Abstract
The rhizospheric microbiome is comprised of many microbes, some of which reduce the virulence of their phytopathogenic neighbors; however, the mechanisms underlying these interactions are largely unknown. Rice soil isolate Pseudomonas chlororaphis EA105 strongly inhibits Magnaporthe oryzae’s in vitro growth by restricting fungal diameter as well as inhibiting the formation of the appressorium, required for penetration. We were interested in elucidating M. oryzae’s response to EA105 treatment, and utilized a microarray approach to obtain a global perspective of EA105 elicited changes in this pathogen. Based on this analysis, three genes of interest were knocked out in M. oryzae 70-15, and their sensitivity to EA105 treatment as well as their ability to infect rice was determined. Priming rice plants with EA105 prior to M. oryzae infection decreased lesion size, and the mutants were tested to see if this effect was retained. A null 70-15 mutant in a trichothecene biosynthesis gene showed less susceptibility to bacterial treatment, forming more appressoria than the parental type 70-15. A similar pattern was seen in a null mutant for a stress-inducible protein, MGG_03098. In addition, when this mutant was inoculated onto the leaves of EA105-primed rice plants, lesions were reduced to a greater extent than in 70-15, implicating the lack of this gene with an increased ISR response in rice. Understanding the global effect of biocontrol bacteria on phytopathogens is a key for developing successful and lasting solutions to crop loss caused by plant diseases and has the potential to greatly increase food supply.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exploiting the antifungal activities of natural soil microbes is currently being explored in many crop systems as a way to increase yields and reduce disease. In rice (Oryza sativa), a staple food crop worldwide, crop protection could directly translate to increased food supply. There is evidence that sheath blight caused by Rhizoctonia solani may be diminished using multiple soil microbes including Bacillus subtilis (Mousivand et al. 2012) as well as mycorrhizal fungi from orchids (Mosquera-Espinosa et al. 2013). Bacterial diseases of rice such as seedling rot and seedling blight may be controllable through the use of bacteriophages (Adachi et al. 2012) while leaf blight caused by the bacterial pathogen Xanthomonas oryzae could be reduced using biocontrol strains of Bacillus (Chithrashree et al. 2011). The most devastating pathogen of rice, Magnaporthe oryzae, is the causal agent of fungal blast, and results in the loss of 10–30 % of rice yields (Skamnioti and Gurr 2009). To control it, several microorganisms are currently being investigated, including B. subtilis (Leelasuphakul et al. 2006), B. methylotrophicus (Shan et al. 2013), Streptomyces globisporus (Li et al. 2011), and Pseudomonas fluorescens (De Vleesschauwer et al. 2008; Krishnamurthy and Gnanamanickam 1998).
Although multiple microorganisms are being investigated for their potential to reduce blast symptoms caused by M. oryzae, the specific mechanisms by which they work are largely unknown. Biocontrol bacteria can work either through direct antagonism of the pathogen, or through a mechanism known as induced systemic resistance (ISR). With ISR, the bacteria initiate a response in plants through the signaling of small molecules such as salicylic acid (SA), jasmonic acid (JA) or ethylene (ET) which results in the plants being less susceptible to pathogens (De Vleesschauwer and Hofte 2009). However, even when considering just a direct inhibition of the pathogen by biocontrol bacteria, there is still much to be discovered about the compounds which are playing a role in inhibition and their response on pathogens. The identity of the some of the antimicrobial compounds has been determined, such as hydrogen cyanide (HCN) (Blumer and Haas 2000), 2,4-diacetylphloroglucinol (2,4-DAPG) (Yang and Cao 2012), pyrrolnitrin, pyoluteorin (Dubuis et al. 2007), surfactants, and β-1-3-glucanases (Leelasuphakul et al. 2006), though there are many more to be discovered. Although these compounds have been shown to inhibit phytopathogens, the specific response that they elicit in the pathogens is not well understood. The pathogen’s response to inhibition is a crucial part of the communications, and will impact the composition of compounds secreted by the pathogen into the rhizosphere, which can directly impact other microorganisms as well as the crop plant. Trichothecenes, for example, play a large role in fungal defense response and are potent eukaryotic toxins (Desjardins et al. 1993). Fungi typically synthesize trichothecenes in response to stress, such as inhibition, and these toxins can accumulate in crop plants, rendering them unsafe for consumption (Desjardins et al. 1993; Jonkers et al. 2012). Thus, it is important to understand the broad impact and secondary effects of using biocontrol strains to antagonize phytopathogens.
The transcriptional response of M. oryzae treated with chemical fungicides has revealed a glimpse of how the fungus is reacting to fungicide treatment. Some of the gene expression changes are general responses to stress, while others are specific to the treatment (Mathioni et al. 2011). One of the most well-studied transcriptional responses is the up-regulation of trichothecene biosynthesis genes (Oh et al. 2008). As previously mentioned, trichothecenes are extremely toxic to eukaryotes and function to inhibit protein synthesis at the ribosome (Desjardins et al. 1993; Havrankova and Ovesna 2012; Merhej et al. 2011). In plants, these compounds impede defense responses by interfering with cell wall fortification and callose deposition (Jansen et al. 2005). Specific environmental conditions, such as the presence of sodium bicarbonate, can reduce the low basal expression of trichothecene synthesis (TRI) genes (Roinestad et al. 1994), while the high levels of reactive oxygen species (ROS) or the presence of azole fungicides at sub-lethal levels, greatly increase the expression of TRI genes (Audenaert et al. 2012; Kulik et al. 2012). When respiration is blocked by compounds such as cyanide, antimycin A, or the antifungal compound SSF-126, alternative respiration genes are induced. The alternate oxidase (AOX) gene was one of the first of these genes to be identified in M. oryzae and was shown to be induced by SSF-136 and hydrogen peroxide (Yukioka et al. 1998). Both the mycelial growth and conidial germination depend on AOX genes to survive in the presence of fungicides that block the canonical respiration pathway (Avila-Adame and Koller 2003). The detoxification of ROS is also associated with specific transcriptional events. In yeasts and filamentous fungi there is a conserved mechanism for coping with ROS which is mediated through the induction of specific genes to either detoxify ROS (catalase, superoxidismutase) or to repair ROS damage and maintain cellular homeostasis. The process is coordinated by proteins in the AP-1 family of bZIP activating proteins (Liu et al. 2005). In addition, transporters also play a role in fungicide response/tolerance by promoting the efflux of toxic compounds out of fungal cells. There are two main classes of transporters, the ATP-binding cassette (ABC) transporters which use energy generated by ATP hydrolysis, and the major facilitator superfamily (MFS) which use proton motive force to move substances across membranes. Some transporters have a broad range of substrates, such as Pdr5p which is induced by multiple fungicides, whereas others are more specific (Del Sorbo et al. 2000). For example, Pdr12p transports only C1–C7 organic acids, such as benzoic, sorbic, or propionic acid. Likewise, the transporter encoded by ABC2 is induced by some but not all fungicides. Azoles (including tricyclazole), pyroquilon, carpropamide, and benomyl all induce transcription of ABC2 but little to no transcriptional effect is seen after treatment with other fungicides such as phthalide, isoprothiolane, or kasugamycin (Lee et al. 2005). In addition, ABC1 transcription is also strongly induced in M. oryzae in response to azoles. Fungicides also typically induce genes in the HOG pathway, responsible for general and osmotic stress responses. This pathway is not present in mammals, which makes it a prime target for development of fungicides (Jiang et al. 2011). At least one of the genes in the HOG pathway, which is induced by ROS as well as fungicides such as tricyclazole, also plays a role in trichothecene biosynthesis. As of yet, there are no genome-wide examinations of M. oryzae’s response to fungicides.
