Abstract
Main conclusion
The Ycf46 mutant of Synechocystis showed growth inhibition under low dissolved CO 2 conditions, suggesting a role for the Ycf46 protein in the process of photosynthetic CO 2 uptake and utilization.
Abstract
Hypothetical chloroplast open reading frame Ycf46 proteins are highly conserved in all cyanobacterial lineages and most algal chloroplast genomes, but their exact function is still unknown. In the cyanobacterium Synechocystis sp. PCC 6803, the Ycf46 encoding gene slr0374 is part of an operon (with slr0373 and slr0376) and responds to many environmental stresses. Transcript levels of the slr0373, slr0374 and slr0376 genes were increased under a low concentration of dissolved inorganic carbon (Ci). Compared with the wild type, the mutant lacking slr0374 showed growth arrest under Ci-deficient conditions but not under iron-deficient or low-light conditions. In addition, the mutant grew more slowly than the wild type under pH 6.0 conditions in which CO2 was the dominant Ci source, indicating the mutant cells had weak CO2 uptake and/or utilization ability. Supplying a high concentration of CO2 (5 %, v/v) to the mutant restored its phenotype to the wild type level. The photosynthetic activity of the mutant was inhibited to a lesser extent by a carbonic anhydrase inhibitor than that of the wild type, which specifically blocked CO2 uptake. Inactivation of slr0374 decreased expression of the ecaB gene and reduced carbonic anhydrase activity. A subcellular localization assay indicated that the Ycf46 protein was soluble. By co-immunoprecipitation assay using Slr0374 as a bait-protein, potential interacting proteins in the size range of 30 kDa were identified. These results suggest that the Ycf46 protein plays a role in the regulation of photosynthesis in cyanobacteria, especially in CO2 uptake and utilization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyanobacteria, the only prokaryotic organism that perform oxygenic photosynthesis, have excellent capability to adapt to various environmental stresses. During their long-term evolution, cyanobacteria have developed efficient CO2-concentrating mechanisms (CCMs) by which cells can raise their CO2 concentration up to 1,000-fold around the active site of Rubisco and perform efficient photosynthesis even under dissolved inorganic carbon (Ci or DIC) limiting conditions (Badger and Price 2003). In the past 30 years there has been a rapid increase in our understanding of these mechanisms, especially of the genes and protein components involved in cyanobacterial CCMs. To date, five DIC transport systems have been identified including three HCO3 − transporters and two CO2 transporters: (1) BCT1, an inducible high-affinity Na+-independent HCO3 − transporter encoded by the cmpABCD operon (Omata et al. 1999); (2) SbtA, an inducible high-affinity Na+-dependent HCO3 − transporter encoded by a single sbtA gene (Shibata et al. 2002); (3) BicA, a low-affinity high-flux Na+-dependent HCO3 − transporter encoded by bicA (Price et al. 2004); (4) a constitutive CO2 uptake system based on the NDH-I4 complex and potentially located on the plasma membrane, encoded by ndhD4/ndhF4/cupB (Shibata et al. 2001); and (5) an inducible CO2 uptake system based on the NDH-I3 complex and located on the thylakoid membrane, encoded by ndhD3/ndhF3/cupA (Shibata et al. 2001).
Despite a good understanding of CCM components, our knowledge of how cyanobacterial cells sense Ci-limitation signals and regulate inorganic carbon utilization is still limited. When exposed to Ci-limitation, cyanobacteria enhance the expression of inducible transporters and increase the number of carboxysomes (Price et al. 2008). It has been reported that one or two LysR-type transcription regulators, known as CcmR (NdhR) and CmpR, control the regulation process (Omata et al. 2001; Wang et al. 2004), although the precise signal transduction pathways are still unknown. To further clarify the CCM regulation mechanisms in cyanobacteria, DNA microarray analyses were carried out in the model species Synechocystis sp. PCC 6803 (Wang et al. 2004; Eisenhut et al. 2007). The microarray results indicated that when cells were transferred from high Ci (5 % CO2) to low Ci (0.035 % CO2) conditions, dozens of genes with unknown functions were strongly up-regulated, besides the genes encoding inducible inorganic carbon transporters such as NDH-13, BCT1, and SbtA (Eisenhut et al. 2007).
Using genetic transformation, we knocked out 22 of these unknown genes in Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803), to identify the functional genes involved in inorganic carbon utilization. Among them, a hypothetical chloroplast open reading frame 46 encoding gene ycf46 (slr0374) was previously reported to be responsive to many environmental stresses, but its exact function is still unknown (Singh and Sherman 2002). In this study, ycf46 (slr0374) was characterized as being involved in inorganic carbon utilization.
