Abstract
Main conclusion
We present a comprehensive overview on flavonoid-related phenotypes of A. thaliana tt and tds mutants, provide tools for their characterisation, increase the number of available alleles and demonstrate that tds3 is allelic to tt12 and tds5 to aha10.
Flavonoid biosynthesis is one of the best-studied secondary metabolite pathways in plants. In the model system Arabidopsis thaliana it leads to the synthesis of three phenolic compound classes: flavonol glycosides, anthocyanins and proanthocyanidins (PAs). PAs appear brown in their oxidised polymeric forms, and most A. thaliana mutants impaired in flavonoid accumulation were identified through screens for lack of this seed coat pigmentation. These mutants are referred to as transparent testa (tt) or tannin-deficient seed (tds). More than 20 mutants of these types have been published, probably representing most of the genes relevant for PA accumulation in A. thaliana. However, data about the genes involved in PA deposition or oxidation are still rather scarce. Also, for some of the known mutants it is unclear if they represent additional loci or if they are allelic to known genes. For the present study, we have performed a systematic phenotypic characterisation of almost all available tt and tds mutants and built a collection of mutants in the genetic background of the accession Columbia to minimise effects arising from ecotype variation. We have identified a novel tt6 allele from a forward genetic screen and demonstrated that tds3 is allelic to tt12 and tds5 to aha10.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the dark brown coloured seeds of Arabidopsis thaliana, yellow seeded mutants are easily discovered. Until now, mutations in 18 genes have been characterised on the molecular level, which result in reduced, altered or absent seed coat pigmentation. The oldest term for this class of mutants is transparent testa (tt), which explains the morphological basis for the observed phenotype: impaired pigmentation makes the seed coat transparent and reveals the yellow colour of the underlying cotyledons (Bürger 1971; Koornneef 1981, 1990). Another term for the same class of mutants is tannin-deficient seed (tds; Abrahams et al. 2002) which alludes more to the chemical nature of the defect: lack of the dark brown pigmentation of wild-type seeds, which is due to the accumulation of condensed tannins or proanthocyanidins (PAs) in the seed coat. Defects in seed coat development or in the cells’ ability to produce, modify or transport PAs can be detected by the naked eye as well as various staining techniques (as shown e.g. in Debeaujon et al. 2000). Acidic vanillin or p-dimethylaminocinnamaldehyde (DMACA) was used in screens to identify mutants with more subtle changes in pigmentation (Abrahams et al. 2002). Identification of PA mutants in A. thaliana is facilitated by the fact that all known enzymes of the central flavonoid pathway are encoded by single copy genes, which is not the case in the general phenylpropanoid pathway as well as for the genes encoding flavonol synthases (FLSs) or glycosyltransferases required for the formation of flavonol glycosides and anthocyanins.
Many of the genes and enzymatic functions encoded by the tt loci are known (Fig. 1) and the enzymatic steps of the pathway in A. thaliana were comprehensively described in a recent review by Saito et al. (2013). However, open questions exist with regard to developmental control of pigment formation, the molecular function of some regulatory factors and genes involved in deposition and oxidation of the metabolites. Control of flavonoid biosynthesis mainly occurs at the level of transcription (Weisshaar and Jenkins 1998). Flavonol formation is regulated by the R2R3-MYB proteins PFG1 to 3 (Stracke et al. 2007, 2010), whereas expression of enzyme-coding genes required for anthocyanin and PA biosynthesis is controlled by a regulatory network involving the combined action of MYB, bHLH and WD40 repeat proteins (MBW). A ternary complex of MYB75/PAP1, BHLH042/TT8 or BHLH002/EGL3 and WD40/TTG1 is crucial for anthocyanin accumulation in young A. thaliana seedlings (Gonzalez et al. 2008; Appelhagen et al. 2011a) and a MYB123/TT2, TT8 and TTG1 complex is the major regulator of PA metabolism in seeds (Baudry et al. 2004). Recently, three additional MBW complexes were shown to contribute to the expression of PA biosynthesis genes in specialised tissues of the seed coat (Xu et al. 2013a). The molecular functions of WIP1/TT1, WRKY44/TTG2 and AGL32/TT16 are less clear. During seed development, pigment accumulation occurs in the innermost cell layer of the seed coat, the endothelium. In wild-type seeds, pigmentation is also found in two specialised areas at the base of the seed termed chalaza and micropyle (Debeaujon et al. 2003; Lepiniec et al. 2006; Fig. 2a, b). TT16 encodes the MADS box protein Agamous-like 32 and influences PA biosynthesis by determining the identity of the endothelial layer within the ovule (Nesi et al. 2002). Mutants of TT1 show a phenotype similar to tt16 with PA accumulation in chalaza and micropyle but reduced and irregular PA spots in the endothelium (Sagasser et al. 2002). The WIP-type zinc finger protein encoded by TT1 was found to interact with MBW MYB factors like PAP1 and TT2 (Appelhagen et al. 2011b) and TT1 and TT16 were recently shown to be involved in the regulation of TT8 expression together with TTG2 (Xu et al. 2013b). TTG2 encodes WRKY44, a factor involved in integument cell elongation, seed mucilage production as well as trichome formation in vegetative parts of the plant, whose expression is influenced by MBW complexes (Johnson et al. 2002; Garcia et al. 2005; Ishida et al. 2007).
