Abstract
Main conclusion
RNAi technology was applied to suppress the expression of starch branching enzyme IIa and IIb and to increase amylose content in maize endosperm, and stably inherited high-amylose maize lines were obtained.
Amylose is an important material for industries and in the human diet. Maize varieties with endosperm amylose content (AC) of greater than 50 % are termed amylomaize, and possess high industrial application value. The high-amylose trait is controlled by multi-enzyme reaction and intricate gene–environment interaction. Starch branching enzymes are key factors for regulating the branching profiles of starches. In this paper, we report the successful application of RNAi technology for improving amylose content in maize endosperm through the suppression of the ZmSBEIIa and ZmSBEIIb genes by hairpin SBEIIRNAi constructs. These SBEIIRNAi transgenes led to the down-regulation of ZmSBEII expression and SBE activity to various degrees and altered the morphology of starch granules. Transgenic maize lines with AC of up to 55.89 % were produced, which avoided the significant decreases in starch content and grain yield that occur in high-amylose ae mutant. Novel maize lines with high AC offer potential benefits for high-amylose maize breeding. A comparison of gene silencing efficiency among transgenic lines containing different hpSBEIIRNA constructs demonstrated that (1) it was more efficient to use both ZmSBEIIa and ZmSBEIIb specific regions than to use the conserved domain as the inverted repeat arms; (2) the endosperm-specific promoter of the 27-kDa γ-zein provided more efficient inhibition than the CaMV 35S promoter; and (3) inclusion of the catalase intron in the hpSBEIIRNA constructs provided a better silencing effect than the chalcone synthase intron in the hpRNA construct design for suppression of the SBEII subfamily in endosperm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize is the most important crop cultivated worldwide with higher production than rice or wheat. Maize starch has the most useful chemical components and accounts for 80 % of global starch production. Starch is an important carbohydrate as a source of food for human and raw material for industry. Maize kernel starch consists of approximately 30 % amylose [degree of polymerization (DP) < 5,000], which is essentially polymerized by α-1,4 linkage and approximately 70 % amylopectin (DP > 5,000), which is highly branched using a α-1,6 linkage at the branching points. Amylose is distinguished from amylopectin by its insolubility in water, lower hot viscosity, finer film-forming ability, high gelatinization temperature, stability (Matveev et al. 2001) and its higher ability to form digestion-resistant complexes (Lourdin et al. 1995; Friedman et al. 1999). Due to these properties, amylose is an ideal material for use in confectionery, papermaking, textiles, pharmaceutical products, resistant starch production and the plastics industry (Krogars et al. 2003), especially for producing photolyzable agricultural films, which can replace non-biodegradable plastics, thereby reducing environmental pollution. However, amylose production is limited by its high costs and the low yield of the procedures used for its extraction from normal starch, and by the shortage of germplasm with high-amylose traits. The maize amylose-extender (ae) mutant exhibits an amylose content (AC) of greater than 50 %, and is deficient in the starch branching enzyme (SBE) IIb isoform (Moore and Creech 1972; Garwood et al. 1976). Corn hybrids exhibiting AC of 50 % to nearly 90 % have been generated by conventional breeding methods by using this mutant and modifier genes. However, the high AC traits in these hybrids are accompanied by side-effects caused by the mutant locus, including high water content during kernel development and great loss in total starch and grain yield; it has been proved to be difficult to avoid these side-effects using conventional breeding methods.
Transgenic approaches including antisense inhibition and RNAi technology have greatly improved critical traits in some important crops. hpRNA-mediated RNAi technology is more stable and effective than antisense-induced gene silencing, and it has been speculated that the reason for this is that the mechanism of hpRNA-mediated suppression does not require RNA-dependent RNA polymerase (RdRP) for the production of dsRNA that is required in antisense inhibition (Chuang and Meyerowitz 2000; Béclin et al. 2002; Saurabh et al. 2014). RNAi technology allows the silencing of a specific gene or of members of a multigene family; therefore, this technology can be used as a powerful tool in the breeding of high-amylose varieties. SBEs, which catalyze the cleavage of oligosaccharide chains by hydrolysis of the α-1,4 linkage and transferring it to another glucan chain to form branch points through α-1,6 linkages, are critical for controlling AC in storage organs; thus, SBEs have become the main RNAi target for genetic modifications in crop improvements.
In maize, starch synthesis is catalyzed by ADP-glucose pyrophosphorylase (AGPase), starch synthases (SSs), SBEs and starch debranching enzymes (DBEs) (Martin and Smith 1995; Nelson and Pan 1995). Based on the characterization of waxy maize and amylose-free (amf) potato mutants that contain 100 % amylopectin and no amylose, it has been suggested that granule-bound starch synthase (GBSS) I (encoded by the Wx gene) is responsible for amylose synthesis (Smith 1990; Denyer et al. 2001). Amylopectin synthesis is more complex, involving the participation of multi-isoforms of SBEs and DBEs. Due to the lack of success in the generation of high AC varieties through over-expression of the GBSS I enzyme, amylopectin synthesis suppression has been used to improve amylose content in crop storage organs, such as potato tubers and wheat, barley and rice endosperm, with ACs of over 64 % (Schwall et al. 2000; Hofvander et al. 2004; Regina et al. 2006, 2010; Guo et al. 2008; Butardo et al. 2011; Zhu et al. 2012).
SBEs isoenzymes have been characterized in many important crops, including potato tuber (Poulsen and Kreiberg 1993; Jobling et al. 1999), pea embryo (Burton et al. 1995), rice (Mizuno et al. 1992), wheat, and maize endosperm (Morell et al. 1997). In maize, three SBE isoforms have been isolated: SBEI, SBEIIa, and SBEIIb, which belong to two distinct classes (Boyer and Preiss 1981; Fisher et al. 1996; Gao et al. 1996). Maize SBEIIa and SBEIIb belong to class II or A (also including SBEIIa and SBEIIb from wheat and rice, pea SBE A, and potato SBE A), whereas SBEI belongs to class I or B (also including wheat SBEI, rice SBEI, pea SBE B and potato SBE B). The N-terminal domain of SBE proteins plays a role in determining the length of the transferred chains, whereas the C-terminal domain controls substrate specificity and catalytic activity. The amino acid sequences of SBEI and SBEII subfamilies exhibit greater variations in the N-terminal and C-terminal sequences than in the central portion (Kuriki et al. 1997). Thus, SBEI primarily transfers longer glucan chains and uses amylose as a substrate, whereas SBEII preferentially transfers shorter chains of DP 6–9 and prefers to use amylopectin rather than amylose as substrates (Guan and Preiss 1993; Takeda et al. 1993; Guan et al. 1997). Although SBEI is expressed moderately in vegetative organs and is strongly expressed in the late development stage of endosperms (Gao et al. 1996), amylopectin chain-length distribution profiles in both transient and storage starches are unaffected in SBEI-deficient mutant maize plants (Blauth et al. 2002).