Materials and methods
Strains and culture conditions
The glucose-tolerant strain Synechocystis 6803 was used in this study (Williams 1988). Cyanobacterial mutants were generated by inserting a kanamycin-resistant cassette, C.K2, into the genome of wild-type cells as described previously (Jiang et al. 2010, 2012). The slr0374-flag strain was generated by fusing the FLAG coding sequence (GAC TAC AAG GAC GAC GAC GAC AAG) to the C-terminus of the slr0374 gene. Complete segregation strains were verified by PCR (primers are listed in Supplemental Table S1). The wild-type and mutant strains were cultured in BG11 liquid medium buffered with 20 mM HEPES–KOH (pH 8.0) at 30 °C under continuous fluorescent cool white lights (40 µmol photons m−2 s−1). To prepare strict Ci-limited cultures, Na2CO3 was removed from the BG11 medium. Different pH treatments were conducted using 20 mM 2-(4-morpholino) ethanesulfonic acid (MES) for pH 6.0, and 20 mM 3-(cyclohexylamino)-1-propanesuhinic acid (CAPS) for pH 10.0. The pH value of the growth medium was measured using a pH meter (Mettler Toledo, Shanghai, China). The growth of the cultures was monitored by measuring the optical density at 730 nm (OD730) with a Cary-300 UV/visible light spectrophotometer. Specific growth rates were calculated as follows: [(log ODt2 − log OD t1)/log2]/(t 2 − t 1), where OD t1 is the turbidity (optical density at 730 nm) after t 1 days and ODt2 is the turbidity (optical density at 730 nm) after t 2 days.
RNA isolation and transcriptional level analysis
Synechocystis 6803 cells of the wild-type and mutant strains were collected by centrifugation and total RNA was isolated using the TRIzol Reagent (Invitrogen). To remove DNA contamination, the RNA solutions were digested with 4 units of RNase-free DNase (Promega) according to the manufacturer’s instructions. The digest was extracted with an equal volume of phenol–chloroform–isoamyl alcohol (25:24:1), and the RNA was pelleted by centrifugation after overnight precipitation with 200 mM LiCl and 75 % (v/v) ethanol at −80 °C. For quantitative RT-PCR analysis, first-strand cDNA was synthesized using the M-MLV Reverse Transcriptase (Promega) according to the manufacturer’s instructions. The cDNA products and primers specific for the CCM-related genes cmpA (slr0040), bicA (sll0834), sbtA (slr1512), ndhF3 (sll1732), ndhF4 (sll0026), ccaA (slr1347), ecaB (slr0051) and ccmN (sll1032) were then used for PCR amplification. The RNase P subunit B gene (rnpB) was selected as the internal control for normalizing target transcript abundance. Amplifications were performed in 20-µL reaction mixtures using the SYBR Green Realtime PCR Master Mix (TOYOBO); the primers used in these experiments are shown in Supplemental Table S1. PCR amplification was performed under the following conditions: DNA polymerase activation at 95 °C for 1 min, denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, product extension at 72 °C for 45 s, and signal acquisition at 85 °C for 1 s. After the last cycle of amplification, the rotor temperature was ramped from 55 to 99 °C at 0.2 °C s−1 for melting curve analysis. Relative quantification of transcripts in this study was estimated using the 2−ΔΔCT method described by Livak and Schmittgen (2001): ΔΔC T = (C T,Target − C T,rnpB ) slr1336 mutant − (CT,Target − CT,rnpB )WT.
Cellular location of the slr0374 gene product
The slr0374-flag strain cells were collected by centrifugation, resuspended in 40 mM Tris–HCl (pH 8.0) with 1 mM phenylmethylsulfonyl fluoride (PMSF), and ruptured with a UP200S ultrasonic processor (Hielscher Ultrasound Technology, Germany) in an ice bath for 15 min at 40 % amplitude and a 30 % duty cycle. Cell debris and unbroken cells were removed by centrifugation at 11,000g at 4 °C for 10 min. Total membrane proteins and cytosolic soluble proteins were separated by centrifugation at 103,000g and 4 °C for 30 min. The total membranes and supernatant were then used for western blot detection. SDS-PAGE and western blotting were performed with standard methods. Briefly, equal amounts of the proteins were loaded, separated by 12 % SDS-PAGE, transferred to nitrocellulose filters (Millipore, Ireland), detected with anti-Flag or anti-CP47 antibody, respectively, and visualized with goat anti-mouse or anti-rabbit alkaline phosphatase antibody with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Amresco) as the substrates. The anti-Flag antibody was purchased from the Sigma company and the anti-CP47 antibody was a kind gift from Professor Xudong Xu (Institute of Hydrobiology, Chinese Academy of Sciences) (Gao and Xu 2009).
CA activity assays and oxygen evolution measurements
A CA assay was conducted by measuring the rate of pH change after the injection of a standard amount of CO2-saturated water as described previously in Jiang et al. (2013). Steady-state oxygen evolution (P) was measured with a Clark-type oxygen electrode (Hansatech) at 30 °C under saturating light. Before measurement, the cells were collected and resuspended in fresh BG11 medium containing 20 mM HEPES (pH 8.0) and 1 mM KHCO3, at a chlorophyll concentration of 4 µg mL−1. The measurements were performed by exposing a 2-mL cell suspension to 560 μmol photons m−2 s−1 white light, with 300 µM EZ added to inhibit CA activity. Relative O2 evolution rates were calculated using the following equation: relative O2 evolution rate = (P −EZ − P EZ)/P −EZ.