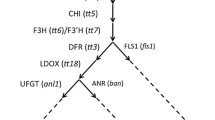
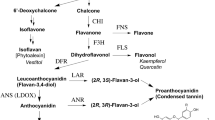
Flavonoid biosynthesis in Arabidopsis thaliana. Black arrows illustrate the pathway in wild-type plants, alternative routes found in f3′h/tt7 mutants are given in grey. Dashed arrows indicate multiple steps. For full names of gene symbols and enzymes see Table 1
transparent testa mutants from Arabidopsis thaliana. a Structure of the seed coat. Cleared wild-type seed at early heart stage viewed with Nomarski optics. b Detection of PAs by vanillin/HCl treatment in a Col-0 wild-type seed of the same age as shown in a. Red colour indicates presence of PAs in the seed coat endothelium (ii1), in chalazal cells and micropylar ii2 cells. c Mature seeds of all characterised transparent testa (tt) mutants. Mutants are classified according to their defects in PA biosynthesis, transport and regulation. Same lines as analysed in Fig. 3. For details on presented alleles and abbreviations see Table 1. oi outer integument, ii inner integument, ch chalaza, mi micropyle. Bar 100 μm
In addition to the unresolved questions with regard to the exact roles of TT1, TT16 and TTG2 in the differential accumulation of PAs in different seed coat tissues during development, it is mainly the steps of transport and polymerisation which occur after the biosynthesis of epicatechin that remain to be elucidated (Fig. 1; Zhao et al. 2010). TT19 for example encodes a glutathione S-transferase (Kitamura et al. 2004) but neither glutathionated cyanidin or epicatechin nor ABC transporter mutants with effects on anthocyanin or PA accumulation were detected in A. thaliana (Klein et al. 2006). Consequently, other roles of TT19 in PA monomer binding for protection and transport have been proposed (Mueller et al. 2000; Kitamura et al. 2010). In line with the hypotheses of these authors, Sun et al. (2012) found that TT19 can physically interact with and increase water solubility of cyanidin and cyanidin-3-O-glucoside and that it can localise to the tonoplast. Francisco et al. (2013) recently showed for the anthocyanin transporter ABCC1 from grape berries that it depends on and co-transports glutathione, rather than anthocyanin-GSH conjugates.
A different class of transporters involved in PA accumulation is represented by TT12, which encodes a multidrug and toxic extrusion protein (MATE; Debeaujon et al. 2001). TT12 was found to transport epicatechin 3′-O-glucoside in yeast microsome assays (Zhao and Dixon 2009) but evidence for an epicatechin-specific glycosyltransferase, as detected in Medicago (Pang et al. 2008, 2013), or a glycosidase that would be required to cleave off sugar before condensation, is still missing (Routaboul et al. 2012). Consistent with TT12-mediated proton gradient-driven PA precursor transport into the vacuole, effects on seed colour were observed in mutants affected in vacuole acidification, which is the proposed function of the P-type ATPase AHA10 (Baxter et al. 2005) and its putative homologue PH5 from Petunia hybrida (Verweij et al. 2008). Membrane-related defects were also proposed to explain the tt phenotype found in the tt15 mutant, which is defective in a UDP-glucose:sterol-glucosyltransferase (DeBolt et al. 2009). This suggestion is remarkable in the light of the very recent findings by Brillouet et al. (2013), who proposed a chloroplast-derived organelle, named tannosome, for the production of condensed tannins, whose formation would require extensive alterations in the properties of various membranes. So far, the only known enzyme acting after biosynthesis of epicatechin is TT10, a laccase with polyphenol oxidase activity (Pourcel et al. 2005). Other enzymes, potentially required for condensation and oxidation, remain to be identified.
Isolation of novel and more detailed analysis of existing but uncharacterised tt or tds mutants may help to find answers to the unresolved questions mentioned above. Some of the reported tt mutants have not been characterised in such detail as to clarify if they represent new loci or novel alleles of known tt genes. Thus, we have performed a systematic phenotypic characterisation of available tt and tds mutants. Previous comparisons of tt mutants indicated significant variation in flavonoid content between Arabidopsis accessions (Routaboul et al. 2006, 2012). To minimise phenotypic differences between mutants arising from ecotype variation and thus increase comparability of the results, we built a mutant collection in the genetic background of the accession Columbia (Col-0; Fig. 2c). A similar approach was recently taken by Bowerman et al. (2012), albeit with a less comprehensive set of mutants. We present an isogenic mutant set covering 17 distinct tt loci, including 14 new alleles, which were isolated from public T-DNA transformant collections. The increased number of available alleles can be applied to analyse allelic series of mutants for a given locus. Together with the description of our isogenic tt mutant collection we provide a set of tools to efficiently characterise novel seed colour mutants that are identified in screens by simple and standardised phenotypic tests. We demonstrated the power and applicability of our approach by presenting a novel tt6 mutant identified in a forward genetic screen and by analysing tds1, 2, 3, 5 and 6 which revealed that tds3 is allelic to tt12 and that tds5 is allelic to aha10.
Materials and methods
tt collection setup and naming of new alleles
To set up a collection of tt mutants in Col-0 genetic background, the SimpleSearch database for GABI-Kat T-DNA insertion lines (http://www.gabi-kat.de/simplesearch; Rosso et al. 2003) was screened for FST hits in all described tt loci. The SALK T-DNA collection (Alonso et al. 2003) was used, if no suitable GABI-Kat allele was available. The genetic background of lines from both, SALK and GABI-Kat, is Col-0 (Bolle et al. 2013). Selected T2 lines were provided by GABI-Kat or T3 SALK lines were ordered from Nottingham Arabidopsis Stock Centre (NASC). Exact insertion positions for the left border were determined in T2 plants (T3 for SALK lines) according to the confirmation process applied to all GABI-Kat lines. A detailed description for confirmation and genotyping of lines from both collections is given in Bolle et al. (2013). Additional information on GABI-Kat lines, including estimated T-DNA copy numbers, can be found in the SimpleSearch database (Kleinboelting et al. 2012). Homozygous T2 plants were identified by PCR genotyping and subsequently used for flavonoid analysis. Information on oligonucleotides used in this study is given in Supplementary Table S1. Newly identified alleles were numbered to the best of our knowledge in continuation of the existing nomenclature. Table 1 provides an overview of the essential characterisation for all lines representing the Col-0 collection as presented in Figs. 2 and 3. Table 1 also contains additional alleles identified in the course of this study that are not included in the figures as they showed identical phenotypes, and the table gives evidence for the proposed numbering. The entire mutant set is available from NASC (the set number for the collection is N2105571).