In maize, the cDNA of ZmSBEIIa and ZmSBEIIb share 67 % overall identity, but they exhibit distinct expression patterns and these proteins play distinct roles in starch branching. In maize leaves, ZmSBEIIa mRNA is accumulated extensively, but the ZmSBEIIb transcript is undetectable. Conversely, in kernels, ZmSBEIIb is expressed abundantly, and its transcript level is about 35 times higher than ZmSBEIIa in endosperm (Gao et al. 1997). In kernels of the sbe2a mutant, the degree of starch branching and composition are unaffected (Blauth et al. 2001), whereas in sbe2b mutant (the ae mutant), kernels contain elongated starch granules, exhibit drastically increased AC and the structure of amylopectin is distinctly altered, exhibiting a lower branching degree, an increased proportion of long B chains and average chain length (Wang et al. 1993; Li et al. 2008). These observations indicate that SBEIIa is mainly responsible for transitory starch branching and is required for starch diurnal cycling in leaf chloroplasts (Yandeau-Nelson et al. 2011), whereas SBEIIb primarily functions in endosperm. Due to the existence of these three SBEs, it is challenging to use mutants that are deficient in single members to illustrate the specific role of each isoform and to elucidate their cooperation in amylopectin synthesis. However, it is time-consuming and labor-intensive to combine the desirable mutant genes and their modifiers to generate high-amylose traits using traditional breeding methods.
In this study, based on an analysis of the impact on increasing AC in maize endosperm by suppressing ZmSBEIIa and ZmSBEIIb genes using RNAi, we obtained transgenic corn plants with high ACs of up to 55.9 % by introducing hpSBEIIRNA constructs into an elite inbred maize line Chang7-2. We also revealed the effects of arm length and specificity, promoter type and the loop intron sequence on hpSBEIIRNA silencing efficiency.
Materials and methods
Cloning of SBEIIRANi fragments and construction of hairpinRNAi vectors
All fragments of ZmSBEIIa (GenBank U65948) or ZmSBEIIb (GenBank EF433557) were cloned by PCR using cDNAs obtained from immature kernels of the maize (Zea mays L.) elite inbred line DH4866 (obtained from Laizhou Academy of Agricultural Science of China) at 16 DAP. These fragments were cloned in both the sense and antisense directions in the vector pFGC5941 using restriction enzymes sites in the 5′ regions of the oligonucleotides. The primers used for cloning these fragments are listed in Supplemental Table S1. The PCR parameters used to amplify fragment 2bc, 2bd, 2bt, 2ac, and 2ad were detailed in Supplemental Materials and methods. Vectors pFGC5941-P35S::2bc-bar, pFGC5941-P35S::2bd-bar, pFGC5941-P35S::2bc-in’-bar, pFGC5941-P35S::2ac-bar, pFGC5941-P35S::2ad-bar, pFGC5941-P27kD::2bc-bar, pFGC5941-P27kD::2bd-bar, pFGC5941-P27kD::2bc-in’-bar, pFGC5941-P27kD::2ac-bar and pFGC5941-P27kD::2ad-bar were constructed as detailed in Supplemental Materials and methods (pFGC5941 GenBank accession no. AY310901; P27kD GenBank Accession No. EF064982).
Maize transformation and transgenic line production
An Agrobacterium-mediated maize shoot-tip genetic transformation was performed as previously described by Li et al. (2011). The maize (Zea mays L.) elite inbred line Chang7-2 was used in this study. The surviving transformed plants after spraying with the herbicide Glufosinate (181.9 mg/l) were chosen for PCR assay. Herbicide-resistant and PCR-positive plants were transferred to the field for harvest. From the progeny of the transformed plants transgenic lines for this study were selected.
PCR analysis and southern blotting
Genomic DNA for PCR analysis was isolated from young maize leaves using the CTAB protocol. The PCR primers used to amplify the bar gene were 5′-ATGAGCCCAGAACGACGCC-3′ (forward primer) and 5′-TCAGATCTCGGTGACGGGC-3′ (reverse primer). The cycle parameters were as follows: 95 °C for 5 min; 35 cycles of 95 °C for 1 min, 56 °C for 1 min, and 72 °C for 1 min; and over-extension for 7 min.
Genomic DNA (30 μg) isolated from PCR-positive T3 transgenic maize leaves was digested for 22 h at 37 °C using XbaI (only one XbaI cleavage site is present within the T-DNA region). Fully digested DNA samples of the transgenic plants and of nontransformed maize were fractionated on 0.8 % agarose gels and transferred to nylon membranes (Roche, Mannheim, Germany). The probe used was labeled with digoxigenin-dUTP (Roche) using the PCR fragment of the bar gene as the template. Probe hybridization and detection were carried out by using the DIG NBT/BCIP detection kit for nucleic acids (Roche) following the manufacturer’s instructions.