Co-immunoprecipitation (Co-IP) and pull-down assay
To screen for proteins interacting with Slr0374, anti-Flag monoclonal mouse antibody (Sigma) and a Protein A/G Plus-Agarose Immunoprecipitation Reagent (SC2003, Santa Cruz Biotechnology) were used for Co-IP assay of the total proteins of the Synechocystis slr0374-flag strain. The Synechocystis slr0374-flag strain was grown to exponential phase in BG11 medium under the standard conditions described above. The cells were collected by centrifugation, resuspended in lysis buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1 % Triton-X-100, 0.1 % SDS] with part of a protease inhibitor cocktail (Roche), and ruptured with a UP200S ultrasonic processor (Hielscher Ultrasound Technology, Germany) in an ice bath for 20 min at 40 % amplitude and a 30 % duty cycle. Cell debris and unbroken cells were removed by centrifugation at 11,000g at 4 °C for 10 min. The supernatant was pre-incubated with about 1 μg normal mouse IgG antibody and 20 μL Protein A/G Plus-Agarose at 4 °C for 30 min, and then centrifuged at 1,000g and 4 °C for 5 min. About 1 mL of the supernatant (including 100–500 μg total proteins) was transferred to a new 1.5 mL Eppendorf tube. About 1 μg anti-Flag antibody (Sigma) was added to the tube and incubated at 4 °C for 1 h, and then 20 μL Protein A/G Plus-Agarose was added and incubated at 4 °C for overnight. The beads were then harvested by centrifugation at 1,000g and 4 °C for 5 min, and washed with lysis buffer three times. After the final wash, the beads were resuspended with 1× SDS loading buffer and analyzed by SDS-PAGE. The bands differing between the Synechocystis slr0374-flag strain and the wild type were cut out for identification by LC–MS/MS analysis. This analysis was performed over a 70-min run on an LTQ-Orbitrap mass spectrometer (Thermo-Fisher Scientific) connected to an Agilent 1200 quaternary HPLC Pump (Pandy and Mann 2000; Xu and Peng 2006). To confirm the identification by LC–MS/MS, the proteins were expressed in the E. coli BL21 (DE3) strain with a His-tag. The His-tag fused proteins from the recombinant E. coli strains together with the proteins from the Synechocystis slr0374-flag strain were pulled down by purification with an Ni-NTA His·Bind® resin (Novagen). The resulting proteins were detected by SDS-PAGE and western blotting with anti-Flag and anti-His-tag antiserum, respectively. SDS-PAGE and western blotting were performed using standard methods. Equal amounts of proteins were loaded, separated by 12 % SDS-PAGE, transferred to nitrocellulose filters (Millipore, Ireland), detected with anti-Flag monoclonal mouse antiserum (Sigma) or anti-His-tag monoclonal mouse antiserum (Sigma), and visualized with goat anti-mouse alkaline phosphatase antibody with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Amresco) as the substrates.
Results and discussion
There are two hypothetical chloroplast open reading frame 46 genes (slr0374 and slr0480) in Synechocystis 6803, but only slr0374 is required for acclimation to Ci-limiting conditions
Twenty-two genes or gene clusters in Synechocystis 6803 were knocked out (gene names are listed in Supplemental Table S2). These genes were chosen according to the DNA microarray results of Wang et al. (2004) and Eisenhut et al. (2007). All of the genes were highly up-regulated by a change from high Ci (5 % CO2) to low Ci (0.035 % CO2) conditions (Wang et al. 2004; Eisenhut et al. 2007). Through genetic transformation and mutant phenotype analyses, we found that all of the mutants had a similar growth phenotype to the wild type when they were cultured in standard BG11 medium. However, when the mutants were transferred to a more Ci-limiting medium, in which Na2CO3 was removed (without shaking and CO2 aeration), the slr0374 mutant showed a much lower growth rate compared with the wild type. Note that ‘Ci-limiting conditions’ in this study refers to culture in BG11 medium without carbonate or bicarbonate addition. Under these conditions, cells can only obtain carbon from the dissolution of atmospheric CO2 into the medium.
The slr0374 gene is annotated as a hypothetical chloroplast open reading frame 46 (ycf46). Another ycf46 homologous gene, slr0480, was also found in the genome of Synechocystis 6803 with 61 % amino acid sequence similarity. Ycf46 homologues are highly conserved in different algae, but missing in green algae and higher plants (Table 1; Supplemental Fig. S1). By investigating the model of Martin et al. (1998) for gene transfer to the nucleus and the evolution of chloroplasts, we found that Slr0374 in Synechocystis 6803 had high similarities with homologues from Porphyra yezoensis (E-value, 1e−94), Porphyra purpurea (E-value, 1e−91), Guillardia theta (E-value, 2e−96), Odontella sinensis (E-value, 2e−95) and Emiliania huxleyi (E-value, 1e−90) (Table 1). Interestingly, no homologues were found in green algae or higher plants, suggesting the loss of the ycf46 gene was a relatively late evolutionary event.