Flavonol glycoside, anthocyanin and proanthocyanidin accumulation in transparent testa mutants. Left column DPBA-stained seedling visualised under UV light. Orange quercetin derivatives, green kaempferol derivatives, blue sinapates. Second column norflurazon-bleached seedlings grown on sucrose for induction of anthocyanin biosynthesis. Third column vanillin/HCl-treated immature seeds as presented in Fig. 2. Fourth column DMACA-stained mature seed. Dark brown/black colour indicates presence of PAs. For untreated mature seeds of all lines see Fig. 2. a Col-0 wild type and mutants of structural flavonoid biosynthesis genes. The presented dfr mutant carries the tt3-1 allele in Landsberg erecta background (marked with asterisk). All other mutants are in Col-0 background. b Mutants in genes related to flavonoid transport. c Mutants of regulatory loci. The shown myb123/tt2 mutant (marked with two asterisks) carries the only available tt2 allele in Col background and does not represent the myb123/tt2 NULL mutant phenotype (see Fig. S2). Bars 0.5 mm for seedlings, 100 μm for vanillin/HCl- and DMACA-treated seeds
tt15-1 sequence analysis
The tt15-1 mutant in Col-2 background was originally isolated by Focks et al. (1999; NASC stock N799994). To identify alterations between tt15-1 and Col-2 sequences, PCR products covering the whole coding sequence of At1g43620 including introns plus 1 kb of upstream sequence were generated on genomic DNA of both genotypes and subjected to sequencing. Oligonucleotides I219/009N were used to amplify and sequence PCR products containing the tt15-1 mutation.
tds allelism tests
All tds lines analysed were kindly provided by Sharon Abrahams and Anthony Ashton. The tds1, tds2 and tds3-1 mutants were isolated from the Feldmann collection in Wassilewskija (Ws) background, tds5 and tds6 are from the Weigel activation tagging collection in Col-7 background (Abrahams et al. 2002). Allelism tests were done by fertilisation of tds mutants with pollen of aha10-6 or tt12-2 plants. Successful crosses were confirmed in F1 by testing for the heterozygous presence of the T-DNA inherited from the pollen donor. Complementation of the seed coat colour phenotype was analysed in F2 seeds gained from F1 plants whose testa is genetically F1. Both T-DNA border positions were determined in tds5 by PCR as described for GABI-Kat alleles with the oligonucleotide pairs I388/036N for right border and 8409/I309 for left border. T-DNA border positions in tds3-1 were addressed using several different oligonucleotides specific for At3g59030 in combination with F-LB102 as left border or F-RB as right border primer, which were successfully used in previous studies to identify T-DNA insertions, without any specific PCR result. PCR reactions also failed on tds3-1 DNA with all oligonucleotide pairs spanning intron1 and the second half of exon1 of At3g59030. Oligonucleotide pair 045N/I263 was subsequently used to specifically detect the tds3-1 allele in comparison to tt12-2.
tt6-5 analyses
The tt6-5 mutant was identified as yellow seeded amongst T2 segregants of GABI-Kat line 352E12. Co-segregation of the yellow seed phenotype with the T-DNA insertion of GK-352E12 in At5g65620 was analysed by PCR and inspection of the colour of mature seeds of T2 plants. Candidate genes for allelism tests were determined by comparison of flavonoid phenotypes to known tt mutants. Allelism to tt6 was tested by crossing tt6-5 with tt6-2, as described for the tds mutants. The TT6 locus was sequenced in tt6-5 with a similar approach as described for tt15-1. Oligonucleotides NP17/RS850 were subsequently used to amplify and sequence PCR products containing the tt6-5 deletion.
Histochemical assays and microscopy
A Leica DM5500 microscope equipped with phase contrast and automated differential interference contrast (DIC) condenser was used for bright field images and Leica filter cube A (340–380 nm excitation filter, 425 nm long-pass suppression filter) for epifluorescence applications. Pictures were taken with Diskus digital imaging software (Technisches Buero Hilgers, Koenigswinter, Germany).
Flavonol glycosides in 5-day-old seedlings were stained using diphenylboric acid 2-aminoethyl ester (DPBA, 0.25 % (w/v), 20 % (v/v) ethanol, 0.01 % (v/v) Triton X-100) and visualised by UV light according to Stracke et al. (2007). Seedlings were grown on filter paper soaked with 3 ppm of the bleaching herbicide norflurazon (NFZ, Supelco PS-1044) to avoid unwanted background fluorescence from chlorophyll.
Anthocyanin accumulation was monitored in 5-day-old seedlings grown on NFZ as described above, but supplemented with 4 % sucrose to induce anthocyanin accumulation. NFZ bleaching was used to improve visibility of the purple anthocyanins. Control plants grown on half strength MS media containing 4 % sucrose without NFZ showed comparable anthocyanin pigmentation.
PA accumulation was addressed using acidic vanillin in developing seeds and DMACA for mature seeds as described previously (Debeaujon et al. 2000; Abrahams et al. 2002). Vanillin/HCl treatment leads to a red adduct; the DMACA reaction causes a blue staining in immature seeds and leads to a black appearance of mature seeds. Both dyes are aromatic aldehydes that are specific to a narrow range of flavanols and dihydrochalcones (Sarkar and Howarth 1976). As the latter are missing in A. thaliana, the assays are indicative of flavan-3,4-diols (leucoanthocyanidins), flavan-3-ols (like epicatechin) as well as polymeric PAs, in this work collectively named vanillin stainable PAs or PA intermediates.
Results and discussion
Effects of tt mutations on the accumulation of the three classes of flavonoids in A. thaliana have been presented previously (e.g. in Shirley et al. 1995; Debeaujon et al. 2000; Bharti and Khurana 2003; Bowerman et al. 2012). However, growth conditions, analytical methods and ecotype backgrounds of the respective partial sets of mutants analysed are not comparable between these studies. Significant variation in secondary metabolite contents between A. thaliana accessions has been described for example by Teng et al. (2005) for anthocyanins or by Routaboul et al. (2012) for seed flavonols and PAs. To provide a homogeneous basis for further comparative studies, we set out to create a complete set of mutants in the Col-0 accession (see Table 1 for AGI codes, protein names and abbreviations) and performed systematic analyses on seeds and plants grown under uniform conditions. Figures 2 and 3 summarise the results of the phenotypic characterisation of tt and tds mutants with respect to their seed colours as well as flavonol, anthocyanin and PA contents.