RNA isolation and real-time RT PCR
Total RNA was isolated from wild type and T2 transgenic maize kernels taken from the middle of ears at 20 DAP using Trizol reagent (Sangon, Shanghai, China) and DNA was digested using DNase I (Takara, Dalian, China). DNase-treated RNA (500 ng) from each sample was reverse-transcribed with random hexamers using a RT reagent kit from Takara following the manufacturer’s instructions. The real-time RT PCR primers used for the SBEIIb gene were designed within the conserved region of ZmSBEIIb. The forward primer sequence was 5′-TTGCCCTTCCTGTTCACG-3′, and the reverse primer sequence was 5′-CACCCATCTTCCAAGTTTCATC-3′. The maize actin gene was used as an internal control. The sense primer for actin was 5′-ATCACCATTGGGTCAGAAAGG-3′, and the antisense primer was 5′-GTGCTGAGAGAAGCCAAAATAGAG-3′. The real-time RT PCR was performed using a ChromoTM 4 (Bio-Rad, Hercules, CA, USA) and SYBR® Premix Ex Taq from Takara. The real-time RT PCR reaction systems and amplification parameters were performed as the manufacturer’s instructions. Fold changes of the ZmSBEII transcripts were calculated according to the \(2^{{ - \varDelta \varDelta C_{\text{t}} }}\) method with three samples and the formula ΔΔC t = (C t,target gene − C t,actin )transgenic − (C t,target gene − C t,actin )WT.
Starch branching enzyme activity determination
Total proteins were extracted from 2–3 seeds harvested at 20 DAP. The SBE activity of the wild type and homozygous SBEIIRNAi transgenic maize kernels was determined according to the method by Zhao et al. (2007) as detailed in Supplemental Materials and methods. The optical density of the final product was measured at 660 nm. The reaction without incubation at 37 °C for 20 min was used as the control. SBE activity was calculated as ΔOD660 (%) = [OD660 (t 0) − OD660 (t 20)]/OD660 (t 0) and one percentage decrease of the starch-iodine-blue was considered as one SBE activity unit.
Measurement of kernel starch and amylose content
Measurement of starch content in mature seed from transgenic lines and WT were performed as previously described (Li et al. 2011). The starch content of the kernels was calculated according to the formula: starch content % = G × 0.9/DW × 100 % (G represents the total glucose content of the eight seeds, and DW represents the dry weight of the eight seeds).
According to the starch content, an amount of powered endosperm equal to 100 mg starch was used for measurement of amylose content by iodine binding method as detailed in Supplemental Materials and methods. The amylose content of each sample was calculated as the ratio of amylose to the total starch content: amylose (%) = G × 100/5/50 (G represents the amylose content of the 5 ml sample of gelatinized starch, as calculated based on the standard curve).
Gel permeation chromatography (GPC) analysis of debranched starches
Maize endosperm starches were isolated from WT and homozygous SBEIIRNAi transgenic maize plants by alkaline protease method and purified maize starches were debranched by isoamylase (Hayashibara Biochemical Laboratories Inc., Okayama, Japan) as described previously (Zhu et al. 2010).
Molecular weight distribution of debranched maize starches were measured with a GPC systerm (PL-GPC 220, Polymer Laboratories Varian Inc., Amherst, MA, USA), equipped with differential refractive index detector and three columns connected in sequence (Phynogel 00H-0646-KO, 00H-0644-KO, 00H-0642-KO columns, Phenomenex, Torrance, CA, USA). The mobile phase was dimethyl sulphoxide (DMSO) containing 5 mM NaNO3 at a flow rate of 0.8 ml/min and the column temperature was 80 °C. A series of dextran standards (American Polymer Standards Corporation, Mentor, OH, USA) were used as standards for calibration curve of retention time versus molecular weight calculations.
Comparison of 100-seed weight and kernel phenotypes of the wild type and transgenic plants
Wild type and homozygous SBEIIRNAi transgenic maize plants were grown in a plot under the same culture condition. Ears were harvested at maturity and five ears of each transgenic line were collected and dried to a constant weight. The weight of 100 seeds of three replicates of each sample was measured. Kernels of SBEIIRNAi transgenic maize plants were compared with those of the wild type and photographed. Transgenic lines with higher 100-grain weights and different grain phenotypes were selected for further research.
Scanning electron microscopic observation of endosperm
The endosperms of mature seeds of the WT and transgenic T3 maize lines were observed using scanning electron microscopy (SEM). Sample pieces of the endosperm were prepared by removing a part of the kernel near the top using a razor blade; the samples were then loaded on SEM specimen stubs with double adhesive tape and sputter-coated with gold (Cressington Sputter Coater 108, Cressington Scientific Instruments Inc., Watford, Hertfordshire, UK). The specimens were scanned and photographed at 5 kV under high vacuum at room temperature using a FEI Quanta FEG 250 environmental scanning electron microscope (ESEM).
Agricultural trait analysis of wild type and SBEIIRNAi transgenic maize in field trials
All yield data were obtained using an experimental field at Lvjia in Jinan (117 29′E, 36 54′N). Wild type and homozygous SBEIIRNAi transgenic T3 lines were chosen for the field trials, and each maize line was sown in a four-row plot (3 m in length, 0.5 m between rows); the seeds were sown at an interval of 20 cm between each plant in April (plant density was approximately 65,000 plants/hectare), and three replicates were planted. Seedlings were thinned at the 3-leaf stage, and 40 plants were retained in each plot. The growth of the SBEIIRNAi transgenic and wild-type lines was monitored during the entire growing period. After pollination, plant height and ear position were measured. Ten plants were harvested from each plot for agricultural trait analysis. Mature ears were dried to constant weight, and ear length, ear weight, 100-grain weight, and starch and amylose contents were recorded.
Results
Construction of hpSBEIIRNA vectors and production of transgenic maize lines
In plants, the effects of hairpinRNA-mediated RNA interference on target genes are influenced by many factors, including the size and sequence of the inverted repeat arms, the intron used between the sense and antisense arms, and the promoter driving the RNAi construct. To obtain the optimal inhibition of ZmSBEIIa and ZmSBEIIb expression using hpRNA constructs, ten hpSBEIIRNA constructs were constructed using different fragments of the conserved or specific sequence of the ZmSBEIIa and/or ZmSBEIIb genes (Fig. 1a). The expression of five hpSBEIIRNA constructs (hp-bc, hp-bd, hp-in’, hp-ac and hp-ad) was driven by the CaMV 35S promoter (P35S) and a maize endosperm-specific 27kD zein promoter (P27kD). Within each set of five hpSBEIIRNA constructs, two constructs (hp-bc and hp-bd) contained 893 nt or 467 nt conserved sequence fragments from ZmSBEIIb; two hpSBEIIRNA constructs (hp-ac and hp-ad) contained 417 nt or 154 nt specific fragments from ZmSBEIIa, which were fused with a 295 nt fragment of the ZmSBEIIb specific region; and one hpSBEIIRNA construct (hp-in’) contained the modified first intron of the castor bean catalase gene CAT-1 instead of the original chalcone synthase intron (CHSA intron) from Petunia in the construct hp-bc.