Generally, conserved chloroplast open reading frame proteins are involved in photosynthesis, but the exact functions of many ycf genes including ycf46 in cyanobacteria remain unknown (Mäenpää et al. 2000). To clarify the exact roles of the two ycf46 genes, we constructed two single mutants (Δslr0374 and Δslr0480) and a double mutant (Δslr0374/slr0480). Complete segregation mutants were obtained after repeatedly growing cells in medium containing antibiotics and were confirmed by PCR on genomic DNA (Fig. 1a). In phenotype analysis, the Δslr0374 mutant, but not the Δslr0480 mutant, showed a significantly reduced growth rate compared with the wild type under Ci-limiting conditions (t test, P < 0.05) (Fig. 1b). The double mutant Δslr0374/slr0480 showed a similar growth rate to the Δslr0374 single mutant (Fig. 1b). These results demonstrated that slr0374 played an important role in the acclimation to Ci-limitation, but the role of the slr0480 gene in this process was negligible. Hence, we focused on only the slr0374 gene in the following research.
PCR analysis (a) and specific growth rate (b) of the Synechocystis 6803 wild type and ycf46 mutants cultured under Ci-limiting conditions. The wild-type and mutant strains were cultured in BG11 medium lacking Na2CO3 and buffered with 20 mM HEPES–KOH (pH 8.0). ycf46 single mutants: Δslr0374 and Δslr0480; double mutant: Δslr0374/slr0480. a Complete segregation of the mutants confirmed by PCR with corresponding primers; b inactivation of one ycf46 gene (slr0374) resulted in growth arrest in Ci-limiting conditions, but inactivation of another ycf46 gene (slr0480) did not change the growth rate. Different letters indicate significant difference (t test, P < 0.05). Mean ± SD (n = 3–4)
The ycf46 gene slr0374 was induced by Ci-limitation
The slr0374 gene, together with slr0373 and slr0376, belongs to the slr0373-slr0374-slr0376 operon (Singh and Sherman 2002). There is no gene named “slr0375” in the genome of Synechocystis 6803, but it has been noted that there is a non-coding RNA (ncRNA) between the slr0374 and slr0376 genes (Voß et al. 2009). The slr0374 gene encodes a putative Ycf46 protein (501 aa) that contains an AAA (ATPases associated with diverse cellular activities) domain. Members of the AAA+-ATPase superfamily are found in all kingdoms of life and are involved in very diverse cellular processes, including protein degradation, membrane fusion and cell division (Bussemer et al. 2009). The upstream gene encodes the protein Slr0373 (126 aa), which contains a helix-turn-helix motif and is a typical ATP-binding domain. The downstream gene slr0376 encodes a putative Ycf35 protein (116 aa), whose function is also unknown.
Although the slr0373-slr0374-slr0376 operon has been reported to be responsive to various environmental stresses, including low Fe (Singh and Sherman 2000), low S, low N, and high NaCl (Singh and Sherman 2002), the exact functions of the encoded proteins are still unknown. More importantly, as conserved chloroplast open reading frames, the functions of Ycf46 (Slr0374) and Ycf35 (Slr0376) in photosynthesis have never been studied. Microarray results showed that transcript levels of the slr0373, slr0374, and slr0376 genes in Synechocystis 6803 were significantly up-regulated when cells were cultured under low Ci (0.03 % CO2) versus high Ci (5 % CO2) conditions (Eisenhut et al. 2007). In the present study, to confirm the linkage of these ycf genes with photosynthesis and CO2 utilization, we compared their transcription levels by RT-PCR from cells grown in BG11 medium with and without carbon source (Na2CO3) addition. The results showed that all three genes were clearly up-regulated when the only inorganic carbon source (Na2CO3) was removed from the culture medium after 24 h (Fig. 2). A decreasing trend in gene transcriptional levels was detected from upstream to downstream genes in the gene cluster (in the order slr0373 > slr0374 > slr0376), which was consistent with the suggestion by Singh and Sherman (2002) that the three genes could be co-transcribed as an operon. These results, including the increased gene transcription in Ci-limiting cells and the growth arrest of the Δslr0374 mutant in Ci-limitation conditions, suggested that the product of the putative ycf46 gene played a positive role in Ci utilization.