Flavonol accumulation in tt mutant seedlings
Naturstoffreagenz A (diphenylboric acid 2-aminoethyl ester, DPBA) is widely used to visualise flavonols, either as a spray reagent on TLC plates or for localisation in planta. Reaction with DPBA confers green fluorescence to kaempferol glycosides whereas quercetin glycosides appear orange after treatment (Sheahan and Rechnitz 1992). Both compounds accumulate in light grown A. thaliana wild-type seedlings, resulting in a combined fluorescence of saturated orange (Peer et al. 2001; Fig. 3). In our collection of mutants, which originated from screens for altered seed colour, only few lines showed alterations of flavonol staining in seedlings. Most prominent was the blue fluorescence of chs/tt4-15 seedlings, which is caused by sinapic acid derivatives (Sheahan and Rechnitz 1993). The characteristic colour is visible due to the complete lack of flavonols in this mutant that is defective in the first enzyme of flavonoid biosynthesis, chalcone synthase (Feinbaum and Ausubel 1988; Koornneef 1990). Seedlings of chi/tt5-2 had a dull yellow appearance under UV after DPBA treatment, which indicates the presence of some flavonols that was already noted by Shirley et al. (1995), Peer et al. (2001) or Routaboul et al. (2006). As CHI catalyses the formation of naringenin chalcone from tetra-hydroxychalcone, its presence in chi/tt5 may be due to spontaneous isomerisation (Jez et al. 2000; Jez and Noel 2002). Interestingly the amounts of this pathway intermediate seem not to be sufficient for the formation of visible anthocyanins or PAs stainable by vanillin. Seedlings of f3h/tt6-2 gave a characteristic red fluorescence after treatment with DPBA, which was clearly distinct from the orange observed in wild type. Owens et al. (2008) proposed that in the absence of F3H activity, F3ʹH may convert naringenin to eriodictyol in f3h/tt6 plants. Eriodictyol can be reduced by DFR and lead to the formation of 3-desoxyflavonoids, which are normally not found in A. thaliana, but could cause the red fluorescence observed in f3h/tt6-2 (Sheahan et al. 1998; Owens et al. 2008). Green fluorescence was found in f3ʹh/tt7-7 seedlings after DPBA treatment indicating the presence of kaempferol derivatives only. This finding is in agreement with the fact that lack of the cytochrome-P450-dependent monooxygenase F3ʹH in the mutant precludes biosynthesis of quercetin glycosides (Fig. 1; Koornneef et al. 1982; Schoenbohm et al. 2000). All other mutants analysed did not show visible differences to wild type with respect to flavonol accumulation, as they are either seed coat specific or affected in steps of flavonoid biosynthesis specifically leading to anthocyanins or PAs. However, when characterising novel yellow seeded mutants, DPBA staining can be used to efficiently determine if they are affected in one of the general steps of flavonoid backbone formation from CHS to F3ʹH.
Anthocyanin accumulation in tt mutant seedlings
Mutants in general steps of flavonoid backbone formation are also impaired in anthocyanin accumulation. Whereas seedlings of chs/tt4-15 and chi/tt5-2 completely lacked anthocyanins, they were visible in f3h/tt6-2. Reduced but detectable anthocyanins were described previously for tt6-1 (Shirley et al. 1995; Albert et al. 1997) and are discussed in a later section. No pigmentation was visible in f3ʹh/tt7-7 seedlings under the growth conditions used. However, it is known that tt7 is capable of producing anthocyanins as derivatives of pelargonidin instead of cyanidin, as expected (Fig. 1; Koornneef et al. 1982). In addition to the four genotypes mentioned above, the mutants tt3-1, tds4-2, tt19-8, tt8-6 and ttg1-22 showed alterations in anthocyanin contents. The TT3 locus encodes dihydroflavonol 4-reductase (DFR; Shirley et al. 1992), which represents a branchpoint in the flavonoid pathway, separating flavonol biosynthesis from anthocyanin- and PA-specific steps. The latter compounds are therefore not present in knock out plants (Fig. 3; Shirley et al. 1995; Routaboul et al. 2006). Unfortunately, no complete null allele for dfr/tt3 is available in the accession Col-0. All Col-0 alleles isolated so far carry T-DNA insertions in the DFR promoter and either do not show a mutant phenotype (GK-212F02, SALK_099848; Bowerman et al. 2012) or are not null (GK-295C10; Fig. S1). For this reason the classical Landsberg allele tt3-1 (Koornneef 1990) is shown in Fig. 3 and Fig. S1 to illustrate the phenotype of a full knock out.
Several mutants exist for leucoanthocyanidin dioxygenase (LDOX), also known as anthocyanidin synthase (ANS), and the glutathione S-transferase GST26, encoded by the TT19 locus (Kitamura et al. 2004). Although both genes are essential for anthocyanin accumulation in plants (Fig. 3), mutant alleles were mainly identified in seed colour screens (e.g. ldox as tds4, tt11, tt17 and tt18; Appelhagen et al. 2011a and references therein). The TTG1 and TT8 loci encode regulatory factors. WD40/TTG1 is an essential part of MBW complexes regulating, among other processes, anthocyanin accumulation in vegetative parts of the plant as well as PA accumulation in the seed (Koornneef 1981; Walker et al. 1999; Baudry et al. 2004). A lack of TTG1 therefore results in seedlings without anthocyanins. BHLH042/TT8 also acts in MBW complexes and is absolutely required for PA accumulation in seeds as complex partner of TTG1 and MYB123/TT2. However, in anthocyanin regulatory complexes with TTG1 and MYB75/PAP1, TT8 can be replaced by the partly redundant factor BHLH002/EGL3, provided that it is expressed. This explains the reduced but detectable anthocyanin accumulation observed in seedlings of bhlh042/tt8-6 plants (Appelhagen et al. 2011a). All remaining mutants in the collection did not show alterations in flavonol and anthocyanin accumulation in vegetative parts of the plants and can thus be classified as affected in seed-specific genes for PA biosynthesis.
Seed coat pigmentation patterns of tt mutants
Differences in PA contents among mature tt and tds seeds or in comparison to wild type are visible to the naked eye (Fig. 2). However, careful investigation of pigment accumulation during seed development has proven useful to unravel the mechanisms involved. Mutants for anthocyanidin reductase (ANR), the first step specific for PA biosynthesis, for example do not produce seeds with a tt phenotype but are even darker than wild type (Fig. 2). In this case, inspection of immature mutant seeds revealed accumulation of anthocyanins in seed coats, which do not occur in this tissue in wild type. This phenotype indicated a redirection in the flux through the flavonoid biosynthetic pathway from PAs to anthocyanins, which was later found to be blocked at ANR in the ban mutant (Fig. 1; Devic et al. 1999; Xie et al. 2003). Detailed analysis of seed pigmentation was also key to understand the tt10 mutation, which results in light brown seeds directly after harvest that darken strongly during storage. As brown seed colour requires oxidative polymerisation of PAs, delayed browning in tt10 seeds could be explained once the mutant was found to be defective in LAC15, encoding an enzyme with polyphenol oxidase activity (Pourcel et al. 2005). These two examples illustrate the importance of a detailed analysis of seed pigmentation patterns over time. For this reason we applied different staining techniques to young and mature seeds, respectively, to characterise our collection. Acidic vanillin was used to visualise accumulation of epicatechin and soluble PAs in seeds carrying embryos of late globular to early heart stage. This developmental stage was chosen, because all relevant cell layers are fully developed until then but no oxidised PAs are visible yet. During maturation a large proportion of PAs gets insoluble by polymerisation, oxidation and reactions with polysaccharides, proteins and other phenolics (Routaboul et al. 2006). Differences in pigmentation among genotypes in mature seeds were visualised using acidic DMACA.