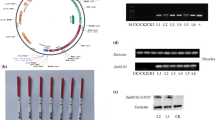
Plasmid construction and molecular characterization of transgenic maize. a T-DNA region of the plasmid pFGC5941-P35S::2bc-bar, pFGC5941-P35S::2bd-bar, pFGC5941-P35S::2bc-in’-bar, pFGC5941-P35S::2ac-bar, pFGC5941-P35S::2ad-bar, pFGC5941-P27kD::2bc-bar, pFGC5941-P27kD::2bd-bar, pFGC5941-P27kD::2bc-in’-bar, pFGC5941-P27kD::2ac-bar and pFGC5941-P27kD::2ad-bar. P35S, the CaMV35S promoter from cauliflower mosaic virus; P27kD, the endosperm-specific promoter from the maize 27-kDa γ-zein; PMAS, a promoter from Agrobacterium tumefaciens; LB left T-DNA border, RB right T-DNA border. b PCR analysis of genomic DNA containing the bar gene: lane 1, nontransformed control Chang7-2; lanes 2–6, transgenic maize lines P35bc, P35in’, P35ad, P27bc, and P27bd; lane 7, the plasmid pFGC5941-PMAS::bar; lanes 8–10, transgenic maize lines P27in’, P27ac and P27ad; line M, the DL2000 marker. c Southern blotting analysis for bar gene. Genomic DNA (30 μg) was digested with XbaI, and hybridized with the bar probe. 1, nontransformed control Chang7-2; 2-9, transgenic maize lines P35bc, P35in’, P35ad, P27bc, P27bd, P27in’, P27ac, and P27ad. M is the λ-EcoT14 I digest DNA marker
Gel permeation chromatography analysis of isoamylase-debranched starch from the transgenic T3 and WT plants. WT, nontransformed maize Chang7-2. Fraction 1 (F1) consisted of amylose; fraction 2 (F2) consisted of high molecular weight chains (containing long B chains); fraction 3 (F3) consisted of low molecular chains (A + B1 chains); Fraction 2 and fraction 3 derived from amylopectin. The area ratio of F1/(F1 + F2 + F3) was treated as the amylose content
These hpSBEIIRNA vectors were used to transform the maize elite inbred line Chang7-2. The corresponding transgenic line name and fragment length of each hpSBEIIRNAi vector were shown in Supplemental Table S2. 10 % of the transformed seedlings survived after herbicide glufosinate screen at the three-leaf stage. The progeny of those Glufosinate-resistant plants were confirmed by PCR analysis for bar gene (Fig. 1b). PCR-positive independent transgenic lines were obtained and further confirmed by Southern blotting analysis for bar gene. As shown in Fig. 1c, signals for exogenous bar gene were detected with specific hybridization patterns. These results showed that the bar gene had been stably integrated into the genomes of the transgenic plants, providing indirect evidence of the integration of the SBEIIRNAi transgenes into the genomes of the transgenic plants. The progeny of the transgenic lines were generated from eight hpSBEIIRNA vectors, in which 6 lines for hp-P35bc, 5 lines for hp-P35in’, 11 lines for hp-P35ad, 5 lines for hp-P27bc, 9 lines for hp-P27bd, 3 lines for hp-P27in’, 7 lines for hp-P27ac and 9 lines for hp-P27ad.
Expression analysis of SBEII in developing kernels of SBEIIRNAi transgenic maize plants
To examine the inhibitory effects of these hpSBEIIRNA constructs on the expression of SBEIIa and SBEIIb in maize endosperm, levels of ZmSBEII transcription and SBE activity were analyzed using total RNA or proteins extracted from kernels of the SBEIIRNAi transgenic lines and wild type at 20 DAP (Supplemental Fig. S1). We observed a significant decrease on the transcriptional level of ZmSBEII in SBEIIRNAi transgenic lines in contrast to the wild type, but the degree of the decrease clearly varied among different transgenic lines. The expression of ZmSBEII in lines P35bc was higher than that in lines P27bc. Within the P27kD transgenic lines, the hp-bc and hp-bd constructs exhibited similarly reduced amplitude on ZmSBEII transcription levels (a fold change of 0.42 compared with WT), while the hp-ac or hp-ad constructs resulted in much lower ZmSBEII transcript levels (approximately 0.33-fold of WT). SBE activity in endosperm from transgenic plants was dramatically decreased to values less than 40 % of that in WT and the SBE activity were concordant with the ZmSBEII transcriptional level. The results indicated that these hpSBEIIRNA constructs could effectively down-regulate the expression of SBEII in the endosperm of SBEIIRNAi transgenic maize plants.
Measurement of amylose content and total starch contents in the transgenic maize kernels
To understand the effects of the down-regulation of ZmSBEIIa and ZmSBEIIb expression on the amylose content (AC) of the endosperms and the total starch amounts in kernels, we measured the AC of mature kernels from the T3 to T5 SBEIIRNAi transgenic lines and the WT using both iodine staining method (in Supplemental Fig. S1) and gel permeation chromatography (GPC) method (in Fig. 2; Table 1). The GPC method could give a more accurate AC and exhibit molecular weight distribution change of isoamylase-debranched starch.