Although M. oryzae’s broad transcriptional response to in vitro stresses has been examined previously (Mathioni et al. 2011), this study, to our knowledge, is the first to examine the global transcriptional responses of M. oryzae to treatment with an antagonistic bacterium with biocontrol potential. In addition, EA105 was originally isolated directly from field-grown rice plants. Survival and assimilation into the rhizospheric microbiome is a crucial factor in the success of a biocontrol bacterium (Bakker et al. 2013), and a natural rice soil isolate is more likely to thrive and perform than isolates which are non-native to rice soil. Our lab has previously shown the ability of EA105 to directly inhibit M. oryzae’s growth and appressoria formation, as well as trigger ISR in rice to reduce blast lesions (Spence et al. unpublished). The objective of this study is to examine the effect of EA105 treatment on the M. oryzae’s transcriptome. We also show the functional significance of some of the key genes in M. oryzae targeted by EA105. These results provide new insights into the importance of functional microbiome in suppressing plant diseases.
Materials and methods
Fungal and bacterial strains and growth conditions
Wild-type M. oryzae 70-15, the sequenced reference strain, was used throughout the experiments. Knockout mutants were constructed in the 70-15 background. For vegetative growth, the fungi were placed on complete medium (CM) containing sucrose (10 g/L), casamino acids (6 g/L), yeast extract (6 g/L), and 1 mL of Aspergillus nidulans trace elements (Per 100 mL: 0.22 g MnSO4·H20, 0.05 g KI, 0.02 g ZnSO4·7H20, 0.01 g H3BO4, 0.1 mL concentrated H2SO4, 0.008 g NiCl2·6H20, 0.007 g CoCl2·6H20) (Mathioni et al. 2011). Oatmeal agar consisting of ground oats (50 g/L) and agar (15 g/L) were used for sporulation. Plates were kept at 25 °C with constant fluorescent light. Bacterial strain EA105, a Pseudomonas chlororaphis, was isolated from rice cultivar M-104 grown in the field by Dr. Venkatesan Sundaresan’s lab from the University of California (Davis). Strain D5 is a cyanide biosynthetic mutant of EA105 (Spence et al. unpublished). P. fluorescens biocontrol strain CHAO was obtained from the Culture Collection of Switzerland. Strain CHA77, an hcnABC mutant in the CHAO background (Laville et al. 1998), was also obtained from the Culture Collection of Switzerland. Bacterial strains were grown in liquid or solid Luria–Bertani (LB) medium at 28–30 °C.
Plant materials and growth conditions
Oryza sativa cultivar M-104 was donated by Dr. Venkatesan Sundaresan from the University of California (Davis). O. sativa Seraceltik, a cultivar of rice that is susceptible to M. oryzae, was maintained in Dr. Nicole Donofrio’s lab at the University of Delaware. Seeds for Oriza sativa Nipponbare were obtained from the United States Department of Agriculture, Agricultural Research Service, from the Genetic Stocks-Oryzae collection. Rice plants were grown in Cornell mix rice soil with 16 h of light (28 °C, 80 % RH) and 8 h of darkness (26 °C, 80 % RH) (Mathioni et al. 2011).
Sample preparation and one-color microarray
A 5 mm plug of M. oryzae 70-15 was placed on a complete medium (CM) agar plate, 4 cm from a 5 μL drop of Luria–Bertani (LB) broth or bacteria in LB (5 × 105 cells/mL). Co-inoculated plates were dried in a laminar flow hood to reduce the motility of EA105. If the bacterial droplet was not given adequate time to dry, EA105 showed enhanced motility, physically interacting with and killing 70-15. At 72 h post-co-inoculation, the fungal mass was scraped from the top of the agar and ground in liquid nitrogen. RNA was extracted using the Qiagen RNeasy Plant Mini Kit. RNA samples were sent on dry ice to Beckman Coulter Genomics where a one-color gene expression microarray was performed using Agilent Magnaporthe (V2) 4X44K slides. Each biological replicate contained tissue from five plates, and there were three biological replicates per treatment.
Microarray analysis
From the data set provided by Beckman Coulter Genomics, data was filtered and excluded from probes that produced intensity values of less than 100 as well as from those that had a p value greater than 0.01. In addition, fold changes were calculated for each sequence ID by comparing intensities over untreated M. oryzae samples. Fold changes smaller than two were also excluded. Heat maps were constructed using the University of Toronto Bar Heatmapper Tool (http://bar.utoronto.ca/ntools/cgi). The Gene Ontology (GO) terms were determined for the top 100 up- and down-regulated genes in each treatment by using the Sequence ID to find the 60-mer probe sequence using a cross-reference file available through NCBI. Then, the probe sequence was put through BLASTn (NCBI) to find the gene accession number for the gene corresponding to the probe. The gene accession number was used to search for previously annotated GO terms based on the version 5 M. oryzae genome sequence (Meng et al. 2009). The data obtained from the microarray were deposited in the Gene Expression Omnibus (GEO) database at http://ncbi.nlm.nih.gov/geo under accession number GSE49597.
Creating gene knockout mutants in M. oryzae
Homologous recombination was used to replace genes of interest with a hygromycin resistance cassette through adaptamer-mediated PCR (Reid et al. 2002). For each gene to be knocked out, a 1.2 kb segment upstream of the 5′ UTR and another 1.2 kb segment downstream of the 3′ UTR were amplified, using primers that would add an adaptor to the 3′ end of the first segment, as well as to the 5′ end of the other segment. The hygromycin resistance cassette was amplified from plasmid pCB1003 using primers that had adaptors complementary to those used in amplifying the upstream and downstream segments. All three segments were combined in a PCR reaction to make the full-length constructs, approximately 3.3 kb. Creation of protoplasts and transformations were conducted following traditional methods (Sweigard et al. 1992). Primers are listed in Online Resource Table S1.
Diffusible and volatile in vitro inhibition assays
For the diffusible assays, a 5 mm fungal plug was placed 4 cm from a 5 μL drop of bacteria (5 × 105 cells/mL in LB) on a solid CM plate. For the volatile assay, compartmentalized plates were used which had four distinct quadrants. The fungal plug was placed in the quadrant opposite of the bacterial drop (Tenorio-Salgado et al. 2013). The diameter of fungal growth was measured every 24 h over the course of 7 days. Three biological replicates of this experiment were performed, each consisting of five plates per treatment.