Using this approach, chs/tt4-15, chi/tt5-2 and dfr/tt3-1 were found to be completely devoid of PAs as expected (Fig. 3). The ectopic anthocyanin accumulation in anr/ban-5 seed coats described above resulted in a diffuse red staining after vanillin treatment and in light brown seeds after DMACA (Fig. 3), each of which confirmed the absence of PAs in this mutant. Further mutants that were easily identified by DMACA staining include tt1-3 and tt16-5. Both resulted in characteristic stainable chalaza and micropyle together with irregular spots in the endothelium. These two mutants are defective in seed coat-specific regulators (Nesi et al. 2002; Sagasser et al. 2002). Vanillin stainable spots in the endothelium have been observed in several mutant alleles of wip1/tt1 and therefore do not indicate leakiness of the mutant but are part of the phenotype (Appelhagen et al. 2011b). The situation is different in the myb123/tt2 allele presented here, which is the only one available in the genetic background of the ecotype Columbia. The spottiness in vanillin as well as DMACA staining observed in tt2-5 (Fig. 3b and S2 c, d) is not seen in the classical allele tt2-1, which is in Landsberg erecta background (Fig. S2 c, d). Sequence characterisation of tt2-5 revealed a T-DNA insertion close to the end of exon 3 of MYB123/TT2. This insertion late in the ORF results in a premature stop codon and leads to reduced transcript levels (Fig. S2b). However, the allele is clearly not NULL (Fig. S2c).
Another mutant with spotty vanillin and DMACA signals was tt15-4. The TT15 locus has been shown to encode UGT80B1, a UDP-glucose:sterol-glucosyltransferase (DeBolt et al. 2009). For our study we also analysed the tt15-1 allele originally described in Focks et al. (1999) on DNA sequence level. This allele is in Col-2 background and we therefore confirmed by sequencing that genomic DNAs of Col-2 and Col-0 are identical from ATG to stop of UGT80B1. In tt15-1, we identified a point mutation in the third exon of UGT80B1 that leads to a premature stop codon most likely resulting in a complete loss of function (Fig. 4d). In accordance with the initial characterisation by Focks et al. (1999), no obvious phenotypic differences to wild type were observed in the mutant with respect to flavonols and anthocyanins. PA accumulation in seed coats, however, was altered in all relevant tissues and resulted in irregular vanillin staining (Fig. 4a, e). DeBolt et al. (2009) describe a reduced content of (acetyl)sterol-glycosides in the mutant resulting in additional pleiotropic effects besides altered seed pigmentation. As sterol-glycosides are compounds of lipid membranes it is tempting to speculate that lack of these may lead to the observed mislocalisation of PAs in tt15 via effects on membrane properties resulting in altered transport (Pourcel et al. 2007). The recent work by Viotti et al. (2013) can be seen in line with this hypothesis as the authors propose that provacuole formation occurs in a subdomain of the smooth ER that differs in lipid composition. If such ER subdomains exist and if they are altered in tt15 remains to be shown.
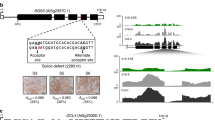
Proanthocyanidin accumulation in NULL mutants of structural flavonoid biosynthesis genes. a Vanillin/HCl-treated immature seeds as shown in Fig. 2. Genotypes as indicated. b Endothelial cells of the same lines as shown in a with higher magnification. Single cells are outlined with dotted lines. Only wild-type cells show large central vacuoles stained by vanillin/HCl. c Anthocyanin accumulation in norflurazon-bleached seedlings. Cells show anthocyanin-filled purple central vacuoles and do not reflect the aberrant PA accumulation pattern in testa cells. d Gene model of tt15 alleles characterised in this study. Exons are shown as open boxes. T-DNA insertions are marked as red triangles. Left border positions are marked with small triangles. A point mutation in tt15-1 (red dash) causes a premature stop codon in exon 3. e Vanillin/HCl-treated seeds as shown in a with focus on chalaza and micropyle. The base of tt15 seeds appears dark (compare with DMACA picture in Fig. 3) but also shows aberrant pigmentation in the vanillin assay. Bars 100 μm for whole seed and 20 μm for close-up pictures in a; 10 μm in b and c; 20 μm in e
Defects in biogenesis of the central vacuole have also been described for all tt mutants involved in PA transport processes including tt19, tt12 and aha10 (Baxter et al. 2005; Kitamura et al. 2010). Our vanillin and DMACA tests also revealed aberrant staining patterns for f3h/tt6-2, f3ʹh/tt7-7 and ldox/tds4-2 (Fig. 3). In developing f3h/tt6-2 seeds, vanillin treatment yielded red staining in all cell types that contain PAs in wild type. However, staining was reduced compared to wild type and did not fill the cells entirely as if the central vacuoles were fragmented or not fully developed (Fig. 4a, b). Vacuole defects were not observed in hypocotyl cells accumulating anthocyanins (Fig. 4c) but in the PA-containing cells of the endothelium as well as in chalaza and micropyle and resulted in a dark speckled appearance of mature f3h/tt6-2 seed coats after DMACA treatment. Flavanone 3-hydroxylase (F3H) belongs to the class of 2-oxoglutarate-dependent dioxygenases. The classical tt6-1 allele encodes a protein that lacks a carboxy-terminal fragment containing conserved amino acid positions essential for Fe(II) binding (Owens et al. 2008). Protein variants carrying mutations in these positions exhibited no F3H activity in vitro (Lukacin and Britsch 1997), suggesting that flavonoids observed in tt6-1 should not result from an F3H protein with residual activity. Instead it was proposed that other 2-oxoglutarate-dependent dioxygenases could also have F3H function (Owens et al. 2008). Such multifunctionality of dioxygenases was discussed for other steps in flavonoid biosynthesis (Martens et al. 2010) and has been shown by Stracke et al. (2009) for FLS1 and LDOX. The pigments detected in tt6 seeds may therefore result from activities of these co-expressed enzymes, whereas pigments in ldox/tds4-2 seeds (Figs. 3, 4) may be due to FLS1 or F3H.
