expression levels were lower than in the wild type (Fig. 7b, c). In addition, we analyzed the expression of YSS1 during leaf development. YSS1 achieved the highest expression levels in wild-type leaves at the L3 stage, and accumulated more in newly expanded leaves but decreased with leaf aging (Fig. 7d). These observations suggested that YSS1 might play an important role at the early leaf development stage, but with a relatively minor role in the mature leaf.
Expression analysis of YSS1. a Expression levels of YSS1 transcript in culms (C), flag leaves (FL), panicles (P), and leaf sheaths (LS) at the booting stage, as well as young leaves (YL) and roots (YR) at the seedling stage of wild-type plants grown in a paddy field. b Diagram of L3 stage rice seedling in which the leaf 3 was fully expanded. L3-1, L3-2, L3-3, and L3-4 represent the first, second, third, and fourth leaf, respectively. SB (shoot base), a 5 mm piece from the bottom of the shoot. P0–P6 indicates the developmental stages of leaf formation, respectively. c Real-time RT-PCR analysis of YSS1 transcript in SB, L3-2, L3-3, and L3-4 of the wild-type and yss1 seedlings at the L3 stage grown in a paddy field. d Expression levels of YSS1 in leaves of wild-type plants at different stages grown in a paddy field. L2-2, L3-2, L3-3, etc. indicate different leaves (for example, L3-2 represents leaf 2 of the L3 stage), respectively. e YSS1 expression analysis during greening of etiolated seedlings by light. After 10-day darkness treatment, etiolated wild-type seedlings grown in a growth chamber under constant 30 °C (C30) were illuminated for 3, 6, 9, 12, 15, 18, or 24 h, respectively. Seedlings grown under continuous darkness were used as a control. The YSS1 gene was normalized using an ubiquitin gene as an internal reference. Data are mean ± SD of three replicates
To identify whether YSS1 expression was induced by light, we tested the alteration of YSS1 transcripts during light-induced greening of wild-type seedlings. The YSS1 mRNA levels increased after exposure to light, peaked after 9 h of illumination, and then gradually decreased and tended to be stable after 15 h of illumination (Fig. 7e). This indicated that light might play an important role in regulating YSS1 expression. Furthermore, we measured the transcript levels of YSS1 and yss1 in yss1 mutant leaves under different temperature conditions and at the L5 stage or booting stage by real-time RT-PCR. Total YSS1 transcripts accumulated less in the yss1 mutant than those in the wild type under different temperature conditions, and reduced as temperatures decrease. yss1 transcripts in yss1 mutant seldom accumulated at C30, but increased under L30/D20 conditions, and accumulated to high levels at C20. Correspondingly, normal YSS1 transcripts lessened as temperatures decrease (Fig. 8a). This corresponded to the observed yss1 phenotypes at different temperatures (Fig. 2a–d). Similarly, total YSS1 expression levels were lower in the leaves of yss1 mutant than wild type at the L5 stage or booting stage (Fig. 8b). We also detected that yss1 transcripts in the yss1 mutant were abundantly present in leaves 2, 3, and 4, but extremely seldom in leaf 5 at the L5 stage and flag leaves at the booting stage. In contrast, in the yss1 mutant plants, the normal YSS1 transcript increased in leaf 5 at the L5 stage, potentially explaining the phenotypic alternation at this stage. These data further supported the notion that decreased accumulation of YSS1 transcripts caused the yss1 phenotypic alternations at the seedling stage.
Real-time RT-PCR detection of transcriptional levels of YSS1 and yss1 transcripts. a Expression levels of YSS1 and yss1 transcripts in the wild-type and yss1 mutant seedlings grown in a growth chamber with 12 h of light/12 h of darkness under C30, L30/D20, and C20. C30 constant 30 °C, L30/D20 12 h of light at 30 °C/12 h of darkness at 20 °C, C20 constant 20 °C. b Expression levels of YSS1 and yss1 transcripts in different leaves at the L5 stage and flag leaves (FL) at the booting stage grown in a paddy field. L5-2, L5-3, L5-4, L5-5 indicate the second, third, fourth, and fifth leaf at the L5 stage, respectively. c Expression levels of YSS1 and yss1 transcripts in the wild-type and yss1 mutant seedlings grown in a growth chamber with short day (SD) and long day (LD). LD represents 15 h of light at 30 °C/9 h of darkness at 25 °C; SD represents 10 h of light at 30 °C/14 h of darkness at 25 °C. “YSS1” and “yss1” indicate normal and abnormal transcript, respectively. The ubiquitin gene was used as an internal control. Data are mean ± SD of three repeats
The yss1 phenotype was affected by some external factors; therefore, we further examined whether the YSS1 expression levels were also regulated. Consistent with the phenotype observed in Fig. 2e, YSS1 transcripts in the yss1 mutant accumulated more under long day than short day (Fig. 8c). However, under nutrient element deficiency conditions, YSS1 expression showed no significant difference in the yss1 mutant compared with the control (Suppl. Fig. S1b). These data further provide evidence that YSS1 expression levels were closely associated with the yss1 mutant phenotypic defects.
