Abstract
Previous research showed that auxin, ethylene, and nitric oxide (NO) can activate the expression of iron (Fe)-acquisition genes in the roots of Strategy I plants grown with low levels of Fe, but not in plants grown with high levels of Fe. However, it is still an open question as to how Fe acts as an inhibitor and which pool of Fe (e.g., root, phloem, etc.) in the plant acts as the key regulator for gene expression control. To further clarify this, we studied the effect of the foliar application of Fe on the expression of Fe-acquisition genes in several Strategy I plants, including wild-type cultivars of Arabidopsis [Arabidopsis thaliana (L.) Heynh], pea [Pisum sativum L.], tomato [Solanum lycopersicon Mill.], and cucumber [Cucumis sativus L.], as well as mutants showing constitutive expression of Fe-acquisition genes when grown under Fe-sufficient conditions [Arabidopsis opt3-2 and frd3-3, pea dgl and brz, and tomato chln (chloronerva)]. The results showed that the foliar application of Fe blocked the expression of Fe-acquisition genes in the wild-type cultivars and in the frd3-3, brz, and chln mutants, but not in the opt3-2 and dgl mutants, probably affected in the transport of a Fe-related repressive signal in the phloem. Moreover, the addition of either ACC (ethylene precursor) or GSNO (NO donor) to Fe-deficient plants up-regulated the expression of Fe-acquisition genes, but this effect did not occur in Fe-deficient plants sprayed with foliar Fe, again suggesting the existence of a Fe-related repressive signal moving from leaves to roots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The main characteristic of Strategy I plants is that they need to reduce Fe(III), the most abundant form of iron in soils, to Fe(II), prior to uptake. The Fe(III) reduction is mediated by a ferric reductase (EC 1.16.1.7), encoded by the FRO gene, and Fe(II) uptake is mediated by a transporter encoded by the IRT1 gene (Walker and Connolly 2008; Ivanov et al. 2011). Both genes are up-regulated under Fe deficiency and are activated by specific bHLH transcription factors, which are also up-regulated by Fe deficiency. These bHLH transcription factors include AtFIT, AtbHLH038, and AtbHLH039 in Arabidopsis (Yuan et al. 2008; Ivanov et al. 2011); SlFER is the FIT homolog in tomato (Brumbarova and Bauer 2005). Besides these genes, there are other important Fe-related genes also up-regulated under Fe deficiency, such as AtAHA2, AtAHA7, and CsHA1, encoding H+-ATPases (EC 3.6.3.6) involved in H+ extrusion (Santi et al. 2005; Santi and Schmidt 2009; Ivanov et al. 2011); AtNAS1 and AtNAS2, encoding nicotianamine synthase enzymes (NAS; EC 2.5.1.43) involved in the synthesis of the Fe(II) chelating agent nicotianamine (NA) (Klatte et al. 2009); AtFRD3, encoding a protein of the multidrug and toxin efflux (MATE) family, responsible for the loading into the xylem of citrate, an Fe chelator, which is essential for the correct distribution of Fe throughout the plant tissues (Durrett et al. 2007; Roschzttardtz et al. 2011); AtCCCL1-3, with similarities to nodulins, probably involved in vacuolar Fe transport (García et al. 2010); and AtMYB72, encoding a transcription factor involved in Fe responses and induced systemic resistance (García et al. 2010). The pathways that regulate the expression of these Fe-related genes are not totally known, but several published reports suggest a role for different hormones, like auxin, ethylene, and nitric oxide (NO) (Lucena et al. 2006; Graziano and Lamattina 2007; Waters et al. 2007; García et al. 2010, 2011; Chen et al. 2010; Bacaicoa et al. 2011; Lingam et al. 2011; Meiser et al. 2011; Ramírez et al. 2011; Romera et al. 2011; Wu et al. 2012). Auxin, ethylene, and NO can up-regulate the expression of Fe-acquisition genes in plants grown with low levels of Fe (or without Fe), but have almost no effect in plants grown with high levels of Fe (Lucena et al. 2006; Graziano and Lamattina 2007; Chen et al. 2010; García et al. 2011). This suggests that the up-regulation of Fe-acquisition genes does not solely depend on hormones (auxin, ethylene and NO), that act as activators, but also on Fe availability (Lucena et al. 2006; Romera et al. 2011).