Epicatechin is the main building block for PA polymerisation in wild-type seeds (Xie et al. 2003). PAs extracted from f3ʹh/tt7 seeds yield pelargonidin instead of cyanidin after hydrolysis, indicating pigmentation by polymers based on epiafzelechin (Abrahams et al. 2002; Routaboul et al. 2006). Total PA content in tt7 seeds is reduced when compared to wild type, potentially due to reduced affinity of one or several of the PA-specific enzymes for intermediates that lack 3ʹ-hydroxylation (Abrahams et al. 2002; Routaboul et al. 2006). DMACA staining of f3ʹh/tt7-7 resulted in seeds with a dotted appearance as already shown by (Abrahams et al. 2002; Fig. 3). Vanillin staining revealed a clearer and finer fragmentation of the vacuoles as observed for f3h/tt6-2 in cells of endothelium, chalaza and micropyle (Fig. 4). Such structures have not been described for these mutant genotypes before and may indicate a similar mechanism leading to their formation. Small vesicular structures were also found in the endothelium, but not the chalaza, of ldox/tds4-2 (Fig. 4). Using membrane- and vacuole-specific dyes it was shown that these are rather compartments surrounded by membranes (Abrahams et al. 2003) than subvacuolar inclusions as observed in anthocyanin-containing vacuoles of cotyledons (Pourcel et al. 2010). In gst26/tt19 mutants, a TT12-GFP fusion protein localising to the tonoplast was found to co-localise with typical vanillin stainable structures (Kitamura et al. 2010). Although LDOX and TT19 act in both, anthocyanin as well as PA biosynthesis, vacuole defects are only observed in PA accumulating but not in anthocyanin-containing tissues (Fig. 4). It was therefore proposed that PA intermediates themselves could be involved as signals in vacuole biogenesis or maintenance (Abrahams et al. 2003). Why accumulation of the same intermediates that leads to defective vacuoles in ldox/tds4 seed coats does not alter vacuole formation in vegetative tissues, however, needs to be clarified.
Efficient phenotypic characterisation allows directed analysis of tt and tds mutants
Having established the described methods for flavonoid-specific staining as well as the tt mutant collection characterised with these as a reference framework, we applied our approach to analyse a mutant line that was discovered in the GABI-Kat population based on its phenotype. In the course of a forward genetic screen, yellow seeds were found in the progeny of GABI-Kat line 352E12. Genotyping by PCR and evaluation of the colour of mature seeds of T2 plants revealed no co-segregation of the yellow seed phenotype with the T-DNA insertion of GK-352E12 in At5g65620. Detailed analysis of flavonoid accumulation in the mutant and comparison to the known tt mutants led us to the assumption that F3H/TT6 was the most likely gene to be affected in the mutant. Test crosses to tt6-2 yielded F1 plants producing yellow seeds (Fig. S3), indicating that F3H/TT6 was defective in both parents. Sequencing various PCR products from GK-352E12 finally revealed a deletion of one bp at position 709 on genomic level relative to the ATG start codon. The resulting frame shift and subsequent premature stop codon change and truncate the sequence in the region encoding the iron-dependent oxygenase domain (InterPro: IPR005123, containing Pfam motif 03171) including the conserved Fe2+ coordinating residue His275 and thus very likely lead to a complete NULL. In continuation of the existing nomenclature, the novel allele was named tt6-5.
Encouraged by this result we also applied the described strategy to the set of tds mutants for which no molecular functions were known so far. The tds mutants 1, 2, 3, 5 and 6 showed no prominent alterations in flavonol and anthocyanin contents, when compared to the corresponding wild types (Ws for tds1, 2 and 3 and Col-7 for tds5 and 6; Fig. 5a). Treatment with vanillin/HCl, however, revealed a weak and irregular red colouration of endothelium and micropylar cells in young tds3 seeds with no signals in the chalazal area. As this phenotype was most comparable to tt12-2 (Fig. 5b; Debeaujon et al. 2001), we performed testcrosses of tds3-1 to tt12-2 and analysed mature seeds from F1 plants. No complementation was observed based on the results from DMACA staining (Fig. 5c), which indicated that tds3-1 was allelic to tt12. Using several combinations of T-DNA and gene-specific primers, we were not able to identify a T-DNA insertion at the tt12 locus At3g59030 in tds3-1 by PCR. Thus, we tried to determine the DNA sequence of the entire genomic region. Several primer combinations that were successfully used in wild type to amplify a region containing the 3′-end of the first exon together with the first intron failed on DNA isolated from tds3-1. Given the fact that the T-DNA insertion in tt12-1 was located in the same region at the beginning of intron 1 (Debeaujon et al. 2001) and that tt12-1 as well as tds3-1 originated from the Feldmann collection, it is tempting to speculate that both mutants in fact may contain the same allele. In the remaining tds mutants 1, 2, 5 and 6 we observed a combination of reduced vanillin stainable PAs in endothelium resulting in lighter colour after DMACA staining together with a clearly pigmented chalaza that resembled the phenotype of aha10-6 (Figs. 3, 5a). We performed systematic test crosses and observed no complementation in tds5 × aha10-6 progeny (Fig. 5c). Using PCR with gene- and T-DNA-specific primers and subsequent sequencing of the PCR products we found that tds5 contains a T-DNA insertion in intron 19 of AHA10. Our identification of tds5 as novel aha10 mutant increases the number of available alleles to address open questions with regard to AHA10 function in flavonoid transport.