The AC of the SBEIIRNAi transgenic lines were greatly increased to 41.86–55.89 % compared with 29.07 % in WT (Fig. 2; Table 1,) and these increases were negatively correlated with the changes in ZmSBEII transcriptional level and SBE activity. Consistent with the pooled field experiment data (Table 2), the largest increase was observed in the SBEIIRNAi P27ac transgenic lines, which exhibited an AC of 55.89 % of total starch, nearly twice the AC of the WT. Line P27ad exhibited an AC of 51.95 %, which was distinctly higher than that exhibited by lines P27bc and P27bd (46.24, 44.78 %). Lines P27bc, P27in’, and P27ad exhibited higher ACs (46.24, 48.26, 51.95 %, respectively) than the corresponding P35bc, P35in’, and P35ad lines (41.86, 45.31, 46.83 %, respectively). The ACs of lines P35in’ and P27in’ (45.31, 48.26 %; the hpRNA constructs contained the catalase intron) were slightly higher than that of lines P35bc and P27bc (41.86 and 46.24 %), which contained the chalcone synthase intron. In addition, the GPC results showed that in starches from SBEIIRNAi transgenic plants, F1 consisted of much more amylose molecules with higher molecular weight and the ratios of F2/F3 were greater than those from wild-type plants, suggesting that transgenic maize starches contained more long chains in amylopectin. After several generations of self-pollination, we measured the ACs of the T5 progeny of each transgenic line (by iodine staining method). The ACs of T5 plants were similar to those of their T3 ancestors, with differences of less than one percent in the values (Table 2). In addition, the T5 maize plants exhibited similar plant phenotypes and kernel production to those of their T3 ancestors, suggesting that the impact of the hpSBEIIRNA constructs on AC and the high AC trait was stably inherited in the transgenic lines.
However, the total starch content decreased from 66 % in the WT to less than 63 % in the SBEIIRNAi lines, exhibiting a negative correlation with AC enhancement as shown in Fig. 3a and Table 2. Line P27ac, which exhibited the highest AC, had the lowest starch content (90.5 % of that of WT; 66 % → 57.5 %). As an exception, among the P35S set of SBEIIRNAi transgenic plants, line P35in’ exhibited a higher AC but also a slightly higher starch content than line P35bc. These results suggested that the hpSBEIIRNA constructs strongly enhanced the AC of maize endosperm, and slightly affected the starch content and kernel yields.
Starch analysis and kernel yields of the transgenic T3 and WT plants. a Comparison of the starch content in DW of the mature kernels. b Comparison of the 100-grain weights. WT nontransformed maize Chang7-2. Three independent lines were analyzed in each transgenic group, and three independent experiments were conducted for each line. The values presented are the means of the nine replicates ± SD. Different small letters represent statistical significances between the means for each construct (P < 0.05, Duncan’s test)
Morphologic changes in the starch granules of transgenic maize lines
Scanning electron microscopy was used to identify morphological changes in starch granules from the farinaceous endosperms of mature SBEIIRNAi transgenic seeds (Fig. 4b). The starch granule of wild type Chang7-2 is a regular spheroid or ellipsoid with a smooth surface. In contrast, the starch granules in transgenic endosperms with reduced SBE activity and high AC exhibited obvious morphological alterations; i.e., irregular ellipsoidal shapes and nonuniform size. The surface of starch granules from lines targeting the ZmSBEIIb conserved domain (P35bc, P35in’, P27bc, P27bd, and P27in’) exhibited slightly protuberances, whereas starch granules from lines P35ad, P27ac and P27ad exhibited more concave morpha. This result indicates that the alterations in starch composition significantly influenced starch granule morphology and had a further impact on the ratio of farinaceous endosperm to cutin endosperm in kernels.
Morphology of kernels and starch granules from the T3 transgenic maize lines and untransformed Chang7-2. a Comparison of the kernel phenotype. Application of the SBEIIRNAi transgene in endosperm resulted in decreased kernel size. WT nontransformed maize Chang7-2. b Scanning electronic micrographs of endosperm starch granules from SBEIIRNAi transgenic T3 and WT mature maize seeds (×1,200). Bar 10 μm
Agronomic traits of different transgenic maize plants
The effects of the hpSBEIIRNA constructs on the agronomic traits of transgenic plants were carefully examined in the field. The transgenic T3 (lines P35bc, P35in’, P35ad, P27bc, P27bd, P27in’, P27ac, and P27ad) and wild-type plants were grown at an experimental station in Jinan. The plant heights and ear locations were recorded at the pollination stage, and the ear length, ear weight, 100-grain weight, AC in endosperm and starch content of the kernels were measured after the mature ears were dried to a constant weight.
As shown in Table 2, plants of the SBEIIRNAi transgenic lines P35bc, P35in’ and P35ad (152.7–153.5 cm) were slightly shorter than that of wild-type plants (156.4 cm), and the differences of ear location among the various lines were not significant. The ear length of the SBEIIRNAi lines was not significantly different or was just slightly shorter than that of WT. However, there were remarkable differences in kernel characteristics among the SBEIIRNAi transgenic lines and the WT. The ear weight, 100-grain weight and starch content of kernels from the transgenic lines were lower than those of the WT, to different extents (Figs. 3b, 4a; Table 2). Among the transgenic lines, the P35bc plants exhibited the lowest AC (approximately 41.86 %) and the highest starch content and exhibited a 100-grain weight 5 % lower than that of the WT; Line P27ac exhibited the greatest decrease in the ear weight and 100-grain weight (101.8 ± 1.9 and 17.0 ± 1.0 g, respectively). For comparison, those of the WT were 108.3 ± 2.5 and 20.2 ± 0.7 g. These results indicated that the AC enhancement in endosperm had a negative impact on total starch content, kernel weight and kernel yield per plant; however, the negative effects were small compared with those seen for the ae mutant. Moreover, the agronomic traits of the transgenic plants, such as AC in endosperm, total kernel starch content and kernel weight, were steadily inherited.
Discussion
The high-amylose trait holds significant value for related industries and human health, and is controlled by complex multi-enzyme catalysis and gene interactions, as well as sophisticated genetic background. Genetic engineering including gene silencing technology, has been extensively applied in high-amylose crop breeding, effectively avoiding the side-effects of the ae mutant on starch synthesis and crop yield (Schwall et al. 2000; Regina et al. 2006, 2010; Zhu et al. 2012); however, less than ideal results were obtained in maize (Guan et al. 2011). In this study, we found that stably inherited transgenic maize lines containing high ACs of up to 55.89 % with slight losses of starch content and/or grain yield were produced using RNAi technology for co-suppression of SBEIIa/b.