It is still an open question as to how Fe acts to repress gene expression and which pool of Fe (e.g., root, phloem, intracellular, apoplastic, etc.) is monitored by the plant to mediate this control. Maas et al. (1988) proposed phloem Fe as an inhibitor of Fe deficiency responses in Strategy I plants. These authors found more Fe in the phloem of Fe-sufficient Ricinus plants (traveling as a Fe complex) than in Fe-deficient plants. Additionally, they also found that the application of Fe-EDTA to leaves decreased some of the Fe deficiency responses, such as proton extrusion and ferric reductase activity (Maas et al. 1988). Based on these results, they proposed that leaves could modulate Fe deficiency responses through phloem Fe, leading to suppression of gene expression under Fe-sufficient conditions (Maas et al. 1988). Several other reports also indicated that application of Fe (as FeSO4 or Fe-citrate) to leaves of Fe-deficient plants suppressed root Fe deficiency responses (Venkatraju and Marschner 1981; Romera et al. 1992; Enomoto et al. 2007).
In contrast with phloem Fe, it is unlikely that total Fe in the roots is involved in the repression of Fe-acquisition genes since some mutants, like the Arabidopsis frd3 mutant, show constitutive activation of Fe-responsive genes even though they accumulate high levels of Fe in their roots (Rogers and Guerinot 2002). The frd3 mutant, affected in xylem Fe transport (Durrett et al. 2007; Roschzttardtz et al. 2011), is chlorotic when grown under Fe-sufficient conditions (Rogers and Guerinot 2002), but it becomes green, and its Fe-acquisition genes are down-regulated, when its leaves are sprayed with Fe (Lucena et al. 2006). These results are again suggestive of the role of phloem Fe, specifically the phloem movement of Fe from shoot to root, in inhibiting the expression of the Fe-acquisition genes (Lucena et al. 2006; García et al. 2011).
Besides the Arabidopsis frd3 mutant, there are other mutants of Arabidopsis (opt3-2, nas4x-1), pea (dgl, brz), and tomato (chloronerva: chln), that show constitutive activation of Fe-acquisition genes when grown under Fe-sufficient conditions (Scholz et al. 1985; Grusak et al. 1990; Kneen et al. 1990; Grusak and Pezeghi 1996; Pich et al. 2001; Stacey et al. 2008; Klatte et al. 2009). The Arabidopsis mutant opt3-2 harbors a T-DNA insertion in the AtOPT3 promoter resulting in reduced AtOPT3 expression (Stacey et al. 2008). AtOPT3, the expression of which is enhanced under Fe deficiency, belongs to the oligopeptide transporter (OPT) family, involved in peptide transport (Stacey et al. 2006, 2008). This mutant is able to accumulate high levels of intracellular Fe in both shoots and roots as demonstrated by wild-type levels of chlorophyll and AtFER1 (ferritin) transcripts under Fe-sufficient conditions (Stacey et al. 2008). However, this mutant presents lower levels of Fe in seeds than wild-type plants. Since the movement of Fe to non-transpirating organs is believed to occur exclusively via phloem transport, it was suggested that the opt3-2 mutation is involved in the transport of a peptide Fe chelator via the phloem (Stacey et al. 2008). Like the Arabidopsis mutant opt3-2, the pea mutants dgl and brz accumulate high levels of Fe in roots and shoots when grown under Fe-sufficient conditions and do not show chlorosis, but symptoms of Fe toxicity (Grusak et al. 1990; Kneen et al. 1990; Grusak and Pezeghi 1996). The specific genes defined by the dgl and brz mutants have not been identified yet. However, in the case of the dgl mutant, it is known, by studies with reciprocal shoot:root grafts, that the constitutive expression of the Fe deficiency responses depends on the genotype of the shoot (Grusak and Pezeghi 1996). Moreover, Marentes et al. (1997) and Marentes and Grusak (1998) found that Fe was bound as part of a molecular complex (Fe-binding peptide) in the phloem of the wild-type pea, but that this complex was not formed in the phloem of the dgl mutant. These results suggest that the dgl mutant phenotype may also be related to defects in phloem Fe transport. The tomato mutant chln was identified as a spontaneous mutation in the cultivar Bonner Beste, later on related to a NAS gene (Ling et al. 1999). The chln mutant does not produce detectable amounts of NA and its constitutive expression of Fe deficiency responses is reduced upon exogenous application of NA (Pich et al. 2001). The nas4x-1 mutant is a quadruple mutant with mutations in the four NAS genes present in Arabidopsis and, consequently, produces very low levels of NA, similar to the tomato mutant chln (Klatte et al. 2009). Some years ago, it was suggested that Fe traveled in the phloem as a Fe–NA complex (Stephan and Scholz 1993). Later on, this idea was discarded, but NA is still thought to play a role in Fe loading and, probably, unloading of the phloem (Schmidke et al. 1999). The fact that all the above cited mutants accumulate high levels of Fe in their roots, when grown under Fe-sufficient conditions, suggests that total Fe in roots is not a key factor in the regulation of Fe-acquisition genes. On the other hand, the fact that most of these mutants (opt3-2, dgl, chln, nas4x-1), either directly or indirectly, are likely affected in the transport of Fe in the phloem suggests that phloem Fe is a key factor in Fe regulation.