Spore germination and appressoria formation assays
Plastic coverslips were sterilized in ethanol and used as hydrophobic surfaces to encourage the germination of conidiospores and subsequent appressoria formation. The 70-15 spores were grown on oatmeal agar for 10 days prior to being scraped into CM broth and filtered through Miracloth. Each coverslip was inoculated with a 50 μL drop containing a final concentration of 105 spores/mL, and 105 bacterial cells/mL. Spores were suspended in CM and bacteria were in LB. Controls contained 105 spores/mL with equal amounts of CM and LB (Tenorio-Salgado et al. 2013). These experiments were repeated in water with similar results. The coverslips were placed in petri dishes with wet filter discs in the center to promote humidity. Plates were sealed and placed in the dark. Germination percentages were calculated after 3 h incubation, and appressoria formation was determined after 24 h. Coverslips were imaged using a Zeiss Axioscope 2 light microscope. Five images were taken per coverslip, and 5 coverslips were used per treatment. Two biological replicates were examined.
In planta infection assays
Rice plants of cultivars M-104, Nipponbare, and Seraceltik were grown in soil for 3 weeks. To check the effect of bacterial priming, overnight cultures of bacteria were washed in water and re-suspended to 0.5 OD. For each plant, 2 mL of bacteria were dispensed onto the soil surface at the base of the plant. At 24 h post-bacterial treatment, the second youngest leaf cut and affixed to a large 15 cm diameter petri dish, on top of moistened paper towels and treated with 70-15 spores. Spores were grown on oatmeal agar for 10 days, and were subsequently scraped into sterile water with .02 % gelatin. Spore concentration was adjusted to 105 spores/mL. On each cut leaf, a total of 4–30 μL droplets of spores were placed along the length of the leaf. Plates were incubated in the dark for 24 h at 25 °C, after which time the spore droplets were wicked away. Plates were then kept in cycles of 16 h light/8 h darkness for 5 days at 25 °C. On the 5th day, the length and width of lesions were measured (Mathioni et al. 2011). A minimum of 8 leaves with 4 droplets per leaf were included per replicate. Three biological replicates were completed.
Sample preparation and confocal imaging of M. oryzae on rice
Rice plants were grown in soil for 3 weeks. To test the effect of bacterial treatment on the plants, some plants were primed as described above, 24 h prior to removing the leaf sheath. The innermost leaf sheath was then removed and cut into an approximately 6 cm segment. The shape of the leaf sheath created a tube into which approximately 200 μL of spores (105 cells/mL in water) were pipetted. The leaf sheaths were kept in a container with moist paper towels in the dark at 25 °C for 18 h. The leaf sheaths were then further dissected to obtain a thin layer around 2–3 cells thick) of the leaf sheath epidermis as per the previously published protocol (Kankanala et al. 2007). Samples were fixed in 4 % paraformaldehyde. Unstained samples were visualized using a Zeiss 510 NLO multiphoton confocal microscope with a C-Apochromat 40× water objective. Samples were excited with an argon laser at 488 nm. At least four plants were examined per replicate, and three biological replicates were completed.
Statistical analysis
Statistical analyses of the results were performed using the statistical software JMP 10. To compare across treatments, the Tukey’s HSD test was used and results were considered to be statistically different when p < 0.05.
Results
Global transcriptional changes in M. oryzae 70-15 following treatment with soil bacteria
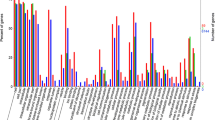
Previously, we have shown that M. oryzae 70-15 (hereafter 70-15) is drastically inhibited in vitro by rice soil isolate EA105, a strain of Pseudomonas chlororaphis isolated from field-grown O. sativa cv. M-104 (Spence et al. unpublished). To further examine the effects of EA105 treatment on 70-15, we used microarray analysis to examine transcriptional changes in 70-15 at 72 h post-EA105 treatment. The bacteria was placed 4 cm from the fungal plug, and plates were kept dry to prevent the bacteria from spreading and overtaking the fungal plug. For comparison, we also included known biocontrol agent P. fluorescens CHAO, as well as the corresponding hcnABC mutant CHA77, to determine the contribution of cyanide to transcriptional changes in 70-15. We have also previously shown that both CHAO and CHA77 inhibit 70-15 but to a lesser extent than EA105. Approximately half of M. oryzae’s genes were significantly down-regulated after treatment with EA105, compared to untreated controls (Table 1). Treatment with CHAO or CHA77 led to down-regulation of only 463 or 80 genes, respectively. EA105 only significantly up-regulated 44 genes, less than the 133 and 124 genes which were seen, respectively, with CHAO or CHA77 treatments (Table 1). When considering the 44 genes that are up-regulated with EA105 treatment, 36 were also up-regulated with CHAO treatment, 7 were up-regulated by CHAO and CHA77 treatment, and only one gene was uniquely up-regulated with EA105 treatment (Fig. 1). This gene, MGG_04034, is a putative NAD-dependent formate dehydrogenase. Due to the large number of 70-15 genes down-regulated with EA105 treatment, the top 100 down-regulated genes from each treatment were considered. Of these, EA105 and CHAO treatments resulted in the unique down-regulation of approximately 30 genes, whereas CHA77 was able to uniquely down-regulate twice as many. Only 14 of these genes were commonly down-regulated with all three bacterial treatments (Fig. 1).
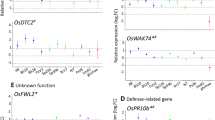
The vast majority of the most highly differentially expressed genes were of unknown function, based on GO term annotations (Online Resource Figs. 1–2). However, we examined several GO categories and generated heat maps to visualize the expression patterns seen with the different bacterial treatments (Fig. 2). When looking at genes involved in mycelium development, there are several that show a similar expression pattern with EA105 or CHAO treatment, but little change with CHA77 treatment. There are about six of these genes that are down-regulated with EA105 treatment, and to a lesser extent CHAO treatment, but not with CHA77 treatment. Genes related to pathogenesis were largely unaffected by any of the bacterial treatments, except MGG_10315 which was significantly down-regulated after EA105 and CHAO treatment (Fig. 2). Similarly, in the transporter category that included both ABC and MFS transporters, only one gene, MGG_06794, showed significant gene expression change. This gene was significantly down-regulated by both EA105 and CHAO treatment, but not CHA77 treatment. Genes involved in detoxification of ROS were unchanged with all three treatments. As expected, genes involved in cyanide detoxification were up-regulated by cyanide producers EA105 and CHAO, but not induced by cyanide mutant CHA77. A major fungal defense response is the production of trichothecenes, compounds that are toxic to eukaryotic organisms (Bennett and Klich, 2003). Genes involved in trichothecene biosynthesis were largely unaffected by EA105 treatment, while the same genes were induced by CHAO and CHA77 treatment (Fig. 2). A selection of 13 genes from multiple categories with varying expression patterns were examined using RT-PCR and expression patterns were similar to data obtained in the microarray (Online Resource Fig. 3).