SBEs are important for regulating the branching profiles of starch and different isoforms play distinct roles in controlling the number of branching points, chain-length distribution and the ratio of amylose to amylopectin (Jeon et al. 2010). Successfully applied genetic manipulation strategies of SBE family for high-amylose breeding can be grouped into two ways: simultaneous inhibition of both SBEI and SBEII subfamilies as reported in potato and rice (Schwall et al. 2000; Zhu et al. 2012) or suppression of only SBEII subfamily as reported in wheat and barley (Regina et al. 2006, 2010). Considering that loss of SBEI function resulted in indistinguishable changes in starch structure (Blauth et al. 2002), in this study, the second way was applied to simultaneously down-regulate SBEIIa and SBEIIb activity in maize endosperm. We obtained high-amylose lines with 55.89 % AC, however, the AC was not as high as that of transgenic high-amylose potato (75 %), wheat or barley (greater than 64 %). The possible reasons were speculated that co-suppression of SBEIIa and SBEIIb might result in different effects on AC in different species for their diverse starch structures and characteristics of SBE isoforms. In wheat endosperm, SBEIIa and SBEIIb isoenzymes accounts for equal levels of SBE activity (Regina et al. 2005); however, AC was not affected when only SBEIIb was suppressed, whereas SBEIIa and SBEIIb co-suppression resulted in AC distinctly increased to over 70 % (Regina et al. 2006). Similar cases were observed in barley (Sun et al. 1998; Regina et al. 2010). These studies indicate that in wheat and barley, SBEIIa and SBEIIb play complementary roles in controlling amylose content. In maize SBEIIb deficiency and the heavy suppression of SBEIIa together did not result in the AC much higher than that of ae mutant, in which SBEIIa activity is unaffected (Hedman and Boyer 1983). It can be inferred that SBEIIa to SBEIIb exhibit functional complementarity in maize endosperm but not as significantly as in wheat or barley. From another aspect, in developing maize endosperm, starch biosynthetic enzymes are physically coordinated and both SBEIIa and SBEIIb coprecipitate with SSI (starch synthase I). Association of these enzymes into complexes might provide an environment for the ordered construction of glucan polymers destined for crystallization and starch granule formation (Hennen-Bierwagen et al. 2008). The organization and features of enzymes in complexes might have direct effects on substrate binding and on the position of branching due to the spatial locations of the SS and BE active sites relative to each other. This might explain why the suppressive effects of SBEIIa/b on AC were not as high as those in wheat and barley.
In maize, cDNA sequences of the central portion of ZmSBEIIa (from 381 to 2,443 nt in GenBank U65948) and ZmSBEIIb shared 78 % similarity, and the 5′ end fragment (0–380 nt) and the 3′ noncoding region (2,443–2,763 nt) are divergent, with only 29 and 38 % similarity, respectively (Gao et al. 1997). In this study, we adopted two strategies for the suppression of SBEII subgroup isoforms: (1) the conserved region of ZmSBEII genes were cloned from ZmSBEIIb and used as hpRNA arms; (2) the specific regions of both ZmSBEIIa and ZmSBEIIb were fused and used as inverted repeat arms. The results show that both strategies resulted in a range of silencing efficacies in transgenic lines, as inferred from the decreased degree of ZmSBEII transcription levels and SBE activity, and distinct ACs in endosperm.
A strategy similar to strategy (1) was successfully applied in wheat and barley (Regina et al. 2006, 2010), based on an hp-SBEIIa construct containing the SBEII homologous region. In this study, using strategy (1), ACs increased to 41.86–45.31 % in transgenic lines from 29.07 % in wild type Chang7-2. Guan et al. (2011) obtained transgenic maize exhibiting an increased AC (35.2 %, compared with 21.1 % in non-transgenic inbred H99) using a 562-nt homologous region from ZmSBEIIa as the hpRNA arm (sharing 83.6 % similarity with the corresponding region of ZmSBEIIb). The difference between the two results might be caused by (a) the 893-nt or 467-nt ZmSBEIIb conserved regions used in our report shared 84.2 or 83.3 % similarity with the corresponding region of ZmSBEIIa and had two to three regions with 23–24 nt of 100 % identity to ZmSBEIIa mRNA sequence; however, the 3′-terminal of the 562-nt ZmSBEIIa fragment in Guan’s report only contained one 22-nt fragment exhibiting 100 % identity to ZmSBEIIb mRNA; (b) in Guan’s study, only the P35S promoter was used, whereas in this study, the hpSBEIIRNA constructs were separately driven by P35S and the endosperm-specific promoter P27kD; furthermore, P27kD exhibited higher silencing efficacy; (c) genetic background of H99 and Chang7-2 might have resulted in different suppression efficiencies for ZmSBEIIa/b. A comparison of the SBEIIRNAi lines showed that strategy (2) of using the specific regions of both ZmSBEIIa and ZmSBEIIb was more effective than strategy (1). Due to the lack of a trustworthy tool to predict the precise Dicer cutting site and the sequence of the processed mature siRNA for a given sequence in plants, we could only speculate that using strategy (2), a number of mature 21-nt siRNAs with complete base-pairing to the target mRNAs could be generated by cleavage of the precursor molecules.
Target region, intron sequence, and promoter type were key factors that determine silencing efficacy (Hirai and Kodama 2008). As reported by Wesley et al. (2001), stable and effective silencing is achievable using RNAi containing hpRNA arms of 98–853 nt in plants. In our study, this finding was confirmed, in that when the arm length of hpRNA constructs was 893 nt (bc) or 467 nt (bd) within ZmSBEII homologous domain, lines P27bc and P27bd showed similar silencing efficacy judging from ZmSBEII transcription levels, SBE activity and AC in endosperm. However, when the gene-specific region was targeted, the P27ac line exhibited lower ZmSBEII transcript levels and SBE activity, and higher AC (55.89 %) than the P27ad line (51 %). The difference between these results was caused by the arm length of 417 nt (c region) or 154 nt (d region) from the ZmSBEIIa-specific region. An arm length of 712 nt (hp-ac, 295 nt + 417 nt) appears to be more effective than an arm length of 449 nt (hp-ad, 295 nt +154 nt). In this case, the size and sequence specificity of the inverted repeat arm of the hpRNA constructs determined the silencing efficiency. This is consistent with results obtained in GBSSIRNAi transgenic potatoes (Heilersig et al. 2006).