The suggestion that the plant monitors phloem Fe to control iron homeostasis (Maas et al. 1988) is not sufficient, however, to fully explain the up-regulation of Fe-acquisition genes caused by ethylene, auxin, and NO in plants grown under low Fe conditions (Lucena et al. 2006; Graziano and Lamattina 2007; Waters et al. 2007; Chen et al. 2010; García et al. 2010, 2011; Bacaicoa et al. 2011; Romera et al. 2011; Wu et al. 2012). To address this issue, Lucena et al. (2006) proposed a model that implicates both phloem Fe and ethylene in the regulation of Fe-acquisition genes by Strategy I plants. Accordingly, ethylene, the level of which increases under Fe deficiency, would act as an activator of AtFIT (or SlFER) expression, and consequently of FRO and IRT1, while phloem Fe would act to repress their expression. Very recently, García et al. (2011) extended this model to more Fe-related genes and also included NO as an activator (already proposed by Graziano and Lamattina 2007) in conjunction with ethylene of Fe-acquisition genes.
One objective of this work was to study the effect of foliar application of Fe on the expression of Fe-acquisition genes in wild-type and mutant plants that constitutively express Fe-acquisition genes, namely the Arabidopsis frd3 and opt3-2 mutants, the pea dgl and brz mutants, and the tomato chln mutant. Another objective was to study the interaction between the foliar application of Fe and ethylene (and NO) on Fe-acquisition genes. Taken together, the results confirm that ethylene and NO activate the expression of Fe-acquisition genes, while Fe (probably as a Fe-related repressive signal coming from the shoot) inhibits it, consistent with earlier reports (Lucena et al. 2006; Graziano and Lamattina 2007; García et al. 2010, 2011).
Materials and methods
Plant material, growth conditions, and treatments
To analyze the effect of foliar application of Fe on the regulation of Fe-acquisition genes, we used wild-type Arabidopsis [Arabidopsis thaliana (L.) Heynh ecotype Columbia], pea (Pisum sativum L. cv Sparkle), and tomato (Solanum lycopersicon Mill. cv Bonner Beste) plants, and some of their mutants that show constitutive up-regulation of Fe-acquisition genes even when grown under Fe-sufficient conditions. Among these mutants, we used the Arabidopsis opt3-2 and frd3-3, the pea dgl (Sparkle [dgl,dgl]) and brz, and the tomato chloronerva (chln). For some studies, we also used wild-type cucumber (Cucumis sativus L. cv Ashley) plants. Arabidopsis, pea, tomato, and cucumber plants were grown on aerated nutrient solution as previously described (Lucena et al. 2006, 2007). When appropriate, plants were transferred to the different treatments.
The treatments imposed were as follows: +Fe: nutrient solution with Fe-EDDHA; +Fe + foliarFe: same as +Fe treatment, but with FeSO4 application to leaves; −Fe: nutrient solution without Fe; −Fe + foliarFe: same as −Fe treatment, but with FeSO4 application to leaves; −Fe + ACC: −Fe treatment with ACC addition during the last 24 h; −Fe + ACC + foliarFe: same as −Fe + ACC treatment, but with FeSO4 application to leaves; −Fe + GSNO: −Fe treatment with GSNO addition during the last 24 h; −Fe + GSNO + foliarFe: same as −Fe + GSNO treatment, but with FeSO4 application to leaves. FeSO4 was dissolved in deionized water (0.05 or 0.1 % w/v) and Tween 20 was added as surfactant. For treatments with FeSO4, leaves were sprayed once at day until total moistening. A stock solution of ACC (Sigma, St Louis, MO, USA) was prepared in deionized water. The stock solution of GSNO was prepared as in García et al. (2010). After treatments, root ferric reductase activity was determined as described previously (Lucena et al. 2006). Finally, the roots were collected and kept at −80 °C for subsequent analysis of mRNA levels.
RT-PCR analysis
Roots were ground to a fine powder in a mortar and pestle in liquid nitrogen. Total RNA was extracted using the Tri Reagent solution (Molecular Research Center, Inc. Cincinnati, OH, USA) according to the manufacturer’s instructions. M-MLV reverse transcriptase (Promega, Madison, WI, USA) was used to generate cDNA with 3 µg of total RNA from roots as the template and random hexamers as the primers. Negative controls included all reaction components except M-MLV enzyme. One tenth of each RT reaction was used as PCR template.