Phenotypes of tannin-deficient seed (tds) mutants and allelism tests. a Flavonol glycoside, anthocyanin and PA accumulation as presented in Fig. 2. The background of tds5 and 6 is Col-7, which is identical to Col-0 (see Fig. 3). b Bases of tds3-1 and tt12-2 seeds in higher magnification. Chalazal cells of both mutants show no vanillin signals. c DMACA-stained mature F2 seeds (with testa from F1 mother plant) of crosses as indicated. Seeds of tds3-1 × tt12-2 and tds5 × aha10-6 crosses show PA deficiency. ch chalaza, mi micropyle. Bars 0.5 mm for seedlings and 100 μm for vanillin/HCl- and DMACA-treated seeds in a; 50 μm in b
Conclusions and outlook
The availability of a comprehensive set of mutants in nearly all aspects of flavonoid biosynthesis makes A. thaliana an invaluable tool for genetic and biochemical elucidation as well as for biotechnological manipulation of this important pathway in plants. As a basis for further investigations, we have built and systematically characterised a collection of tt mutants in the genetic background of the accession Col-0. Our findings in general confirmed characterisations of other alleles of the respective tt or tds mutants. In addition, for several mutants we observed features that were not described previously. The combination of visible phenotypic differences with staining techniques as presented here yields a nearly unique combination of features for every tt mutant known so far. Placing a newly identified mutant with such a phenotype into this grid allows efficient mutant classification, tentative identification of candidate genes for sequencing and reduces the number of required crosses in allelism tests. We have demonstrated the power of this approach by identifying the novel f3h allele tt6-5, which represents the first mutant that resulted from a forward genetic screen in the GABI-Kat collection. Additional T-DNA insertion lines isolated based on altered seed colour are currently being characterised. Furthermore, we showed that tds3 is allelic to tt12 and that tds5 is allelic to aha10. Since another aha10 allele may be represented by tt13 (own results and mentioned in Saito et al. 2013) the list of published seed colour mutants to be characterised could be reduced to tt9, which was described by Koornneef (1990) and mapped by Shirley et al. (1995), as well as the tds mutants 1, 2 and 6. We hope that the collection presented here will be a valuable resource to address open questions in flavonoid biosynthesis, transport, polymerisation and oxidation e.g. by functional analyses which require crosses with other ecotypes or characterisation of allelic series of mutants.
Abbreviations
- AGI:
-
Arabidopsis Genome Initiative
- Col:
-
Columbia
- DMACA:
-
p-dimethylaminocinnamaldehyde
- DPBA:
-
Diphenylboric acid 2-aminoethyl ester
- EGL3:
-
ENHANCER OF GLABRA 3
- MBW:
-
MYB-bHLH-WD40 repeat protein
- NFZ:
-
Norflurazon
- PA(s):
-
Proanthocyanidin(s)
- PAP1:
-
Production of Anthocyanin Pigment 1
- PFG:
-
Production of Flavonol Glycosides
- tds :
-
tannin-deficient seed
- TLC:
-
Thin-layer chromatography
- tt :
-
transparent testa
- Ws:
-
Wassilewskija
References
Abrahams S, Tanner GJ, Larkin PJ, Ashton AR (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol 130:561–576
Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35:624–636
Albert S, Delseny M, Devic M (1997) BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J 11:289–299
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Appelhagen I, Huep G, Lu GH, Strompen G, Weisshaar B, Sagasser M (2010) Weird fingers: functional analysis of WIP domain proteins. FEBS Letters 584:3116–3122
Appelhagen I, Jahns O, Bartelniewoehner L, Sagasser M, Weisshaar B, Stracke R (2011a) Leucoanthocyanidin dioxygenase in Arabidopsis thaliana: characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene 484:61–68
Appelhagen I, Lu GH, Huep G, Schmelzer E, Weisshaar B, Sagasser M (2011b) TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J 67:406–419
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102:2649–2654
Bharti A, Khurana J (2003) Molecular characterization of transparent testa (tt) mutants of Arabidopsis thaliana (ecotype Estland) impaired in flavonoid biosynthetic pathway. Plant Sci 165:1321–1332
Bolle C, Huep G, Kleinbolting N, Haberer G, Mayer K, Leister D, Weisshaar B (2013) GABI-DUPLO: a collection of double mutants to overcome genetic redundancy in Arabidopsis thaliana. Plant J 75:157–171
Bowerman PA, Ramirez MV, Price MB, Helm RF, Winkel BS (2012) Analysis of T-DNA alleles of flavonoid biosynthesis genes in Arabidopsis ecotype Columbia. BMC Res Notes 5:485
Brillouet JM, Romieu C, Schoefs B, Solymosi K, Cheynier V, Fulcrand H, Verdeil JL, Conéjéro G (2013) The tannosome is an organelle forming condensed tannins in the chlorophyllous organs of Tracheophyta. Ann Bot (Lond) 112:1003–1014
Buer CS, Sukumar P, Muday GK (2006) Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol 140:1384–1396
Bürger D (1971) Die morphologischen Mutanten des Göttinger Arabidopsis-Sortiments, einschließlich der Mutanten mit abweichender Samenfarbe. Arabidopsis Inf Serv 8:36–42
Debeaujon I, Leon-Kloosterziel K, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122:403–413
Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13:853–872
Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15:2514–2531
DeBolt S, Scheible WR, Schrick K, Auer M, Beisson F, Bischoff V, Bouvier-Navé P, Carroll A, Hematy K, Li Y, Milne J, Nair M, Schaller H, Zemla M, Somerville C (2009) Mutations in UDP-glucose:sterol glucosyltransferase in Arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiol 151:78–87
Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19:387–398
Feinbaum RL, Ausubel FM (1988) Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol 8:1985–1992
Focks N, Sagasser M, Weisshaar B, Benning C (1999) Characterization of tt15, a novel transparent testa mutant of Arabidopsis thaliana (L.) Heynh. Planta 208:352–357
Francisco RM, Regalado A, Ageorges A, Burla BJ, Bassin B, Eisenach C, Zarrouk O, Vialet S, Marlin T, Chaves MM, Martinoia E, Nagy R (2013) ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25:1840–1854
Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17:52–60
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Gonzalez A, Mendenhall J, Huo Y, Lloyd A (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Developmental Biology 325:412–421
Gonzalez A, Mendenhall J, Huo Y, Lloyd A (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Developmental Biology 325: 412-421 Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, Okada K, Wada T (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19:2531–2543
Jez JM, Noel JP (2002) Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences. J Biol Chem 277:1361–1369
Jez JM, Bowman ME, Dixon RA, Noel JP (2000) Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat Struct Biol 7:786–791
Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14:1359–1375
Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37:104–114