Due to the lack of data regarding the promoter strength of P35S and P27kD in maize endosperm, we designed two sets of hpSBEIIRNA constructs promoted by P35S or P27kD. The expression data presented in Supplemental Fig. S1a indicate that in developing maize endosperm, the P27kD::hpRNA constructs suppressed ZmSBEII expression much more strongly than P35S, and P27kD::hpRNA lines exhibited higher ACs (up to 55.89 %) than the P35S lines (up to 46.83 %). In addition, P27kD::hpRNA lines exhibited normal plant morphology, whereas plant height was slightly lower in the P35S lines than in the WT. These results suggest that the P27kD::hpRNA constructs were more efficient at inducing ZmSBEII genes silencing in endosperm. Similar cases were previously reported that the embryo-specific promoters were better in embryo (Schmidt and Herman 2008; Schmidt et al. 2011).
In conclusion, we successfully applied RNAi technology to increase amylose content in maize endosperm and obtained stably inherited transgenic maize lines with ACs of up to 55.89 %, mimicking the high AC trait of ae mutant. In an earlier report (Garwood et al. 1976), seven independently occurring ae maize alleles (named B1, B2, i1, i2, M1, M2, and Ref) were backcrossed into W64A and W23 inbred lines to produce high-amylose corns, but collapsed kernel, lackluster endosperm, and about 20 % decrease in yield appeared. The elite inbred line Chang7-2 in this paper had superiority in combining ability and fine kernel characteristics and had been extensively used in conventional breeding in China. In our study, the Chang7-2 SBEIIRNAi transgenic maize plants showed relatively normal kernel morphology and just slight decrease in starch content and yield. We speculated that the activity of SBEIIb were not completely knocked out in the endosperm of transgenic lines and residuary SBEIIb would maintain amylopectin synthesis at a low level in kernels; while in ae mutant, ZmSEBIIb expression could not be detected (Gao et al. 1997). Although the AC needs to be increased further, these new SBEIIRNAi maize germplasm lines could be used in high-amylose variety breeding. By using RNAi technology we could directly obtain high-amylose germplasm from the elite inbred line. In developing the various SBEIIRNAi lines of maize, pFGC5941-P27kD::2ac-2bt-bar structure was ascertained to be the most efficient hpSBEIIRNA construct for inhibiting amylopectin synthesis.
Abbreviations
- AC:
-
Amylose content
- ae:
-
Amylose-extender
- AGPase:
-
ADP-glucose pyrophosphorylase
- CTAB:
-
Cetyltrimethylammonium bromide
- DP:
-
Degree of polymerization
- DAP:
-
Days after pollination
- GBSS:
-
Granule-bound starch synthetase
- RNAi:
-
RNA interference
- SBE:
-
Starch branching enzymes
- DBE:
-
Starch debranching enzymes
- SS:
-
Starch synthetases
- WT:
-
Wild type
References
Béclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12:684–688
Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guiltinan MJ (2001) Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol 125:1396–1405
Blauth SL, Kim K-N, Klucinec J, Shannon JC, Thompson D, Guiltinan M (2002) Identification of Mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol Biol 48:287–297
Boyer CD, Preiss J (1981) Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol 67:1141–1145
Burton RA, Bewley JD, Smith AM, Bhattacharyya MK, Tatge H, Ring S, Bull V, Hamilton W, Martin C (1995) Starch branching enzymes belonging to distinct enzyme families are differentially expressed during pea embryo development. Plant J 7:3–15
Butardo VM, Fitzgerald MA, Bird AR, Gidley MJ, Flanagan BM, Larroque O, Resurreccion AP, Laidlaw HK, Jobling SA, Morell MK (2011) Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA-and hairpin RNA-mediated RNA silencing. J Exp Bot 62:4927–4941
Chuang C-F, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Nat Acad Sci USA 97:4985–4990
Denyer K, Johnson P, Zeeman S, Smith AM (2001) The control of amylose synthesis. J Plant Physiol 158:479–487
Fisher DK, Gao M, Kim K-N, Boyer CD, Guiltinan MJ (1996) Allelic analysis of the maize amylose-extender locus suggests that independent genes encode starch-branching enzymes IIa and IIb. Plant Physiol 110:611–619
Friedman RB, Deboer ED, Delgado GA, Furcsik SL, Qvyjt F, Tenbarge FL (1999) Targeting of an appropriate amylose type starch for specific product applications. Macromol Symp 140:81–91
Gao M, Fisher DK, Kim K-N, Shannon JC, Guiltinan MJ (1996) Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol Biol 30:1223–1232
Gao M, Fisher DK, Kim K-N, Shannon JC, Guiltinan MJ (1997) Independent genetic control of maize starch-branching enzymes IIa and IIb: isolation and characterization of a Sbe2a cDNA. Plant Physiol 114:69–78
Garwood DL, Shannon J, Creech R (1976) Starches of endosperms possessing different alleles at the amylose-extender locus in Zea mays L. Cereal Chem 53:355–364
Guan H, Preiss J (1993) Differentiation of the properties of the branching isozymes from maize (Zea mays). Plant Physiol 102:1269–1273
Guan H, Li P, Imparl-Radosevich J, Preiss J, Keeling P (1997) Comparing the properties of Escherichia coli branching enzyme and maize branching enzyme. Arch Biochem Biophys 342:92–98
Guan S, Wang P, Liu H, Liu G, Ma Y, Zhao L (2011) Production of high-amylose maize lines using RNA interference in sbe2a. Afr J Biotechnol 10:15229–15237
Guo Z, Zhang J, Wang D, Chen Z (2008) Using RNAi technology to produce high-amylose potato plants. Sci Agric Sin 41:494–501
Hedman KD, Boyer CD (1983) Allelic studies of the amylose-extender locus of Zea mays L.: levels of the starch branching enzymes. Biochem Genet 21:1217–1222
Heilersig H, Loonen A, Bergervoet M, Wolters A, Visser R (2006) Post-transcriptional gene silencing of GBSSI in potato: effects of size and sequence of the inverted repeats. Plant Mol Biol 60:647–662