Primer pairs for Arabidopsis, tomato, and cucumber genes were designed as previously described (Lucena et al. 2006; Waters et al. 2007; García et al. 2010). Primer pairs for pea PsFRO1 and PsRIT1 were as follows: PsFRO1F (GCA AAA CAC CAA ACA TTG TTC); PsFRO1R (ACT ACC AGG TGA AAC TGA TTG); PsRIT1F (GAG ATC AAG AGA TGG GTG CT); and PsRIT1R (CAT CAA TAA CTT CAA GCC CA).
18S cDNA was amplified using QuantumRNA Universal 18S Standards primer set (Ambion, Austin, TX, USA) as the internal control. The thermal cycler program was one initial cycle of 94 °C, 5:00; followed by cycles of 94 °C, :45; 55 °C, :45; 72 °C, 1:00 with 30 cycles for all genes all followed by a final 72 °C elongation cycle of 7:00.
57Fe determination
In some experiments, 57FeSO4 (0.05 %, w/v) was sprayed onto shoots of different genotypes grown under Fe-sufficient conditions and, after two additional days, roots were harvested to analyze their 57Fe content as described by Rodriguez-Castrillón et al. (2008). For Fe determinations, samples were dried and digested with nitric acid in a microwave oven. Total Fe and 57Fe were determined using Inductively Coupled Plasma Mass Spectrometry (ICP-MS), model Agilent 7500c. The concentration of 57Fe in roots (coming from the 57FeSO4 applied to leaves) was calculated taking into account that approximately 2.2 % of the 57Fe in roots is due to its natural abundance.
Results
Effect of foliar application of Fe on ferric reductase activity and expression of Fe-related genes
As shown in Fig. 1, the application of FeSO4 to leaves of Fe-deficient Arabidopsis Columbia plants greatly decreased ferric reductase activity as well as the expression of AtFRO2, AtIRT1, and the bHLH transcriptional regulatory genes AtFIT1, AtbHLH038, and AtbHLH039. However, the FeSO4 treatment did not significantly affect either ferric reductase activity or gene expression in Fe-deficient Arabidopsis opt3-2 plants (Fig. 1). Similar to Arabidopsis Columbia plants, the application of FeSO4 to leaves of Fe-deficient pea Sparkle and tomato Bonner Beste plants greatly decreased their ferric reductase activity and the expression of Fe-acquisition genes (Figs. 2, 4). By contrast, the FeSO4 treatment did not decrease, but slightly increased both ferric reductase activity and expression of Fe-acquisition genes in Fe-sufficient pea dgl plants (Fig. 3). In the case of the brz and dgl mutants, we did the experiments with plants grown with low levels of Fe because it has been shown that these mutants present higher ferric reductase activity when grown under these conditions (Grusak et al. 1990; Grusak and Pezeshgi 1996).
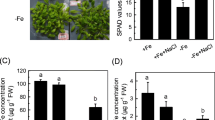
Effect of the foliar application of Fe on ferric reductase activity and expression of Fe-acquisition genes in Fe-deficient Arabidopsis Columbia and opt3-2 plants. Plants were grown in nutrient solution with 20 µM Fe and transferred to nutrient solution without Fe (−Fe). After 1 day, half of the −Fe plants were sprayed with FeSO4 (0.05 % w/v; −Fe + fol Fe). One day later, the ferric reductase activity was determined and total root RNA extracted. a Notice the color of the reduction assay in a Columbia (drastically inhibited) and an opt3-2 (not inhibited) plant treated with foliar Fe. b Ferric reductase activity (values are the mean ± SE of six replicates). c Expression of Fe-acquisition genes. RT-PCR was performed using total RNA from roots as template and gene-specific primers to amplify partial cDNAs of AtFRO2, AtIRT1, AtFIT, AtbHLH038, and AtbHLH039. 18S cDNA was amplified as positive control
Effect of the foliar application of Fe on ferric reductase activity and expression of Fe-acquisition genes in Fe-deficient pea Sparkle plants. Plants were grown in nutrient solution with 20 µM Fe (+Fe) and some of them transferred to nutrient solution without Fe (−Fe). On this and the following day, half of the −Fe plants were sprayed with FeSO4 (0.1 % w/v; −Fe + fol Fe). One day later, the ferric reductase activity was determined and total root RNA extracted. a Ferric reductase activity (values are the mean ± SE of six replicates). b Expression of Fe-acquisition genes. RT-PCR was performed using total RNA from roots as template and gene-specific primers to amplify partial cDNAs of PsFRO1 and PsRIT1. 18S cDNA was amplified as positive control
Effect of the foliar application of Fe on ferric reductase activity and expression of Fe-acquisition genes in Fe-sufficient pea brz and dgl plants. Plants were grown in nutrient solution with 3 µM Fe (+Fe). Half of the +Fe plants were sprayed with FeSO4 (0.1 % w/v) during 2 days (+Fe + fol Fe). One day later, ferric reductase activity and expression of Fe-acquisition genes were determined as in Fig. 2. a Notice the lower intensity of the red color, corresponding to a lower reductase activity, in the reduction assay of the brz plant treated with foliar Fe and the inverse in the dgl plant not inhibited by this treatment. b Ferric reductase activity. c Expression of Fe-acquisition genes