yss1 is defective in plastid transcription and synthesis of chloroplast proteins
Given that YSS1 is a chloroplast nucleoid-localized protein, we speculated that it may be responsible for plastid transcription. Next, we examined the plastid gene expression patterns in wild type and yss1 mutant using quantitative real-time RT-PCR method. Expression of PEP-dependent photosynthesis genes (psaA, psaB, psbA, and rbcL) was clearly down-regulated in the yss1 mutant compared to wild type. On the contrary, NEP-dependent housekeeping genes (rpoA, rpoB, and rpoC1) were up-regulated in the yss1 mutant (Fig. 9a). This is a typical plastid gene expression pattern resulting from impaired PEP transcription.
Real-time RT-PCR expression of chloroplast genes and immunoblot analysis of chloroplast proteins. a Transcript abundance of PEP-dependent (psaA, psaB, psbA, and rbcL) and NEP-dependent genes (rpoA, rpoB, and rpoC1). Total RNA was extracted from leaf 3 of wild-type and yss1 mutant seedlings at the L3 stage under a paddy field. The ubiquitin gene was used as an internal control. Data are mean ± SD of three repeats. “**”and “*” separately indicate significance at P = 0.01 and P = 0.05 by Student’s t test. b Western blot analysis of plastid-encoded and nuclear-encoded proteins in WT and yss1 plants. Total proteins were extracted from fully emerged leaf 3 of wild-type and yss1 seedlings at the L3 stage grown in a paddy field. Antibodies for the following chloroplast proteins were used: psbA, psbD, psaA, rpoB, rpoC2, AtpB, rpl2, NADH4, NADH5, NADH7, CP29, ATPase, OEEP1, OEEP3, POR, and OspTAC3. HSP82 was used as a loading control. c Quantification of the band intensity of chloroplast proteins in WT (×0.5 and ×0.25) and yss1 mutant (×1) compare to WT (×1) corresponding to b
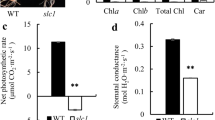
Next, we performed Western blot analyses to detect whether the synthesis of chloroplast proteins were affected in the yss1 mutant. The contents of all tested plastid-encoded proteins, including photosystem-II subunits (psbA and psbD), photosystem-I subunit (psaA), RNA polymerase subunits (rpoB and rpoC2), ATP synthase CF1 β subunit (AtpB), ribosomal protein L2 (RPL2), and NADH dehydrogenase subunits (NADH4, NADH5, and NADH7), were reduced in yss1 seedlings. Similarly, the contents of nuclear-encoded proteins CP29 (chlorophyll a/b binding protein), ATPase (ATP synthase β subunit), OEEP1, and OEEP3 (oxygen-evolving enhancer proteins 1 and 3), were also decreased. However, we found no clear reductions in nuclear-encoded proteins POR (protochlorophyllide oxidoreductase proten) and OspTAC3 (plastid transcriptionally active 3) in the yss1 mutant (Fig. 9b, c).