Magnaporthe oryzae genes were categorized and heat maps were generated to visualize expression patterns. Numbers indicate log2 of fold changes for each treatment compared to untreated samples. Each treatment was done in biological triplicate, and differentially expressed genes had intensity values of at least 100, fold changes of at least 2, and a p value <0.01
In vitro characterization of M. oryzae deletion mutants
Based on the microarray data analysis, several genes of interest were chosen for further examination. Genes of interest included those which had a unique expression pattern with EA105 treatment compared to the other bacterial treatments, as well as genes with large expression changes with all three bacterial treatments. MGG_08440 was the only gene involved in trichothecene biosynthesis which was up-regulated by EA105 treatment. It was also the most highly up-regulated trichothecene biosynthesis gene with CHAO and CHA77 treatments. When EA105 treatment was compared to CHAO treatment, only one gene, MGG_03098, was significantly (2.2-fold) up-regulated. This gene codes for a stress-inducible protein which is highly conserved across kingdoms. Compared to untreated M. oryzae, it is actually down-regulated with CHAO and CHA77 treatments and only very slightly up-regulated with EA105 treatment. A third gene of interest, MGG_17822, codes for a carboxypeptidase and is very highly up-regulated in 70-15 treated with EA105 and CHAO, but not CHA77 (Online Resource Fig. 4). For each of these genes, full deletion mutants were created in the 70-15 background. Mutants had vegetative growth rates equivalent to 70-15 (Online Resource Fig. 5), appeared normal when grown on CM plates, and retained the ability to germinate and form appressoria in planta (Fig. 3). Some of the appressoria 70-15ΔMGG_08440 appeared to be elongated, particularly on M-104 leaves (Fig. 3c) but the majority of appressoria appeared normal. Spore germination and appressoria formation on plastic hydrophobic coverslips were similar between the mutants and 70-15, with a slight reduction in spore germination with 70-15ΔMGG_08440 (Supplemental Fig. 6). In addition, all three mutants retained the ability to form lesions in rice cultivars M-104, Seraceltik, and Nipponbare (Fig 4).
M. oryzae 70-15 and the three mutants were tested for their ability to form lesions in a Seraceltik, b M-104, and c Nipponbare. Error bars indicate standard error. A means comparison was done within lesion length and within lesion width for all pairs with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05). Twelve leaves were examined per treatment, in each of three biological replicates
The mutants were tested for their response to bacterial treatments in an in vitro diffusible assay in which a fungal plug and bacterial droplet were co-inoculated on a CM plate, 4 cm apart. Susceptibility to EA105, naturally isolated from O. sativa cv. M-104 soil, D5, a cyanide non-producing mutant in the EA105 background, and CHAO, a known P. fluorescens biocontrol strain, were tested. At 7 days post-co-inoculation, the degree of mutant inhibition in response to EA105, D5, and CHAO was similar to that seen in 70-15. However, 70-15ΔMGG_17822 was significantly less susceptible to D5 treatment compared to EA105 treatment. In all the other M. oryzae strains there was no significant difference in the inhibition by EA105 and D5 (Fig. 5a). A similar assay was set up to test the contribution of bacterial volatile compounds to the inhibition of M. oryzae. In this assay, compartmentalized plates were used so that the fungal plug and bacterial droplet could only share headspace within the dish, but could not exchange diffusible compounds through the media. In 70-15 and all three mutants, EA105 and D5 have equivalent inhibitory activities, which are stronger than that seen with CHAO (Fig. 5b). However, mutants 70-15ΔMGG_03098 and 7015ΔMGG_17822 were more susceptible to CHAO treatment than wild-type 70-15 (Fig. 5b).
Susceptibility of fungal strains to EA105, D5, and CHAO was tested using a diffusible plate assay in which a 5 mm fungal plug was placed 4 cm from a droplet of bacteria on a solid plate (a), as well as a volatile component only assay in which quadrant plates were used to physically separate the media on which the bacterium and fungus were grown (b). Error bars indicate standard error. A means comparison was done for all pairs with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05)
The effect of bacterial treatment on the M. oryzae mutants’ abilities to germinate and form appressoria was also examined. Bacterial treatment with EA105, D5, and CHAO had no effect on the germination percentage of 70-15 or 70-15ΔMGG_17822; however, EA105 was able to slightly reduce germination in 70-15ΔMGG_08440, while D5 marginally reduced germination in 70-15ΔMGG_03098 (Fig. 6a). Appressoria formation is almost completely halted in 70-15 treated with EA105 or D5. The M. oryzae deletion mutants also show a similar pattern, although the reduction in appressoria formation is not as drastic as the parental 70-15. With 70-15ΔMGG_03098, D5 was not able to restrict appressoria formation to the same extent as EA105, as was the case with all the other M. oryzae strains (Fig. 6b). CHAO also reduces appressoria formation in 70-15, and to a lesser extent in 70-15ΔMGG_17822. However, CHAO was unable to significantly reduce appressoria formation in mutants 70-15Δ08440 and 70-15Δ03098 (Fig. 6b).
Fluorescent pseudomonads such as EA105, D5, and CHAO produce multiple antimicrobial compounds that play a role in inhibiting phytopathogens in the soil. The production of cyanide is common amongst these biocontrol bacteria and has a broad range of action. At soil pH, cyanide is typically present as HCN, a gaseous form (Blumer and Haas 2000). HCN interferes with cellular respiration by binding and inhibiting the activity of an enzyme of the electron transport chain, cytochrome C oxidase, interfering with the transfer of electrons to oxygen, and therefore preventing the cell from aerobically producing ATP (Blumer and Haas, 2000). When 70-15 and the mutants were treated in the diffusible and volatile compartment plate assays with cyanide at concentrations up to 500 μM, fungal growth was not significantly inhibited (Fig. 7a, b). Another antibiotic compound frequently produced by biocontrol pseudomonads is 2,4-diacetylphloroglucinol (2,4-DAPG) which is a diffusible compound. When tested against the M. oryzae strains, concentrations of 25 μM and above significantly inhibited vegetative growth (Fig. 7c). However, there were no differences in the degree of inhibition between wild-type 70-15 and the mutant strains.