RT-PCR analysis of the slr0373, slr0374 and slr0376 gene transcript levels in Synechocystis 6803 in BG11 media with Na2CO3 (+Ci) and without Na2CO3 (–Ci). Two independent experiments showed identical results. The housekeeping gene rnpB was used as a reference gene. PCR using RNA as a template confirmed the RNA solution was not contaminated by genomic DNA
The ycf46 gene (slr0374) was independent from the regulation of the known CCM transcription regulators CcmR and CmpR
Considering its potential roles in Ci utilization or CCM, we were interested in whether slr0374 was regulated by the LysR-type transcription regulators CcmR and CmpR, which are the main transcription regulators functioning in CCM (Omata et al. 2001; Wang et al. 2004). CcmR is a transcription repressor and can repress the transcription of SbtA and Ndh-I3 when cells are cultured in high Ci conditions, while this repression is removed when cells are transferred back to Ci-limitation environments (Omata et al. 2001). CmpR functions as a transcription activator and activates the transcription of the Cmp operon, which encodes a HCO3 − transporter, BCT1 (Wang et al. 2004). We constructed mutants of the two transcription regulators CcmR (Sll1594) and CmpR (Sll0030) and cultured them in Ci-limitation conditions with the wild type as a control. The transcript level of the ycf46 gene (slr0374) was monitored by RT-PCR in the wild type and the two mutants (Supplemental Fig. S2). The results from two independent experiments showed that there was no significant change in the expression level of slr0374 in the wild type and the two mutants, suggesting that the gene was not regulated by these two transcription regulators (Supplemental Fig. S2). Considering that the ycf46 gene (slr0374) was up-regulated under various environmental stresses, we speculated that other transcription regulators responsive to environmental stresses might regulate its transcription.
The ycf46 mutant is impaired in CO2 utilization
To clarify the functions of the putative Ycf46 and Ycf35 proteins, detailed physiological experiments were performed with the Δslr0374 mutant, Δslr0374/slr0376 mutant (slr0374, ncRNA, and slr0376 deletion) and wild type (Fig. 3a, c). The PCR results confirmed that the mutants were fully segregating (Fig. 3b, d). Although the transcriptional levels of slr0374 or slr0376 changed under various stresses such as iron starvation and oxidative stress (Singh and Sherman 2002), the ycf46 mutant did not show obvious growth differences compared with the wild type when the cells were grown under a low iron concentration and low-light conditions (Supplemental Fig. S3). It also seemed to be even more tolerant to methyl viologen than the wild type (Supplemental Table S3). Since the pH value can change the ratio of Ci sources (CO2, HCO3 − and CO3 2−) in liquid medium, we cultured the mutant and the wild type with standard BG11 medium under different pH conditions (pH 6.0, 8.0 and 10.0). In pH 6.0, 8.0 and 10.0 conditions the dominant Ci sources are CO2, HCO3 −, and CO3 2−, respectively (Golterman et al. 1978). When the cells were cultured in the pH 8.0 medium (standard conditions), the specific growth rates of the two mutant strains (1.29 ± 0.05 for Δslr0374 and 1.30 ± 0.01 for Δslr0374/slr0376) were similar to that of the wild type (1.31 ± 0.03) (t test, P > 0.05) (Fig. 3f). However, under pH 6.0 conditions, both the Δslr0374 and Δslr0374/slr0376 mutants showed significantly lower growth rates (0.63 ± 0.02 and 0.68 ± 0.04, respectively) than the wild type (1.17 ± 0.01) (t test, P < 0.05) (Fig. 3e). pH 10.0 conditions are a strict stress on cyanobacterial cells, but the mutants showed similar growth rates (0.98 ± 0.01 for Δslr0374, 0.91 ± 0.02 for Δslr0374/slr0376) to the wild type (0.99 ± 0.06) (t test, P > 0.05) (Fig. 3g). These results suggested that the inactivation of Ycf46 and/or Ycf35 probably decreased the cells’ capability to utilize CO2, rather than HCO3 − or CO3 2−. As there were no differences in growth rates between the Ycf46 single mutant and the Ycf46/Ycf35 double mutant, these two genes were probably co-transcribed in the same operon as suggested by Singh and Sherman (2002).
Construction (a, c), PCR analysis (b, d), and growth characteristics under different pH conditions (e–g) of the Synechocystis 6803 Δslr0374 and Δslr0374/slr0376 mutants. A kanamycin-resistant cassette, C.K2, was inserted at different loci of the gene cluster to obtain Δslr0374 and Δslr0374/slr0376 mutants (a, c). PCR results confirmed that the mutants were completely segregating (b, d). PCR was carried out with the P1/P2 or P1/P3 primer pairs. P1, P2 and P3 represent the primers Slr0374-1, Slr0374-2 and Slr0374-3 in Table S1, respectively. The growth curves for the Synechocystis strains were obtained in standard BG11 medium buffered at pH 6.0, 8.0 or 10.0, respectively (e–g)
Supplying a high concentration of CO2 (5 %, v/v) to the ycf46 mutant restored its phenotype under pH 6.0 conditions to the wild type level (Supplemental Fig. S4), which further confirmed that the ycf46 mutant was impaired in CO2 utilization. A complementation strain of the ycf46 mutant, in which the ycf46 gene was expressed with a highly efficient psbA2 promoter (Jiang et al. 2012), grew even better than the wild type, suggesting that the increased copies of ycf46 were helpful to cells grown in low pH conditions (Supplemental Fig. S4).