Discussion
In recent years, a number of Chl-deficient mutants have been reported in rice. Some of them are seedling-specific and gradually become normal during later growth stages, and some affect the plants for the entire life cycle. For example, young seedling albino (ysa) mutant plants show obvious albino phenotypes before the L3 stage but gradually turn green and recover to normal wild-type appearance after the L6 stage (Su et al. 2012). ylc1 is a young leaf chlorosis mutant that exhibits a chlorotic phenotype in seedlings or young leaves (Zhou et al. 2013). In addition, ygl1 and vyl are reported to affect rice growth throughout the whole developmental stages. Leaves of the ygl1 mutant display a yellow-green leaf phenotype and contain less chlorophyll than the wild type throughout life cycle (Wu et al. 2007). The vyl mutant also shows decreased Chl levels throughout all developmental stages (Dong et al. 2013). Here, we identified a novel young seedling stripe mutant, yss1, which produced striated leaves before the L5 stage. Unlike the above mutants, the chlorotic leaves in yss1 mutant was detected only in the second, third, and fourth leaves and did not become green (Fig. 1b, c). Phenotypic analyses demonstrated that yss1 is a low temperature-sensitive mutant (Figs. 2a, Fig. 3e). The v1, v2, v3, st1, and ylc1 mutants were also reported to exhibit more extreme phenotypes at C20 (Kusumi et al. 1997, 2011; Sugimoto et al. 2004, 2007; Yoo et al. 2009; Zhou et al. 2013), potentially due to the blocked translation of plastid proteins (Grennan and Ort 2007; Rogalski et al. 2008; Liu et al. 2010). However, not all Chl-deficient mutants are to be temperature-sensitive; for example, the vyl mutant exhibits a similar chlorotic phenotype across a range of temperatures (Dong et al. 2013).
Previous studies showed that mutations in the splicing site usually abolished the splicing of impaired intron and generated an elongated transcript, such as wsl2 and vyl (Mao et al. 2012; Dong et al. 2013). In contrast, in addition to the 8th intron-retained transcript, the yss1 mutant was still able to generate wild-type transcript and displayed a stripe leaf phenotype at the early seedling stage. We speculated that point mutations abolishing the splicing of the 8th intron would lead to more severe or even lethal phenotypic defects. Supporting the notion, dramatically down-regulated expression of wild-type YSS1 transcript as temperature decreased led to more severe leaf color phenotype and disrupted chloroplast development (Figs. 2a–d, 3, 8a). Quantative analysis using real-time RT-PCR showed that the mutation in yss1 mutant led to significantly reduced accumulation of total YSS1 transcripts (Fig. 5a), possibly due to RNA quality control mechanisms (Isken and Maquat 2007). Furthermore, our transgenic experiments demonstrated that downregulation of YSS1 transcript potentially conferred the leaf color phenotype in yss1 mutant (Fig. 5b). Supporting the notion, overexpression of the aberrant yss1 transcript under the control of the same UBIQUITIN promoter failed to phenocopy the yss1 mutant defects (Fig. 5e, f), thus also ruling out the possibility that yss1 caused yss1 mutant phenotypes potentially through modified inhibitory effect on wild-type YSS1 functions. Interestingly, we observed that yss1 mutant phenotypic alternations were significantly enhanced as temperature decreased, and quantative analysis using real-time RT-PCR showed that expression of wild-type YSS1 transcript was dramatically down-regulated as temperatures decrease (Fig. 8a), which is a possible dosage effect, similar to the previously reported Wx gene for regulating amylase content (Wang et al. 1995; Isshiki et al. 1998).
The reason why many Chl-deficient mutants exhibit seedling-specific chlorotic phenotypes but subsequently develop normally remains unknown. Previous assumptions were that paralogs may compensate the functions during the later stages, or such genes play essential roles during early leaf stages but are dispensable at later growth stages (Su et al. 2012; Zhou et al. 2013). As for the question how a generally down-regulated condition increased abnormal transcript only in specific leaf, one possible explanation is that low temperature can disrupt the amendment of YSS1 transcript in yss1 mutant, thus producing more abnormal yss1 transcript, and vice versa at high temperature. The enhanced accumulation of YSS1 transcript in leaf 5 at the L5 stage may explain the reason why the yss1 phenotype disappeared in leaf 5. However, as for stage-specific absence of yss1 phenotype in other stages, such as the booting stage, it is possible that YSS1 might be sufficient to maintain the wild-type phenotype, although it has lower expression (Fig. 8b). Alternatively, other regulatory factors may also facilitate the phenotypic recovery at these stages.