The effect of cyanide on the M. oryzae strains was tested using a diffusible (a) and volatile only (b) assay. A Tukey’s HSD test was done within each strain, ns not significant. The effect of various concentrations of 2,4-DAPG was examined in the diffusible assay (c). Error bars indicate standard error. A means comparison was done for all pairs with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05)
Ability of rhizobacteria to indirectly protect rice against wild-type and mutant M. oryzae infection
The ability of the M. oryzae mutants to form lesions on rice leaves was examined in rice cultivars M-104, Seraceltik, and Nipponbare. All mutants were capable of forming lesions in these cultivars. However, 70-15ΔMGG_17822 formed lesions that were slightly but significantly smaller in width than 70-15 lesions in cultivars M-104, Seraceltik, and Nipponbare (Fig. 4). Previously, we have shown that pre-treating rice roots with EA105 prior to M. oryzae infection reduces the number of lesions on rice plants (Spence et al., unpublished). Here, we show that the size of lesions is also reduced when plants were primed with EA105. When M-104 plants were primed with EA105, the size of lesions from the M. oryzae mutants was reduced, except for 70-15ΔMGG_17822. For this mutant, priming with EA105 significantly reduced lesion width, but lesion length was not significantly reduced in cultivar M-104 (Fig. 8). In Seraceltik, lesion length was modestly but significantly reduced, but there was no change in lesion width (Online Resource Fig. 7) while EA105 priming had no effect on the length or width of lesions that this mutant made on Nipponbare leaves (Online Resource Fig. 8). In all three rice cultivars there is a clear reduction in the ability of EA105 to reduce the size of lesion.
Rice plants of cultivar M-104 were primed with EA105 and subsequently infected with the M. oryzae strains. a Lesion length and b lesion width were measured at 5 days post-infection and c images were taken post-priming with EA105 and M. oryzae treatments. C control, UP unprimed, P primed. Error bars indicate standard error. A means comparison was done for all pairs with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05)
Discussion
Rice blast infections caused by M. oryzae ruin 10–30 % of rice crops (Skamnioti and Gurr 2009), which poses a threat to the food security of a staple crop consumed worldwide. This devastating plant pathogen has natural resistance to many fungicides, and can evolve resistance to newly encountered fungicides in as quickly as two growing seasons (Hamada et al. 1967; Sakurai et al. 1976; Takagaki et al. 2004). Further, many of these fungicidal chemicals also pose a risk to the environment and human health (Nomoto and Mori 1997; Yoon et al. 2011) and may also reduce soil fertility by harming beneficial microbes (Aktar et al. 2009). More recently, soil microbes are being evaluated as a more sustainable and effective method to control diseases in many plants, including rice, but none are currently being used which control M. oryzae. We have isolated a bacterium, EA105, from rice soil which shows direct in vitro inhibition of M. oryzae vegetative growth as well as an ability to interfere with the formation of appressoria, a structure that is critical during M. oryzae’s invasion of rice. When rice roots were pre-treated with EA105 prior to M. oryzae infection, the length and width of blast lesions were reduced. However, the mechanisms by which EA105 can directly antagonize M. oryzae, as well as reduce blast lesion size in rice, are unknown. We have used global transcriptional profiling to better understand M. oryzae’s response to EA105 treatment.
The most striking aspect of the transcriptional analysis is the down-regulation of nearly 6,000 M. oryzae genes following EA105 treatment. Likely, the decrease in expression of many of these genes is a secondary effect of the drastic growth inhibition that is seen with EA105 treatment. Although EA105, CHAO, and CHA77 all significantly restrict M. oryzae growth, only 14 of the 100 most highly down-regulated genes were common to all three treatments, which could be indicative either of EA105’s stronger antifungal activity that leads to down-regulation of a larger number of genes, or of a different modes of action for these bacteria against the fungal pathogen. Genes involved in routine cellular processes such as growth and metabolism were amongst those generally down-regulated. Few M. oryzae genes were significantly up-regulated following EA105 treatment, with only one being uniquely up-regulated by EA105. The putative function of this gene, MGG_04034, is a NAD-dependent formate dehydrogenase. Typically these enzymes catalyze the oxidation of formate to bicarbonate, transferring electrons to NAD+, and they are crucial for metabolizing C1 compounds (Popov and Lamzin 1994). The up-regulation of this gene with EA105 treatment but not the other two bacterial treatments suggests that EA105 may be producing a one-carbon compound that is not produced by the other two bacteria.
There was similarity in the expression pattern of many genes following EA105 and CHAO treatment, which were unchanged with CHA77 treatment. With CHA77 being a non-producer of cyanide in CHAO background, these expression changes are likely a response to cyanide. As expected, multiple cyanide hydratases were up-regulated with EA105 and CHAO but not CHA77 treatment. A subset of genes involved in mycelium development were down-regulated in this pattern, as well as one transporter gene, MGG_06794, and one pathogenesis-related gene, MGG_10315. Aside from these examples, there was largely no change in the expression of genes involved in pathogenesis, transport, or ROS detoxification, contrary to what was expected. ATP-binding cassette (ABC) transporters are transmembrane proteins responsible not only for transferring substrates, including toxins, out of the cell, but also can function in DNA repair (Higgins 1992). In addition, multi-drug resistance is typically the result of over expression of a variety of ABC transporters which remove antibiotics from within the cell. However, there was no change in expression of ABC transporter genes with any of the bacterial treatments. Similarly, there was no change in expression of major facilitator superfamily (MFS) transporters, secondary carriers that move small solutes across chemiosmotic gradients (Del Sorbo et al. 2000). There was also no change in the expression of peroxidases, catalases, and superoxide dismutases associated with ROS detoxification, suggesting that none of the bacterial treatments led to ROS accumulation in M. oryzae.
An integral part of the defense response in fungi is the production of potent toxins called trichothecenes. Knowledge regarding trichothecene production in M. oryzae is limited; however, the production of these compounds in other fungi such as Fusarium graminearum has been studied extensively (Desjardins et al. 1993; Havrankova and Ovesna 2012; Merhej et al. 2011). Secretion of trichothecenes is linked to increased virulence in some phytopathogenic fungi (Desjardins et al. 1996), and increased trichothecene biosynthesis occurs when fungi are challenged with antifungal compounds at sub-lethal levels (Kulik et al. 2012). Interestingly, CHAO and CHA77 treatment increased transcripts of trichothecene biosynthesis gene MGG_09777 but EA105 did not. Similarly, AMG07075 was induced nearly twice as much with CHAO and CHA77 treatment as compared to EA105. CHAO and CHA77 may be producing an antifungal that is triggering higher up-regulation of these biosynthetic genes; however, these two bacteria are not able to inhibit M. oryzae to the extent that EA105 does. EA105 may possess a mechanism for partially evading detection by M. oryzae, which prevents defense responses such as trichothecene biosynthesis.