Hennen-Bierwagen TA, Liu F, Marsh RS, Kim S, Gan Q, Tetlow IJ, Emes MJ, James MG, Myers AM (2008) Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol 146:1892–1908
Hirai S, Kodama H (2008) RNAi vectors for manipulation of gene expression in higher plants. Open Plant Sci J 2:21–30
Hofvander P, Andersson M, Larsson CT, Larsson H (2004) Field performance and starch characteristics of high-amylose potatoes obtained by antisense gene targeting of two branching enzymes. Plant Biotechnol J 2:311–320
Jeon J-S, Ryoo N, Hahn T-R, Walia H, Nakamura Y (2010) Starch biosynthesis in cereal endosperm. Plant Physiol Biochem 48:383–392
Jobling SA, Schwall GP, Westcott RJ, Sidebottom CM, Debet M, Gidley MJ, Jeffcoat R, Safford R (1999) A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterisation of multiple forms of SBE A. Plant J 18:163–171
Krogars K, Heinämäki J, Karjalainen M, Niskanen A, Leskelä M, Yliruusi J (2003) Enhanced stability of rubbery amylose-rich maize starch films plasticized with a combination of sorbitol and glycerol. Int J Pharm 251:205–208
Kuriki T, Stewart DC, Preiss J (1997) Construction of chimeric enzymes out of maize endosperm branching enzymes I and II: activity and properties. J Biol Chem 272:28999–29004
Li L, Jiang H, Campbell M, Blanco M, J-l Jane (2008) Characterization of maize amylose-extender (ae) mutant starches. Part I: relationship between resistant starch contents and molecular structures. Carbohydr Polym 74:396–404
Li N, Zhang S, Zhao Y, Li B, Zhang J (2011) Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 233:241–250
Lourdin D, Valle GD, Colonna P (1995) Influence of amylose content on starch films and foams. Carbohydr Polym 27:261–270
Martin C, Smith AM (1995) Starch biosynthesis. Plant Cell 7:971–985
Matveev YI, Van Soest J, Nieman C, Wasserman L, Protserov V, Ezernitskaja M, Yuryev V (2001) The relationship between thermodynamic and structural properties of low and high amylose maize starches. Carbohydr Polym 44:151–160
Mizuno K, Kimura K, Arai Y, Kawasaki T, Shimada H, Baba T (1992) Starch branching enzymes from immature rice seeds. J Biochem 112:643–651
Moore CW, Creech RG (1972) Genetic fine structure analysis of the amylose-extender locus in Zea mays L. Genetics 70:611–619
Morell MK, Blennow A, Kosar-Hashemi B, Samuel MS (1997) Differential expression and properties of starch branching enzyme isoforms in developing wheat endosperm. Plant Physiol 113:201–208
Nelson O, Pan D (1995) Starch synthesis in maize endosperms. Annu Rev Plant Biol 46:475–496
Poulsen P, Kreiberg JD (1993) Starch branching enzyme cDNA from Solanum tuberosum. Plant Physiol 102:1053–1054
Regina A, Kosar-Hashemi B, Li Z, Pedler A, Mukai Y, Yamamoto M, Gale K, Sharp PJ, Morell MK, Rahman S (2005) Starch branching enzyme IIb in wheat is expressed at low levels in the endosperm compared to other cereals and encoded at a non-syntenic locus. Planta 222:899–909
Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M (2006) High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Nat Acad Sci USA 103:3546–3551
Regina A, Kosar-Hashemi B, Ling S, Li Z, Rahman S, Morell M (2010) Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J Exp Bot 61:1469–1482
Saurabh S, Vidyarthi AS, Prasad D (2014) RNA interference: concept to reality in crop improvement. Planta 239:543–564
Schmidt MA, Herman EM (2008) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1:910–924
Schmidt MA, Barbazuk WB, Sandford M, May G, Song Z, Zhou W, Nikolau BJ, Herman EM (2011) Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiol 156:330–345
Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi Y-C, Gidley MJ, Jobling SA (2000) Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat Biotechnol 18:551–554
Smith AM (1990) Evidence that the “waxy” protein of pea (Pisum sativum L.) is not the major starch-granule-bound starch synthase. Planta 182:599–604
Sun C, Sathish P, Ahlandsberg S, Jansson C (1998) The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley. Plant Physiol 118:37–49
Takeda Y, Guan H, Preiss J (1993) Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr Res 240:253–263
Wang Y, White P, Pollak L, Jane J (1993) Characterization of starch structures of 17 maize endosperm mutant genotypes with Oh43 inbred line background. Cereal Chem 70:171–179
Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27:581–590
Yandeau-Nelson MD, Laurens L, Shi Z, Xia H, Smith AM, Guiltinan MJ (2011) Starch-branching enzyme IIa is required for proper diurnal cycling of starch in leaves of maize. Plant Physiol 156:479–490
Zhao FM, Qi X, Xiao J, Wang XZ (2007) Improved method for determining starch branching enzyme activity. Plant Physiol Comm 6:1167–1169
Zhu L, Liu Q, Sang Y, Gu M, Shi Y (2010) Underlying reasons for waxy rice flours having different pasting properties. Food Chem 120:94–100
Zhu L, Gu M, Meng X, Cheung SC, Yu H, Huang J, Sun Y, Shi Y, Liu Q (2012) High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol J 10:353–362
Acknowledgments
We thank Liu, Q., Zhang, C., and Man, J. (Key Laboratories of Crop Genetics and Physiology of the Jiangsu Province and Plant Functional Genomics of the Ministry of Education, Yangzhou University) for assistance in GPC analysis. We thank Cong, H. (Life Science College of Shandong University) for help on performing SEM for observing the morphology of starch granules. We thank Li, S. (Life Science College of Shandong University) for the maize shoot-tip genetic transformation and cultivation. This work was supported by the Hi-Tech Research and Development (863 Program of China, 2012AA10A306).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Y., Li, N., Li, B. et al. Reduced expression of starch branching enzyme IIa and IIb in maize endosperm by RNAi constructs greatly increases the amylose content in kernel with nearly normal morphology. Planta 241, 449–461 (2015). https://doi.org/10.1007/s00425-014-2192-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2192-1