Taken together, the above results clearly show that the application of Fe to leaves of Fe-deficient wild-type cultivars greatly decreased both ferric reductase activity and the expression of Fe-acquisition genes. In the same way, although less drastically, foliar Fe application also inhibited the ferric reductase activity and the expression of Fe-acquisition genes in the pea brz mutant and in the tomato chln mutant (Figs. 3, 4). However, the application of foliar Fe to the opt3-2 and dgl mutants, which are presumably affected in the transport of a Fe-related repressive signal in the phloem, did not inhibit Fe responses. In comparison with the “phloem Fe” mutants opt3-2 and dgl, we also examined the Arabidopsis frd3 mutant impaired in xylem Fe transport (Durrett et al. 2007; Roschzttardtz et al. 2011). When FeSO4 was applied to leaves of the frd3 mutant (either grown with Fe or without Fe), both the ferric reductase activity and the expression of Fe-acquisition genes were greatly decreased (Fig. 5) as occurred in the wild-type cultivar Columbia (Fig. 1), confirming previous results (Lucena et al. 2006). In addition to the above Fe-acquisition genes, we also found that foliar Fe application to Columbia and frd3 plants also inhibited the expression of other Fe-related genes (AtFRD3, AtNAS1, AtNAS2, AtCCCL1, AtCCCL2, AtMYB72, At2OGFe; Fig. 6), identified as ethylene (NO)-responsive genes by García et al. (2010). This is in contrast to opt3-2 plants, where the expression of all these genes remained constitutively high (Fig. 6). Since the three bHLH transcription factors (AtFIT1, AtbHLH038, and AtbHLH039) are key regulators of Fe-acquisition genes (Yuan et al. 2008), these results suggest that the tight control of Fe-acquisition responses is abolished in the opt3-2 mutant and that the expression of many Fe-related genes are de-regulated in this mutant (Fig. 6).
Effect of the foliar application of Fe on ferric reductase activity and expression of Fe-acquisition genes in Fe-deficient tomato Bonner Beste and chloronerva plants. Plants were grown in nutrient solution with 20 µM Fe (+Fe) and some of them transferred to nutrient solution without Fe (−Fe). On this and the following day, half of the −Fe plants were sprayed with FeSO4 (0.1 % w/v; −Fe + fol Fe). One day later, the ferric reductase activity was determined and total root RNA extracted. a Ferric reductase activity (values are the mean ± SE of six replicates). b Expression of Fe-acquisition genes. RT-PCR was performed using total RNA from roots as template and gene-specific primers to amplify partial cDNAs of SlFRO1, SlIRT1, and SlFER. 18S cDNA was amplified as positive control
Effect of the foliar application of Fe on ferric reductase activity and expression of Fe-acquisition genes in Fe-deficient and Fe-sufficient Arabidopsis frd3-3 plants. Plants were grown in nutrient solution with 20 µM Fe (+Fe) and half of them transferred to nutrient solution without Fe (−Fe). After 1 day, half of the –Fe and +Fe plants were sprayed with FeSO4 (0.05 % w/v; −Fe + fol Fe; +Fe + fol Fe). One day later, ferric reductase activity and expression of Fe-acquisition genes were determined as in Fig. 1. a Notice the lack of red color in the reduction assay of the frd3-3 plant treated with foliar Fe, corresponding to inhibition of the reductase activity. b Ferric reductase activity. c Expression of Fe-acquisition genes
Effect of the foliar application of Fe on the expression of Fe-related genes in Fe-deficient Arabidopsis Columbia, frd3-3, and opt3-2 plants. Experimental conditions as in Fig. 1. RT-PCR was performed using total RNA from roots as template and gene-specific primers to amplify partial cDNAs of AtFRD3, AtNAS1, AtNAS2, AtCCCL1, AtCCCL2, AtMYB72, and At2OGFe. 18S cDNA was amplified as positive control
To test whether the inhibitory effect of foliar-applied Fe was related to Fe moving from leaves to roots, we used 57Fe to check the movement of Fe in the different wild-type and mutant genotypes. All the genotypes treated with foliar 57Fe showed accumulation of this isotope in roots higher than the 2.2 % expected from natural abundance (data not shown), which means that 57Fe moved from leaves to roots in all cases. The concentration of 57Fe in roots (after subtracting the 2.2 % expected from natural abundance) was similar in the different mutants when compared to their respective wild-type cultivars (Table 1).