To further examine the role of trichothecene biosynthetic gene MGG_08440, a knockout mutant was created in 70-15. This mutant showed a slight defect in germination, but was otherwise similar to 70-15 in morphology and growth. There were no differences in the mutant’s susceptibility to EA105, D5 (a cyanide mutant in EA105), or CHAO treatment in regards to vegetative growth. However, EA105 was able to reduce spore germination in 70-15∆MGG_08440 but not in 70-15. Spore germination is a critical process that only occurs under favorable conditions (Osherov and May 2001). It is possible that in 70-15∆MGG_08440, which may lack a gene in antibacterial defense, EA105 is at an advantage and is able to create an environment which is unfavorable for spore germination. Contrastingly, EA105 and D5 were less effective in reducing appressorium formation in this mutant as compared to 70-15, and CHAO also failed to inhibit appressorium formation in this mutant. The response of 70-15∆MGG_08440 to cyanide and 2,4-DAPG did not differ from 70-15, and are not likely to be the bacterially produced compounds that target induction of this gene in 70-15. There was no difference in this mutant’s ability to form lesions on rice leaves, and EA105 priming reduced 70-15ΔMGG_08440 lesion size to a similar extent as 70-15, indicating that this gene is unlikely to be involved in EA105’s mechanism of inducing ISR. This finding is particularly interesting, as this gene of interest had been identified through a direct antagonistic interaction between the bacteria and fungus yet it may also be playing a role in a process where the bacterium indirectly reduces disease symptoms, mediated through the plant.
Another gene involved in M. oryzae defense response is MGG_03098, which codes for a stress-inducible protein. Interestingly, this was the only gene that was significantly up-regulated with EA105 treatment compared to CHAO treatment, suggesting that it is induced by a compound produced by EA105 but not CHAO. This gene is highly conserved in both prokaryotes and eukaryotes, and is involved in thiazole biosynthesis. Thiazole is commercially used as a fungicide, but is also necessary for the synthesis of thiamine. Thiamine accumulation in plants has been associated with an ISR-like defense response and disease reduction in soybean (Abdel-Monaim 2011), pearl millet (Pushpalatha et al. 2011), tobacco (Malamy et al. 1996), Arabidopsis, thaliana (Ahn et al. 2005, 2007), and rice (Wang et al. 2006). The A. thaliana ortholog, THI1, has been implicated in DNA repair (Machado et al. 1996), but there are no reports of fungal homologs with this function. The Fusarium oxysporum homolog, STI35, is responsive to oxidative stress specifically, (Ruiz-Roldan et al. 2008) yet there was no change in expression of ROS detoxification genes after EA105 treatment. To examine the importance of this gene in M. oryzae’s response to EA105, a knockout mutant was created. This mutant, 70-15∆MGG_03098, does not appear to have any phenotypic differences from 70-15 nor growth defects. It also does not differ in its susceptibility to EA105, D5, or CHAO treatment in either the diffusible or volatile vegetative growth assays. In addition, 70-15∆MGG_03098 germinates and forms appressoria similarly to 70-15. As with 70-15, bacterial treatment had no effect on spore germination, but CHAO was unable to inhibit appressoria formation in this mutant, and D5 was impaired in its ability to reduce appressoria formation, which may indicate a role of this gene as a target for bacteria to evade recognition by M. oryzae. When this gene is missing, perhaps bacteria are recognized sooner, and have less of a detrimental effect on appressoria formation. EA105 still reduces 70-15∆MGG_03098 appressoria formation to almost the same extent seen with 70-15, perhaps due to an alternative strategy involving a combination of cyanide and other bacterial components which are not present in CHAO. There were no differences in the susceptibility of this mutant to cyanide or 2,4-DAPG, again indicating that these are not likely to be the bacterial products targeting this gene. In planta, 70-15∆MGG_03098 results in similar-sized lesions as 70-15, but there is a more drastic reduction in lesion length following priming with EA105. Reduced lesion size has previously been correlated with reduced blast disease severity and reduced virulence (Cacique et al. 2012; Mentlak et al. 2012; Tredway et al. 2003).
One of the most highly up-regulated M. oryzae genes following EA105 and CHAO treatment was MGG_17822, which encodes a putative carboxypeptidase. There have not been any reports of the function of this gene in M. oryzae. A BLASTn search revealed similarity to zinc carboxypeptidases, Type A digestive carboxypeptidases, and soluble (rather than anchored) carboxypeptidases. In addition, it was similar to carboxypeptidases which must be cleaved to become active, and those which are involved in proteolysis. Due to the lack of information regarding this gene, it is difficult to speculate on its function in M. oryzae and further functional investigations are needed. One explanation is that it may be involved in the proteolysis of a bacteria-produced antifungal protein, resulting in its up-regulation with EA105 and CHAO treatment. However, this gene also appears to be responsive to cyanide, since it was highly up-regulated in the presence of EA105 and CHAO, but not with CHAO’s cyanide-deficient mutant, CHA77. A full deletion mutant for this gene, 70-15∆MGG_17822 showed no phenotypic changes or growth defects. In addition, conidiospores germinated and formed appressoria normally. This mutant was significantly less susceptible to D5 treatment than EA105 in the diffusible assay, but not the volatile assay, implicating a role for diffusible cyanide in the inhibition. However, there were no differences in this mutant’s susceptibility to cyanide or 2,4-DAPG. Bacterial treatments did not affect 70-15∆MGG_17822 germination, but led to a similar but smaller reduction in appressoria formation than in 70-15. In planta, 70-15∆MGG_17822 lesions were not as wide as 70-15 lesions, and lesion length was not significantly decreased with EA105 treatment. Surprisingly, this gene which is targeted by EA105 in a direct interaction between bacteria and fungus also appears to be playing a critical role in ISR since the lack of this gene in M. oryzae renders EA105 unable to reduce blast lesion length. Three rice cultivars were tested, with differing susceptibilities to rice blast. Even so, the trends seen with EA105 priming as well as the ability of the mutant M. oryzae strains to infect were similar across cultivars.
The antagonistic interaction between EA105 and M. oryzae is a complicated two-way communication which becomes more complex when examining their indirect communications mediated through rice. However, the details of these complex communications are important for understanding the ways in which biocontrol bacteria effectively reduce disease. Using microbes to combat plant disease has many advantages over chemical fungicides including sustainability and reduced environmental impact, but most importantly this method promotes a lasting effectiveness due to the natural ability of microbes to co-evolve with their phytopathogen counterparts. Another important consideration in examining bacteria for biocontrol potential is their viability in the field. EA105 was isolated from rice soil, and is, therefore, more likely to thrive in rice field conditions than non-native bacteria. Thus far, investigations have mainly focused on the compounds secreted by biocontrol bacteria, but little is known about their transcriptional and functional effects in the pathogen. Global transcriptional profiling provides insight into the pathogen’s response to treatment, complementing studies which identify bacterially produced antifungal compounds. The studies pertaining to understanding the systemic defense response in plants subjected to biocontrol bacteria will also give insights to transcriptional changes in plants associated with beneficial microbiome. Biocontrol of plant diseases is a promising alternative to traditional disease management strategies, but there is much to discover about the relationships and communications that occur between biocontrol bacteria, pathogens, and other residents of the plant microbiome.