The ycf46 mutant showed decreased expression of the ecaB gene and reduced carbonic anhydrase (CA) activity
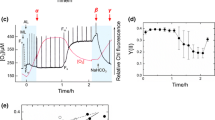
Quantitative reverse transcription PCR (qRT-PCR) was used to evaluate the transcript levels of several CCM component genes in the wild-type and Δslr0374/slr0376 mutant strains. The cells used for qRT-PCR analysis were cultured in standard growth conditions (pH 8.0). Most CCM component genes showed no significant difference in their transcriptional levels between the wild-type and mutant cells, while ecaB, which encodes a possible non-carboxysome carbonic anhydrase (EcaB) (So et al. 1998; Anthony et al. 2002), showed a decreased transcriptional level in the mutant (Table 2). The total carbonic anhydrase activity of the Δslr0374/slr0376 mutant was 43.6 ± 27.0 WAU (mg Chl a)−1, which was significantly lower than that of the wild type, 176.5 ± 8.3 WAU (mg Chl a)−1 (t test, P < 0.05; Fig. 4a). This result was consistent with the lower transcriptional level of ecaB in the mutant cells. EcaB is thought to be an external carbonic anhydrase outside of the carboxysome (So et al. 1998). CA is absent from the cytosol in cyanobacteria, which means that the enzyme is located either in the periplasm, or on the outer surface of the plasma membrane or S-layer (Kupriyanova et al. 2007, 2011). Although limitations in methodology restricted our identification of the subcellular location of the Slr0374 protein in the periplasmic space, we hypothesize that the Ycf46 protein in Synechocystis is involved in the regulation of CCM either directly or indirectly, which might be mediated by a non-carboxysome carbonic anhydrase.
CA activity (a) and inhibition of a specific CA activity inhibitor (EZ) on O2 exchange rates (b) of the Synechocystis 6803 wild type (WT) and the Δslr0374/slr0376 mutant. The relative rate shows the O2 exchange rates with EZ versus without EZ. Exponential cells were measured with a Clark-type oxygen electrode (Chlorolab 2, Hansatech)
The photosynthetic activity of the ycf46 mutant was less sensitive to a carbonic anhydrase inhibitor compared with the wild type
The O2 evolution rates of the wild-type and mutant strains were measured in the presence of 300 µM EZ (ethoxyzolamide), which is a membrane-permeable inhibitor of CA and specifically inhibits active CO2 uptake (Price and Badger 1989). The two strains showed similar O2 evolution rate (wild type, 4.33 ± 0.71 μmol O2 mg−1 Chl a min−1; ycf46 mutant, 3.54 ± 1.21 μmol O2 mg−1 Chl a min−1) under normal growth conditions without EZ treatment. The EZ treatment results showed that 25 % of the relative O2 evolution rate was supported by the CO2 uptake system in wild-type cells. While in the Δslr0374/slr0376 mutant cells, the O2 evolution rate was less inhibited by EZ (about 17 %) (Fig. 4b), indicating a relatively lower CO2 uptake capability in the mutant (P < 0.05, t test). This result was consistent with the growth arrest in pH 6.0 medium (Fig. 3e), and further indicated that inactivation of the slr0374-slr0376 genes decreased the CO2 uptake capability and carbonic anhydrase activity.
The Ycf46 protein Slr0374 is a soluble protein
Because of the difficulty in obtaining a heterologous recombinant protein, we introduced a FLAG tag into the end (upstream of the stop codon) of the Slr0374 protein in Synechocystis 6803 and obtained a transgenic slr0374-flag strain. After isolating the total membrane and soluble fractions of the slr0374-flag strain, we used SDS-PAGE and western blotting to reveal the location of the Slr0374-Flag protein with an anti-Flag antibody. The results showed that Slr0374 was located in the soluble fraction, but not in the membrane fraction (Fig. 5). An antibody to the thylakoid membrane protein CP47 was used as a control marker. These results confirmed that the Ycf46 (Slr0374) protein was a soluble protein, not a thylakoid membrane protein as previously expected (Singh and Sherman 2002).