Effect of the interaction of foliar-applied Fe with ethylene and NO on ferric reductase activity and expression of Fe-acquisition genes
Besides ethylene, NO has also been implicated in the up-regulation of Fe-acquisition genes (Graziano and Lamattina 2007; Chen et al. 2010; García et al. 2010, 2011; Ramírez et al. 2011; Romera et al. 2011). Both ethylene and NO up-regulate the expression of Fe-acquisition genes in plants grown with low levels of Fe (or without Fe), but barely in those grown with high levels of Fe (Lucena et al. 2006; Graziano and Lamattina 2007; Chen et al. 2010; García et al. 2011). To examine this further, we studied the expression of Fe-acquisition genes in Fe-deficient Arabidopsis and cucumber plants treated with either ACC (ethylene precursor) or GSNO (NO donor) and with foliar application of Fe. The results showed that either ACC or GSNO up-regulated the expression of Fe-acquisition genes (and ferric reductase activity) when applied to plants growing without Fe (Fig. 7; it should be noted that plants were grown without Fe only for 24 h to avoid the induction of genes by the Fe deficiency itself). However, neither ACC nor GSNO up-regulated the expression of Fe-acquisition genes (and ferric reductase activity) when applied simultaneously with foliar Fe (Fig. 7). Similar patterns of expression were observed for the Fe-responsive genes CsFRO1, CsIRT1, and CsHA1 in cucumber (Fig. 8). However, CsHA2, the expression of which is independent of Fe availability (Santi et al. 2005), was not affected by ethylene, NO, or foliar Fe treatment (Fig. 8), which suggests that these treatments are rather specific to Fe-responsive genes.
Effect of the interaction between the foliar application of Fe and the treatments with either ACC or GSNO on ferric reductase activity and expression of Fe-acquisition genes in Fe-deficient Arabidopsis Columbia plants. Plants were grown in nutrient solution with 20 µM Fe (+Fe) and some of them transferred to nutrient solution without Fe (−Fe). Some of the −Fe plants were treated with either GSNO 100 µM (−Fe + GSNO) or ACC 1 µM (−Fe + ACC) and half of the −Fe + GSNO and −Fe + ACC plants were sprayed with FeSO4 (0.05 % w/v; −Fe + GSNO + folFe and −Fe + ACC + folFe). One day later, ferric reductase activity and expression of Fe-acquisition genes were determined as in Fig. 1. a Ferric reductase activity. b Expression of Fe-acquisition genes
Effect of the interaction between the foliar application of Fe and the treatments with either ACC or GSNO on ferric reductase activity and expression of Fe-acquisition genes in Fe-deficient cucumber plants. Plants were grown in nutrient solution with 20 µM Fe (+Fe) and some of them transferred to nutrient solution without Fe (−Fe). Some of the −Fe plants were treated with either GSNO 100 µM (−Fe + GSNO) or ACC 1 µM (−Fe + ACC) and half of the −Fe + GSNO and −Fe + ACC plants were sprayed with FeSO4 (0.1 % w/v; −Fe + GSNO + folFe and −Fe + ACC + folFe). One day later, the ferric reductase activity was determined and total root RNA extracted. a Ferric reductase activity (values are the mean ± SE of six replicates). b Expression of Fe-acquisition genes. RT-PCR was performed using total RNA from roots as template and gene-specific primers to amplify partial cDNAs of CsFRO1, CsIRT1, CsHA1, and CsHA2. 18S cDNA was amplified as positive control
Discussion
Although detached roots can respond to Fe deficiency (Bienfait et al. 1987; Enomoto et al. 2007), most published work suggests a key role for the aerial part of the plant in the regulation of Fe deficiency responses (Landsberg 1984; Maas et al. 1988; Grusak and Pezeghi 1996; Li et al. 2000; Lucena et al. 2006; Enomoto et al. 2007; Bacaicoa et al. 2011; García et al. 2011; Wu et al. 2012). A model suggests that shoots suffering from Fe deficiency send promotive signals to the roots, leading to the induction of Fe deficiency responses (Landsberg 1984; Grusak and Pezeghi 1996; Li et al. 2000; Enomoto et al. 2007; Bacaicoa et al. 2011; Wu et al. 2012). Among the promotive signals, several results support a role for auxin, ethylene, and NO, either coming from the shoots or produced by the roots (Landsberg 1984; Li et al. 2000; Han et al. 2005; Lucena et al. 2006; Graziano and Lamattina 2007; Waters et al. 2007; Chen et al. 2010; García et al. 2010, 2011; Bacaicoa et al. 2011; Romera et al. 2011; Wu et al. 2012). It should be noted that the production and regulation of auxin, ethylene, and NO are tightly interrelated (Chen et al. 2010; García et al. 2011; Romera et al. 2011). A competing model suggests that Fe-sufficient shoots send repressive signals (phloem Fe) to the roots, which repress the Fe deficiency responses, with this repression released under Fe deficiency (Maas et al. 1988).