It is noted that YSS1-GFP and the Chl autofluorescence signals do not completely merge (Fig. 6a), indicating a possible non-chloroplast second localization of YSS1-GFP. To further identify the nature of YSS1-GFP located compartments, we tried to perform cell fractionation analysis of the YSS1 protein in rice seedling. Unfortunately, we failed to detect the distribution of native YSS1 protein in various cell compartments in wild-type plant, although extensive efforts, most likely due to the poor quality of the anti-YSS1 antibodies. Actually, spotty localization patterns similar to YSS1-GFP have been reported previously and are defined as nucleoids, such as AtHSP21 and AtpTAC3 (Yagi et al. 2012; Zhong et al. 2013). However, the reason why partial spotty nucleoids seemingly located in the periphery of Chl autofluorescence-labeled chloroplast remains unknown. It is reported that the nucleoid-associated proteins might participate in plastid genes expression (Yagi et al. 2012; Zhong et al. 2013). Similarly, decreased accumulation of YSS1 gene also led to the reduced expression levels of PEP-dependent photosynthetic genes (psaA, psaB, psbA, and rbcL) and the increased transcriptional accumulations of genes for NEP (rpoA, rpoB, and rpoC1) (Fig. 9a), implying that PEP activity for plastid transcription was suppressed in yss1 mutant. Furthermore, our results showed that the levels of nuclear-encoded and plastid-encoded chloroplast proteins were generally decreased in yss1 mutant (Fig. 9b, c). These data suggest that YSS1 mutation might influence the chloroplast development potentially through modulating PEP activity during the early leaf development in rice. Given the low temperature influence on yss1 phenotype, it is possible that decreased chloroplast translation and transcription by PEP may be due to oxidative damage resulting from the decreased electron transport efficiency and ROS production (Nishiyama et al. 2006). Because the lack of evidence that YSS1 protein interacts with the PEP complex, we cannot conclude that YSS1 is directly involved in plastid transcription. Additionally, nucleoid-localized nature of YSS1 protein does not mean that it is essentially involved in transcription; this can be other functions such as replication and stability. Further experiments aim at isolating YSS1-interacting partners and functional analyses of these factors are needed to unravel the molecular function of YSS1 in regulating chloroplast development. Nevertheless, this is the first report that a DUF3727 superfamily member participates in regulation of chloroplast development. The mutational yss1 protein lacked an intact DUF3727 domain and thus probably lost it’s, at least partial, function specific for early leaf development in rice. Interestingly, we also recently presented evidence that a DUF1338 superfamily member FLO7 functions as a plant unique regulator required for amyloplast development in peripheral endosperm in rice (Zhang et al. 2016).
Author contribution statement
KN Z, YL R, F Z, YH W, X Z, ZJ C, ZF L, and JM W designed research, KN Z, YL R, F Z, Y W, L Z, J L, SL Z, H Z, and JL W conducted experiments, KN Z, WW M, LW W, FQ W, XP G, and CL L analyzed data, KN Z, YL R, CM W, L J, and JM W wrote and modified the paper. All authors read and approved the manuscript.
Abbreviations
- DUF:
-
Domain of unknown function
- GFP:
-
Green fluorescent protein
- NEP:
-
Nuclear-encoded phage-type RNA polymerase
- PEP:
-
Plastid-encoded plastid RNA polymerase
- TEM:
-
Transmission electron microscopy
- yss1 :
-
Young seedling stripe1
References
Allison LA (2000) The role of sigma factors in plastid transcription. Biochimie 82:537–548
Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. BBA-Bioenergetics 1847:761–769
Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein–protein interactions in rice. Mol Plant Pathol 7:417–427
Dong H, Fei GL, Wu CY et al (2013) A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol 162:1867–1880
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Fujiwara M, Nagashima A, Kanamaru K, Tanaka K, Takahashi H (2000) Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase sigma factors in Arabidopsis thaliana. FEBS Lett 481:47–52
Grennan AK, Ort DR (2007) Cool temperatures interfere with D1 synthesis in tomato by causing ribosomal pausing. Photosynth Res 94:375–385
Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16:4041–4048
Hedtke B, Börner T, Weihe A (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277:809–811
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hiratsuka J, Shimada H, Whittier R et al (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217:185–194
Hricova A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141:942–956
Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T (2005) A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42:133–144
Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Gene Dev 21:1833–1856
Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H (1997) Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domain sigma 70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA 94:14948–14953
Isshiki M, Morino K, Nakajima M, Okagaki RJ, Wessler SR, Izawa T, Shimamoto K (1998) A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J 15:133–138