References
Abdel-Monaim MF (2011) Role of riboflavin and thiamine in induced resistance against charcoal rot disease of soybean. Afr J Biotechnol 10:10842–10855
Adachi N, Tsukamoto S, Inoue Y, Azegami K (2012) Control of bacterial seedling rot and seedling blight of rice by bacteriophage. Plant Dis 96:1033–1036
Ahn IP, Kim S, Lee YH (2005) Vitamin B-1 functions as an activator of plant disease resistance. Plant Physiol 138:1505–1515
Ahn IP, Kim S, Lee YH, Suh SC (2007) Vitamin B-1-induced priming is dependent on hydrogen peroxide and the NPR1 gene in Arabidopsis. Plant Physiol 143:838–848
Aktar W, Sengupta D, Chowdhury A (2009) Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol 2:1–12
Audenaert K, Monbaliu S, Deschuyffeleer N, Maene P, Vekeman F, Haesaert G, De Saeger S, Eeckhout M (2012) Neutralized electrolyzed water efficiently reduces Fusarium spp. in vitro and on wheat kernels but can trigger deoxynivalenol (DON) biosynthesis. Food Control 23:515–521
Avila-Adame C, Koller W (2003) Impact of alternative respiration and target-site mutations on responses of germinating conidia of Magnaporthe grisea to Qo-inhibiting fungicides. Pest Manag Sci 59:303–309
Bakker PA, Berendsen RL, Doornbos RF, Wintermans PC, Pieterse CM (2013) The rhizosphere revisited: root microbiomics. Front Plant Sci 4:165. doi:10.3389/fpls.2013.00165
Bennett JW, Klich M (2003) Mycotoxins. Clin Microbiol Rev 16:497
Blumer C, Haas D (2000) Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol 173:170–177
Cacique IS, Domiciano GP, Rodrigues FA, do Vale FXR (2012) Silicon and manganese on rice resistance to blast. Bragantia 71:239–244
Chithrashree S, Udayashankar AC, Nayaka SC, Reddy MS, Srinivas C (2011) Plant growth-promoting rhizobacteria mediate induced systemic resistance in rice against bacterial leaf blight caused by Xanthomonas oryzae pv. oryzae. Biol Control 59:114–122
De Vleesschauwer D, Hofte M (2009) Rhizobacteria-induced systemic resistance. In: VanLoon LC (ed) Plant innate immunity, vol 51. Academic Press Ltd-Elsevier Science Ltd, London, pp 223–281
De Vleesschauwer D, Djavaheri M, Bakker P, Hofte M (2008) Pseudomonas fluorescens WCS374r-Induced Systemic Resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol 148:1996–2012
Del Sorbo G, Schoonbeek HJ, De Waard MA (2000) Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet Biol 30:1–15
Desjardins AE, Hohn TM, McCormick SP (1993) Trichothecene biosynthesis in Fusarium species—chemistry, genetics, and significance. Microbiol Rev 57:595–604
Desjardins AE, Proctor RH, Bai GH, McCormick SP, Shaner G, Buechley G, Hohn TM (1996) Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol Plant Microbe Interact 9:775–781
Dubuis C, Keel C, Haas D (2007) Dialogues of root-colonizing biocontrol pseudomonads. Eur J Plant Pathol 119:311–328
Hamada M, Hashimoto T, Takahashi S, Yoneyama M, Miyake T, Takeuchi Y, Okami Y, Umezawa H (1967) Antifungal activity of Kasugamycin. J Antibiot B 20:424–426
Havrankova H, Ovesna J (2012) Genes of trichothecene biosynthesis in the Fusarium genus. Chem Listy 106:818–825
Higgins CF (1992) ABC transporters—from microorganisms to man. Annu Rev Cell Biol 8:67–113
Jansen C, von Wettstein D, Schafer W, Kogel KH, Felk A, Maier FJ (2005) Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. P Natl Acad Sci USA 102:16892–16897
Jiang JH, Yun YZ, Fu J, Shim WB, Ma ZH (2011) Involvement of a putative response regulator FgRrg-1 in osmotic stress response, fungicide resistance and virulence in Fusarium graminearum. Mol Plant Pathol 12:425–436
Jonkers W, Dong YH, Broz K, Kistler HC (2012) The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. PLoS Pathog 8:1–18
Kankanala P, Czymmek K, Valent B (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19:706–724
Krishnamurthy K, Gnanamanickam SS (1998) Biol control of rice blast by Pseudomonas fluorescens strain Pf7-14: evaluation of a marker gene and formulations. Biol Control 13:158–165
Kulik T, Lojko M, Jestoi M, Perkowski J (2012) Sublethal concentrations of azoles induce tri transcript levels and trichothecene production in Fusarium graminearum. FEMS Microbiol Lett 335:58–67
Laville J, Blumer C, Von Schroetter C, Gaia V, Defago G, Keel C, Haas D (1998) Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHAO. J Bacteriol 180:3187–3196
Lee YJ, Yamamoto K, Hamamoto H, Nakaune R, Hibi T (2005) A novel ABC transporter gene ABC2 involved in multidrug susceptibility but not pathogenicity in rice blast fungus Magnaporthe grisea. Pestic Biochem Phys 81:13–23
Leelasuphakul W, Sivanunsakul P, Phongpaichit S (2006) Purification, characterization and synergistic activity of beta-1,3-glucanase and antibiotic extract from an antagonistic Bacillus subtilis NSRS 89-24 against rice blast and sheath blight. Enzyme Microb Tech 38:990–997
Li QL, Jiang YH, Ning P, Zheng L, Huang JB, Li GQ, Jiang DH, Hsiang T (2011) Suppression of Magnaporthe oryzae by culture filtrates of Streptomyces globisporus JK-1. Biol Control 58:139–148
Liu HJ, Colavitti R, Rovira II, Finkel T (2005) Redox-dependent transcriptional regulation. Circ Res 97:967–974
Machado CR, deOliveira RLC, Boiteux S, Praekelt UM, Meacock PA, Lenck CFM (1996) Thi1, a thiamine biosynthetic gene in Arabidopsis thaliana, complements bacterial defects in DNA repair. Plant Mol Biol 31:585–593
Malamy J, SanchezCasas P, Hennig J, Guo AL, Klessig DF (1996) Dissection of the salicylic acid signaling pathway in tobacco. Mol Plant Microbe Interact 9:474–482
Mathioni SM, Belo A, Rizzo CJ, Dean RA, Donofrio NM (2011) Transcriptome profiling of the rice blast fungus during invasive plant infection and in vitro stresses. BMC Genomics 12:1–20
Meng SW, Brown DE, Ebbole DJ, Torto-Alalibo T, Oh YY, Deng JX, Mitchell TK, Dean RA (2009) Gene ontology annotation of the rice blast fungus Magnaporthe oryzae. BMC Microbiol 9:1–6
Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, Saitoh H, Terauchi R, Nishizawa Y, Shibuya N, Thomma B, Talbot NJ (2012) Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24:322–335
Merhej J, Richard-Forget F, Barreau C (2011) Regulation of trichothecene biosynthesis in Fusarium: recent advances and new insights. Appl Microbiol Biot 91:519–528
Mosquera-Espinosa AT, Bayman P, Prado GA, Gomez-Carabali A, Otero JT (2013) The double life of Ceratobasidium: orchid mycorrhizal fungi and their potential for biocontrol of Rhizoctonia solani sheath blight of rice. Mycologia 105:141–150