Subcellular localization of the Slr0374 protein in Synechocystis 6803. Total protein was extracted from Synechocystis 6803 slr0374-flag cells, and then centrifuged at 103,000g and 4 °C for 30 min to isolate soluble and membrane protein solutions. The solutions were detected with an SDS-polyacrylamide gel and western blotting with anti-flag antibody. The anti-CP47 antibody was used as a membrane protein marker
Co-immunoprecipitation (Co-IP) results with Slr0374 as a bait-protein
Since a Flag tag was introduced into the genome in the Synechocystis slr0374-flag strain (Fig. 6a), Co-IP was a good way to find proteins that have protein–protein interactions with Slr0374 in vivo. As shown in Fig. 6b, a protein band of about 30 kDa was found in the Co-IP protein solution of the slr0374-flag strain (marked with arrow in Fig. 6b), but not in the wild-type solution. By LC–MS/MS, nine candidate proteins were identified (Table 3). We tried to express these nine proteins fused with a His-tag in Escherichia coli and confirm the protein–protein interactions in vitro by pull-down assay combined with a western blot. Unfortunately, only three proteins were successfully expressed in soluble fractions. The other six proteins were expressed in inclusion bodies, and were not suitable for further protein–protein interaction study. These three recombinant proteins (Slr1051-histag, Slr1665-histag, and Slr0075-histag) were pulled down with an Ni-NTA His·Bind® resin (Novagen) together with the soluble proteins of the Synechocystis slr0374-flag strain. As shown in Supplemental Fig. S5, all three proteins were detected by western blotting with an anti-His-tag antibody, as expected (upper panel). When testing with an anti-Flag antibody, weak Slr0374 protein bands (about 56 kDa) were found in the Slr0075-histag and Slr1665-histag recombinant protein solutions, but not for the Slr1051-histag recombinant protein. These results further suggested that Slr0075 and Slr1665 exhibited protein–protein interaction with Slr0374. Although no CA-encoding protein was recovered by the Co-IP assay, our results provide valuable information about Slr0374 and its regulation mechanism in the cyanobacterial CCM. It is probable that Slr0374 regulates CA activity in an indirect manner. We noticed that Slr0075 is also a conserved chloroplast open reading frame, Ycf16, which is predicted to encode a soluble SufC protein involved in Fe/S cluster assembly. Similar to slr0374, the slr0075 gene also responds to many environmental stresses, such as low iron and oxygenic stress (Singh et al. 2003; Kobayashi et al. 2004). Our unpublished data showed that the transcriptional level of the slr0075 gene was significantly lower in the Δslr0374 mutant, consistent with the suggestion that they have protein–protein interaction with each other. The lower efficiency of Fe–S assembly in the Δslr0374 mutant likely influences the CA activity, and the mechanisms are being investigated by our group and should be elucidated in the future.
Co-IP analysis with Slr0374 as a bait-protein. a Construction of the Synechocystis slr0374-flag strain. A flag tag was fused at the end of slr0374, a C.K2 fragment was inserted between the flag tag and ncRNA for resistance selection, and thus the expression of slr0374 and slr0376 were not influenced. b Co-IP with Slr0374 as a bait-protein. Total protein solutions of the Synechocystis slr0374-flag strain (bait) and wild type (control) were incubated with anti-flag antibody that was coupled with protein A/G. The eluted solutions were electrophoresed on an SDS-PAG, and then dyed with AgNO3
The putative Ycf46 (Slr0374) from the unicellular cyanobacterium Synechocystis 6803 is highly conserved in all lineages of cyanobacteria and a limited number of algal plastid genomes, but has been lost in the green algae and higher plants. Structural analysis of potential functional motif indicated that Ycf46 (Slr0374) has an AAA-type ATPase domain, a putative leucine zipper, and a phosphorylation site. AAA modules are widespread among bacteria and archaea, but their common cellular functions are still not well characterized, besides ATP binding and/or hydrolysis. Based on recent researches, it has been proposed that the AAA domain-containing proteins have chaperone-like function for the maturation of specific protein complexes and regulate a number of cellular processes (Snider and Houry 2006; El Bakkouri et al. 2010). Ycf46 (Slr0374) as well as Ycf35 (Slr0376), may be involved in the regulation of CO2 utilization in cells by some CCM components, particularly carbonic anhydrase. The evidence for the direct interaction between carbonic anhydrase and the Ycf46 protein needs further investigation.