Kitamura S, Matsuda F, Tohge T, Yonekura-Sakakibara K, Yamazaki M, Saito K, Narumi I (2010) Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J 62:549–559
Klein M, Burla B, Martinoia E (2006) The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett 580:1112–1122
Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40:D1211–D1215
Koornneef M (1981) The complex syndrome of ttg mutants. Arabidopsis Inf Serv 18:45–51
Koornneef M (1990) Mutations affecting the testa colour in Arabidopsis. Arabidopsis Inf Serv 27:1–4
Koornneef M, Luiten W, de Vlaming P, Schram AW (1982) A gene controlling flavonoid 3′-hydroxylation in Arabidopsis. Arabidopsis Inf Serv 19:113–115
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Lukacin R, Britsch L (1997) Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3β-hydroxylase. Eur J Biochem 249:748–757
Martens S, Preuß A, Matern U (2010) Multifunctional flavonoid dioxygenases: flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 71:1040–1049
Mueller LA, Goodman CD, Silady RA, Walbot V (2000) AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol 123:1561–1570
Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L (2002) The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14:2463–2479
Owens DK, Crosby KC, Runac J, Howard BA, Winkel BS (2008) Biochemical and genetic characterization of Arabidopsis flavanone 3β-hydroxylase. Plant Physiol Biochem 46:833–843
Pang Y, Peel G, Sharma S, Tang Y, Dixon R (2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105:14210–14215
Pang Y, Cheng X, Huhman DV, Ma J, Peel GJ, Yonekura-Sakakibara K, Saito K, Shen G, Sumner LW, Tang Y, Wen J, Yun J, Dixon RA (2013) Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta 238:139–154
Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126:536–548
Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17:2966–2980
Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I (2007) Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 12:29–36
Pourcel L, Irani NG, Lu Y, Riedl K, Schwartz S, Grotewold E (2010) The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol Plant 3:78–90
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenised population (GABI-Kat) for flanking sequence tag based reverse genetics. Plant Mol Biol 53:247–259
Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224:96–107
Routaboul JM, Dubos C, Beck G, Marquis C, Bidzinski P, Loudet O, Lepiniec L (2012) Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J Exp Bot 63:3749–3764
Sagasser M, Lu G-H, Hahlbrock K, Weisshaar B (2002) A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev 16:138–149
Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem 72:21–34
Sarkar SK, Howarth RE (1976) Specificity of the vanillin test for flavanols. J Agric Food Chem 24:317–320
Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B (2000) Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem 381:749–753
Sheahan JJ, Rechnitz GA (1992) Flavonoid-specific staining of Arabidopsis thaliana. BioTechniques 13:880–883
Sheahan JJ, Rechnitz GA (1993) Differential visualization of transparent testa mutants in Arabidopsis thaliana. Anal Chem 65:961–963
Sheahan JJ, Cheong H, Rechnitz GA (1998) The colorless flavonoids of Arabidopsis thaliana (Brassicaceae). I. A model system to study the orthodihydroxy structure. Am J Bot 85:467–475
Shirley BW, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4:333–347
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50:660–677
Stracke R, De Vos RCH, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, Weisshaar B (2009) Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229:427–445
Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010) Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188:985–1000
Sun Y, Li H, Huang JR (2012) Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol Plant 5:387–400
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F (2008) An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat Cell Biol 10:1456–1462
Viotti C, Kruger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutte Y, Frescatada-Rosa M, Wolfenstetter S, Sauer N, Hillmer S, Grebe M, Schumacher K (2013) The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25:3434–3449
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11:1337–1350
Wangwattana B, Koyama K, Nishiyama Y, Kitayama M, Yamazaki M, Saito K (2008) Characterization of PAP1-upregulated glutathione S-transferase genes in Arabidopsis thaliana. Plant Biotechnol 25:191–196
Weisshaar B, Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1:251–257
Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299:396–399
Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C (2013a) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol 202:132–144
Xu W, Grain D, Le Gourrierec J, Harscoet E, Berger A, Jauvion V, Scagnelli A, Berger N, Bidzinski P, Kelemen Z, Salsac F, Baudry A, Routaboul JM, Lepiniec L, Dubos C (2013b) Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol 198:59–70
Zhao J, Dixon RA (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21:2323–2340
Zhao J, Pang Y, Dixon RA (2010) The mysteries of proanthocyanidin transport and polymerization. Plant Physiol 153:437–443
Acknowledgments
We are grateful to Nadja Parusel for help with tt6-5 mutant analysis and Andrea Voigt for excellent technical assistance. We thank Sharon Abrahams and Anthony Ashton (CSIRO-Plant Industry, Canberra, Australia) for seeds of the tds mutant lines, NASC for providing additional seed stocks and the reviewers for their suggestions to improve the manuscript. This project was funded in part by the German Federal Ministry of Education and Research (BMBF) in the context of the German plant genomics program GABI (Förderkennzeichen 0313855).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special topic: Anthocyanins. Guest editor: Stefan Martens.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2014_2088_MOESM2_ESM.tiff
Fig. S1 Genomic structure and phenotypes of different tt3 alleles. a Gene model of tt3 alleles characterised in this study. Symbols and colours as in Fig. 4. b Left column, vanillin/HCl-treated seeds; right column, DMACA-stained mature seed as in Fig. 3 (TIFF 9696 kb)
425_2014_2088_MOESM3_ESM.tiff
Fig. S2 Genomic structure and phenotypes of different tt2 alleles. a Gene model of tt2 alleles characterised in this study. Symbols and colours as in Fig. 4. b TT2 transcript levels in tt2-5 relative to the expression in Col-0 determined by qRT-PCR. c Vanillin/HCl-treated seeds of the indicated developmental stages. d DMACA-stained mature seed as in Fig. 3. e Mature seeds as in Fig. 2 (TIFF 6376 kb)
425_2014_2088_MOESM4_ESM.tiff
Fig. S3 Genomic structure and phenotypes of different tt6 alleles. Upper panel, gene model of tt6 alleles characterised in this study. Symbols and colours as in Fig. 4. Lower panel, mature seeds as in Fig. 2 (TIFF 3306 kb)
Rights and permissions
About this article
Cite this article
Appelhagen, I., Thiedig, K., Nordholt, N. et al. Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta 240, 955–970 (2014). https://doi.org/10.1007/s00425-014-2088-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2088-0