Kasai K, Kawagishi-Kobayashi M, Teraishi M, Ito Y, Ochi K, Wakasa K, Tozawa Y (2004) Differential expression of three plastidial sigma factors, OsSIG1, OsSIG2A, and OsSIG2B, during leaf development in rice. Biosci Biotechnol Biochem 68:973–977
Kindgren P, Kremnev D, Blanco NE et al (2012) The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J 70:279–291
Kubota Y, Miyao A, Hirochika H, Tozawa Y, Yasuda H, Tsunoyama Y, Niwa Y, Imamura S, Shirai M, Asayama M (2007) Two novel nuclear genes, OsSIG5 and OsSIG6, encoding potential plastid sigma factors of RNA polymerase in rice: tissue-specific and light-responsive gene expression. Plant Cell Physiol 48:186–192
Kühn K, Richter U, Meyer EH, Delannoy E, de Longevialle AF, Borner T, Millar AH, Small I, Whelan J (2009) Phage-type RNA polymerase RPOTmp performs genespecific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21:2762–2779
Kusumi K, Mizutani A, Nishimura M, Iba K (1997) A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J 12:1241–1250
Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K (2011) A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J 68:1039–1050
Liu X, Rodermel SR, Yu F (2010) A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol 10:287
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ (2012) Nucleoiden-riched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158:156–189
Mao B, Cheng Z, Lei C et al (2012) Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 235:39–52
Moreira D, Le Guyader H, Philippe H (2000) The origin of red algae and the evolution of chloroplasts. Nature 405:69–72
Mullet JE (1993) Dynamic regulation of chloroplast transcription. Plant Physiol 103:309–313
Nishiyama Y, Nishiyama Y, Allakhverdiev S, Murata N (2006) A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. BBA-Bioenergetics 1757:742–749
Pfalz J, Pfannschmidt T (2013) Essential nucleoid proteins in early chloroplast development. Trends Plant Sci 18:186–194
Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18:176–197
Quesada V, Sarmiento-Manus R, Gonzalez-Bayon R et al (2011) Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J 68:738–753
Ren YL, Wang YH, Liu F et al (2014) GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicle traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 26:410–425
Reumann S, Inoue K, Keegstra K (2005) Evolution of the general protein import pathway of plastids (review). Mol Membr Biol 22:73–86
Rogalski M, Schottler MA, Thiele W, Schulze WX, Bock R (2008) Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20:2221–2237
Sakamoto W, Miyagishima S, Jarvis P (2008) Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. Arabidopsis Book 6:e0110
Steiner S, Schroter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157:1043–1055
Su N, Hu ML, Wu DX et al (2012) Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol 159:227–238
Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K (2004) The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol 45:985–996
Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K (2007) The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J 52:512–527
Terasawa K, Sato N (2005) Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol 46:649–660
Tozawa Y, Hasegawa H, Terakawa T, Wakasa K (2001) Characterization of rice anthranilate synthase alpha-subunit genes OASA1 and OASA2. Tryptophan accumulation in transgenic rice expressing a feedback-insensitive mutant of OASA1. Plant Physiol 126:1493–1506
Wang ZY, Zheng FQ, Shen GZ, Gao JP, Snustad DP, Li MG, Zhang JL, Hong MM (1995) The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J 7:613–622
Wu ZM, Zhang X, He B et al (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145:29–40
Xu YZ, Arrieta-Montiel MP, Virdi KS et al (2011) MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23:3428–3441
Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T (2012) Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc Natl Acad Sci USA 109:7541–7546
Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC (2009) Rice Virescent3 and Stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150:388–401
Zhang L, Ren YL, Lu BY et al (2016) FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. J Exp Bot 67:633–647
Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C (2013) Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25:2925–2943
Zhou KN, Ren YL, Lv J et al (2013) Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 237:279–292
Acknowledgments
This work was supported by the grants from the Fundamental Research Funds for Excellent Young Scientists of ICS-CAAS (Grant to YR, 2014JB04-009; 1610092015003-08), the 973 Program of China (2011CB100102), the High Technology Program from NDRC ([2012]1961), the 863 Program of China (2014AA10A604-4), and Jiangsu Science and Technology Development Program (BE2014394). We thank Dr. Bing Hu (Nanjing Agricultural University) for assistance in transmission electron microscopy analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
K. Zhou, Y. Ren, F. Zhou and Y. Wang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, K., Ren, Y., Zhou, F. et al. Young Seedling Stripe1 encodes a chloroplast nucleoid-associated protein required for chloroplast development in rice seedlings. Planta 245, 45–60 (2017). https://doi.org/10.1007/s00425-016-2590-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2590-7