Some years ago, Lucena et al. (2006) proposed a model which integrates both promotive and repressive signals in the regulation of Fe-acquisition genes in the roots. According to that model, shoots can send both promotive (ethylene or substances that promote its synthesis, like auxin or ACC) and repressive signals (related to phloem Fe) to the roots. The root can subsequently also produce and amplify the promotive signals, integrate both kind of signals, and “decide” whether to induce or repress Fe-acquisition genes (Lucena et al. 2006). This model is supported by the fact that exogenous application of auxin, ethylene, or NO can up-regulate the expression of Fe-acquisition genes in plants grown under low levels of Fe (or without Fe), but has little effect on plants grown under high levels of Fe, presumably due to the presence of Fe-related repressive signals under these conditions (Lucena et al. 2006; Graziano and Lamattina 2007; Chen et al. 2010; García et al. 2011). The question arises, therefore, as to whether Fe itself, when present in sufficient levels, acts as a repressive signal. Lucena et al. (2006) proposed phloem Fe (or some signal derived from it), rather than total Fe in the root, as the repressive signal of Fe-acquisition genes. Their proposal was based on the fact that there are mutants, like the Arabidopsis frd3 mutant, that show constitutive activation of Fe-acquisition genes when grown under Fe-sufficient conditions, despite the accumulation of high levels of Fe in their roots. The same occurs with the Arabidopsis opt3-2 and nas mutants, the tomato chln mutant, and the pea brz and dgl mutants. The fact that all the above cited mutants accumulate high levels of Fe in their roots, when grown under Fe-sufficient conditions, suggests that total Fe in roots is not a key factor in the regulation of Fe-acquisition genes. On the other hand, the fact that most of these mutants (opt3-2, dgl, chln, nas4x-1), either directly or indirectly, are likely affected in the transport of Fe in the phloem suggests that phloem Fe is a key factor in Fe regulation.
To test the possibility that the repressive signal could be related to Fe recirculating back from leaves to root, as proposed by Maas et al. (1988), we compared the effect of the foliar application of Fe on ferric reductase activity and gene expression in wild-type and mutant plants that constitutively express Fe-acquisition (and other Fe-related) genes. As shown in Figs. 1, 2, 4, and 6, the application of Fe to leaves of Fe-deficient Arabidopsis, pea, and tomato wild-type plants drastically decreased both ferric reductase activity and expression of Fe-acquisition (and other Fe-related; see Introduction) genes, which confirms previous published results (Venkatraju and Marschner 1981; Maas et al. 1988; Romera et al. 1992; Enomoto et al. 2007). The same occurred when Fe was applied to leaves of the pea brz mutant (Fig. 3), the tomato chln mutant (Fig. 4), and the Arabidopsis frd3 mutant (Figs. 5, 6; Lucena et al. 2006). However, when Fe was applied to leaves of the Arabidopsis opt3-2 mutant (Figs. 1, 6) and the pea dgl mutant (Fig. 3), ferric reductase activity and the expression of all genes studied remained high. These results clearly indicate that opt3-2 and dgl plants lack the ability to repress the expression of Fe-acquisition (and other Fe-related) genes in roots upon foliar Fe application. Curiously, both mutants, opt3-2 and dgl, are presumably affected in the transport of a Fe-related repressive signal (Fe-peptide?) in the phloem (Marentes et al. 1997; Marentes and Grusak 1998; Stacey et al. 2008). AtOPT3 belongs to the OPT family related to peptide transport and has been involved in the transport of a peptide Fe chelator via the phloem (Stacey et al. 2006, 2008). In the case of the dgl mutant, although the mutation has not been identified yet, Marentes et al. (1997) and Marentes and Grusak (1998) found a probable Fe-binding peptide in the phloem of the wild-type pea that was not present in the phloem of the dgl mutant.