Mousivand M, Jouzani GS, Monazah M, Kowsari M (2012) Characterization and antagonistic potential of some native biofilm forming and surfactant producing Bacillus subtilis strains against six pathotypes of Rhizoctonia solani. J Plant Pathol 94:171–180
Nomoto S, Mori K (1997) Synthetic microbial chemistry. 30. Synthesis of acetophthalidin, a fungal metabolite which inhibits the progression of the mammalian cell cycle. Liebigs Ann Recl 4:721–723
Oh Y, Donofrio N, Pan H, Coughlan S, Brown DE, Meng S, Mitchell T, Dean RA (2008) Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol 9:R85
Osherov N, May GS (2001) The molecular mechanisms of conidial germination. FEMS Microbiol Lett 199:153–160
Popov VO, Lamzin VS (1994) NAD(+)-dependent formate dehydrogenase. Biochem J 301:625–643
Pushpalatha HG, Sudisha J, Geetha NP, Amruthesh KN, Shetty HS (2011) Thiamine seed treatment enhances LOX expression, promotes growth and induces downy mildew disease resistance in pearl millet. Biol Plantarum 55:522–527
Reid RJD, Sunjevaric I, Kedacche M, Rothstein R (2002) Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast 19:319–328
Roinestad KS, Montville TJ, Rosen JD (1994) Mechanism for sodium bicarbonate inhibition of trichothecene biosynthesis in Fusarium tricinctum. J Agr Food Chem 42:2025–2028
Ruiz-Roldan C, Puerto-Galan L, Roa J, Castro A, Di Pietro A, Roncero MIG, Hera C (2008) The Fusarium oxysporum sti35 gene functions in thiamine biosynthesis and oxidative stress response. Fungal Genet Biol 45:6–16
Sakurai H, Naito H, Fujita S (1976) Sensitivity distribution of phytopathogenic bacteria and fungi to antibiotics. J Antibiot 29:1230–1236
Shan HY, Zhao MM, Chen DX, Cheng JL, Li J, Feng ZZ, Ma ZY, An DR (2013) Biocontrol of rice blast by the phenaminomethylacetic acid producer of Bacillus methylotrophicus strain BC79. Crop Prot 44:29–37
Skamnioti P, Gurr SJ (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol 27:141–150
Sweigard JA, Chumley FG, Valent B (1992) Disruption of a Magnaporthe grisea cutinase gene. Mol Gen Genet 232:183–190
Takagaki M, Kaku K, Watanabe S, Kawai K, Shimizu T, Sawada H, Kumakura K, Nagayama K (2004) Mechanism of resistance to carpropamid in Magnaporthe grisea. Pest Manag Sci 60:921–926
Tenorio-Salgado S, Tinoco R, Vazquez-Duhalt R, Caballero-Mellado J, Perez-Rueda E (2013) Identification of volatile compounds produced by the bacterium Burkholderia tropica that inhibit the growth of fungal pathogens. Bioengineered 4:236–243
Tredway LP, Stevenson KL, Burpee LL (2003) Components of resistance to Magnaporthe grisea in ‘Coyote’ and ‘Coronado’ tall fescue. Plant Dis 87:906–912
Wang GN, Ding XH, Yuan M, Qiu DY, Li XH, Xu CG, Wang SP (2006) Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation. Plant Mol Biol 60:437–449
Yang F, Cao YJ (2012) Biosynthesis of phloroglucinol compounds in microorganisms-review. Appl Microbiol Biot 93:487–495
Yoon MY, Kim YS, Ryu SY, Choi GJ, Choi YH, Jang KS, Cha B, Han SS, Kim JC (2011) In vitro and in vivo antifungal activities of decursin and decursinol angelate isolated from Angelica gigas against Magnaporthe oryzae, the causal agent of rice blast. Pestic Biochem Phys 101:118–124
Yukioka H, Inagaki S, Tanaka R, Katoh K, Miki N, Mizutani A, Masuko M (1998) Transcriptional activation of the alternative oxidase gene of the fungus Magnaporthe grisea by a respiratory-inhibiting fungicide and hydrogen peroxide. BBA Gene Struct Expr 1442:161–169
Acknowledgements
H.P.B. acknowledges the support from National Science Foundation Award PGPR-0923806. We would like to thank Dr. Venkatesan Sundaresan and his lab for the M-104 seeds and for the soil and root samples from which we obtained our isolates. We would also like to thank Dr. Sandra Mathioni for her advice on microarray analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource Fig. 1 Pie charts were constructed to visualize the distribution of the top 100 up-regulated M. oryzae genes with a) EA105, b) CHAO, and c) CHA77 treatments into GO term categories.
Online Resource Fig. 2 Pie charts were constructed to visualize the distribution of the top 100 down-regulated M. oryzae genes with a) EA105, b) CHAO, and c) CHA77 treatments into GO term categories.
Online Resource Fig. 3 RT-PCR was used to validate microarray data for 13 genes of various categories with various expression patterns. Error bars indicate standard error.
Online Resource Fig. 4 Microarray fold change data for three genes of interest which were individually knocked out in M. oryzae 70-15. Error bars indicate standard error. A means comparison was done within each gene with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05).
Online Resource Fig. 5 The growth rate of the mutants was compared to that of wild-type 70-15 over the course of 7 days on CM plates. Error bars indicate standard error.
Online Resource Fig. 6 a) The germination percentage and b) appressoria formation percentage of untreated M. oryzae 70-15 and mutants were tested. Error bars indicate standard error. A means comparison was done for all pairs with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05).
Online Resource Fig. 7 Rice plants of cultivar Seraceltik were primed with EA105 and subsequently infected with the M. oryzae strains. a) Lesion length and b) Lesion width were measured at 5 days post infection and c) images were taken. C = Control; UP = Unprimed; P = Primed. Error bars indicate standard error. A means comparison was done for all pairs with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05).
Online Resource Fig. 8 Rice plants of cultivar Nipponbare were primed with EA105 and subsequently infected with the M. oryzae strains. a) Lesion length and b) Lesion width were measured at 5 days post infection and c) images were taken. C = Control; UP = Unprimed; P = Primed. Error bars indicate standard error. A means comparison was done for all pairs with Tukey’s HSD test. Means with the same letter are not significantly different (p < 0.05).
Rights and permissions
About this article
Cite this article
Spence, C.A., Raman, V., Donofrio, N.M. et al. Global gene expression in rice blast pathogen Magnaporthe oryzae treated with a natural rice soil isolate. Planta 239, 171–185 (2014). https://doi.org/10.1007/s00425-013-1974-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1974-1