Abbreviations
- AAA:
-
ATPases associated with diverse cellular activities
- CA:
-
Carbonic anhydrase
- CCM:
-
CO2-concentrating mechanism
- Ci :
-
Dissolved inorganic carbon
- Co-IP:
-
Co-immunoprecipitation
- EZ:
-
Ethoxyzolamide
- LC–MS/MS:
-
Liquid chromatography with tandem mass spectrometry
- ncRNA:
-
Non-coding RNA
- qRT-PCR:
-
Quantitative reverse transcription PCR
- Ycf:
-
Hypothetical chloroplast open reading frame
References
Anthony KC, So M, John M, Espie GS (2002) Characterization of a mutant lacking carboxysomal carbonic anhydrase from the cyanobacterium Synechocystis PCC 6803. Planta 214:456–467
Badger MR, Price GD (2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 383:609–622
Bussemer J, Chigri F, Vothknecht UC (2009) Arabidopsis ATPase family gene 1-like protein is a calmodulin-binding AAA+-ATPase with a dual localization in chloroplasts and mitochondria. FEBS J 276:3870–3880
Eisenhut M, Wobeser EAV, Jonas L, Schubert H, Ibelings BW, Bauwe H, Matthijs HCP, Hagemann M (2007) Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol 44:1946–1959
El Bakkouri M, Gutsche I, Kanjee U, Zhao B, Yu M, Goret G, Schoehn G, Burmeister WP, Houry WA (2010) Structure of RavA MoxR AAA+ protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity. Proc Natl Acad Sci USA 107:22499–22504
Gao H, Xu XD (2009) Depletion of Vipp1 in Synechocystis sp. PCC 6803 affects photosynthetic activity before the loss of thylakoid membranes. FEMS Microbiol Lett 292:63–70
Golterman HL, Clymo RS, Ohnstad MAM (1978) Methods for chemical analysis of freshwater. Blackwell Scientific Publications, Oxford, p 213
Jiang HB, Kong RQ, Xu XD (2010) The N-acetylmuramic acid 6-phosphate etherase gene promotes growth and cell differentiation of cyanobacteria under light-limiting conditions. J Bacteriol 192:2239–2245
Jiang HB, Lou WJ, Du HY, Price NM, Qiu BS (2012) Sll1263, a unique cation diffusion facilitator protein that promotes iron uptake in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol 53:1404–1417
Jiang HB, Cheng HM, Gao KS, Qiu BS (2013) Inactivation of Ca2+/H+ exchanger in Synechocystis sp. strain PCC 6803 promotes cyanobacterial calcification by upregulating CO2-concentrating mechanisms. Appl Environ Microbiol 79:4048–4055
Kobayashi M, Ishizuka T, Katayama M, Kanehisa M, Pakrasi MB, Pakrasi HB, Ikeuchi M (2004) Response to oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 45:290–299
Kupriyanova EV, Villarejo A, Markelova AG, Gerasimenko L, Zavarzin G, Samuelsson G, Los DA, Pronina NA (2007) Extracellular carbonic anhydrases of the stromatolite-forming cyanobacterium Microcoleus chthonoplastes. Microbiology 153:1149–1156
Kupriyanova EV, Sinetova MA, Markelova AG, Allakhverdiev SI, Los DA, Pronina NA (2011) Extracellular β-class carbonic anhydrase of the alkaliphilic cyanobacterium Microcoleus chthonoplastes. J Photochem Photobiol, B 103:78–86
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Mäenpää P, Gonzalez EB, Chen L, Khan MS, Gray JC, Aro EM (2000) The ycf9 (orf 62) gene in the plant chloroplast genome encodes a hydrophobic protein of stromal thylakoid membranes. J Exp Bot 51:375–382
Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162–165
Omata T, Price GD, Badger MR, Okamura M, Gohta S, Ogawa T (1999) Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc Natl Acad Sci USA 96:13571–13576
Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S (2001) Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol 183:1891–1898
Pandy A, Mann M (2000) Proteomics to study genes and genomes. Nature 405:837–846
Price GD, Badger MR (1989) Ethoxyzolamide Inhibition of CO2 uptake in the cyanobacterium Synechococcus PCC7942 without apparent inhibition of internal carbonic anhydrase activity. Plant Physiol 89:37–43
Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L (2004) Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci USA 101:18228–18233
Price GD, Badger MR, Woodger FJ, Long BM (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59:1441–1461
Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T (2001) Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci USA 98:11789–11794
Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, Kaplan A, Ogawa T (2002) Genes essential to sodium-dependent bicarbonate transport in cyanobacteria. J Biol Chem 277:18658–18664
Singh AK, Sherman LA (2000) Identification of iron-responsive, differential gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803 with a customized amplification library. J Bacteriol 182:3536–3543
Singh AK, Sherman LA (2002) Characterization of a stress-responsive operon in the cyanobacterium Synechocystis sp. strain PCC 6803. Gene 297:11–19
Singh AK, McIntyre LM, Sherman LA (2003) Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 132:1825–1839
Snider JD, Houry WA (2006) MoxR AAA+ ATPases: a novel family of molecular chaperones? J Struct Biol 156:200–209
So AKC, Van Spall HGC, Coleman JR, Espie GS (1998) Catalytic exchange of 18O from 13C 18O-labelled CO2 by wild-type cells and ecaA, ecaB, and ccaA mutants of the cyanobacteria Synechococcus PCC7942 and Synechocystis PCC6803. Can J Bot 76:1153–1160
Voß B, Georg J, Schön V, Ude S, Hess W (2009) Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genom 10:123
Wang HL, Bradley LP, Robert LB (2004) Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J Biol Chem 279:5739–5751
Williams JGK (1988) Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167:766–778
Xu P, Peng JM (2006) Dissecting the ubiquitin pathway by mass spectrometry. Biochim Biophys Acta 1764:1940–1947
Acknowledgments
We are grateful to Professor Xudong Xu (Institute of Hydrobiology, Chinese Academy of Sciences) for kindly providing the anti-CP47 antiserum. This study was funded by the National Natural Science Foundation of China (No. 31100184 and No. 31170309).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, HB., Song, WY., Cheng, HM. et al. The hypothetical protein Ycf46 is involved in regulation of CO2 utilization in the cyanobacterium Synechocystis sp. PCC 6803. Planta 241, 145–155 (2015). https://doi.org/10.1007/s00425-014-2169-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2169-0