Like opt3-2 and dgl, the pea brz mutant and the tomato chln mutant also present constitutive Fe responses and accumulate very high amounts of Fe in their shoots when grown under Fe-sufficient conditions (Scholz et al. 1985; Kneen et al. 1990). However, these mutants, by contrast to opt3-2 and dgl, can still respond to foliar Fe application (Figs. 3, 4). Why do they respond to exogenous applied Fe and not respond to the internal Fe accumulated in their shoots when grown under Fe-sufficient conditions? A possible explanation for this is that perhaps both brz and chln mutants fail in the rate by which the probable Fe-related repressive signal is loaded (or unloaded) into (or out of) phloem; upon foliar Fe application, this rate can increase and phloem can attain the sufficient concentration of Fe-related repressive signal to repress Fe responses. Although the brz mutation has not been identified yet, this mutant was less drastically inhibited than the wild-type cultivar Sparkle upon foliar Fe application (Figs. 2, 3). Similarly, the chln mutant was less drastically inhibited than the wild-type cultivar Bonner Beste (Fig. 4). Although NA has been involved in Fe loading (Schmidke et al. 1999) and the chln mutation is related to a NAS gene (Ling et al. 1999), it is probable that the residual NA produced by other NAS genes (in Arabidopsis, there have been identified 4 NAS genes; Klatte et al. 2009) could be enough for the loading (or unloading) of a sufficient Fe-related repressive signal upon foliar Fe application. Very recently, it has been found that nas mutant, also related to NA as chln, can accumulate Fe in phloem, but there are problems in unloading it out of the phloem (Schuler et al. 2012). In the case of the Arabidopsis frd3 mutant (Figs. 5, 6), the loading of a Fe-related repressive signal into the phloem would not be a problem because this mutant is only affected in xylem Fe transport (Durrett et al. 2007; Roschzttardtz et al. 2011).
Since some of the mutants used in this work (opt3-2, dgl, chln) are probably affected in the transport of Fe in the phloem, the question arises as to whether the Fe-related repressive signal is the whole Fe moving in the phloem or not. To further clarify this, we applied 57Fe to leaves of opt3-2, dgl, brz, and chln mutants and found that all of them accumulated similar levels of 57Fe in their roots than their respective wild-type cultivars (Table 1). Since no statistical significant differences between the values of Fe translocation between mutants and WT were found, it is obvious that Fe applied to leaves achieves the roots in all mutants. This result emphasizes that whole Fe in the phloem is not the inhibitor of Fe responses, and suggests that the mutants might be impaired in the transport of a specific Fe compound (or a Fe-related signal). This opens the way to two possibilities: (i) foliar-applied Fe generates specific repressive signal(s) or (ii) foliar-applied Fe restricts the sending of promotive signal(s) to the roots. This second possibility is difficult to assume for at least two reasons. First, if OPT3 were able to transport a promotive signal, then the opt3-2 mutant, with lower abundance of this transporter (Stacey et al. 2008), should transport much less of this promotive signal and the Fe responses would be abolished more drastically than in the wild-type Columbia upon foliar Fe application, but this does not occur (Figs. 1, 6). Second, the results presented in Figs. 7 and 8 show that the addition of known promotive signals (ethylene, NO) to Fe-deficient plants greatly enhanced both ferric reductase activity and expression of Fe-acquisition genes when foliar Fe was not applied, but not when it was. These results suggest that Fe-sprayed leaves send repressive signal(s) to roots that counteract the promotive effects of ethylene and NO. The first possibility suggesting that foliar-applied Fe generates specific repressive signal(s) agrees with the fact that opt3-2 and dgl are presumably affected in the transport of a Fe-peptide in the phloem (Marentes et al. 1997; Marentes and Grusak 1998; Stacey et al. 2008), which could be the actual repressive signal. It is also possible that the repressive signal could be a non-peptide compound or a Fe-unrelated compound (i.e., a hormone, a small RNA,…). In any case, the repressive signal should be transported through the OPT3 transporter; if not, it would enter into (or unload out of) the phloem of the opt3-2 mutant and should repress Fe responses.
In conclusion, the results obtained in this work suggest that Fe-sprayed leaves send repressive signal(s) to roots and imply that ethylene and NO by themselves are not sufficient to induce Fe-acquisition genes until these repressing signal(s) are released. These results support the model proposed by our group considering both promotive (auxin, ethylene, NO) and repressive (Fe-related compound in phloem) signals in the regulation of Fe-acquisition genes (Lucena et al. 2006; García et al. 2011; Romera et al. 2011). The combinatorial control of Fe responses by promotive (ethylene, NO,..) and repressive (Fe-related compounds in phloem) signals would confer Fe specificity to the system, avoiding the induction of Fe responses by increases of the promotive signals originated by other causes besides Fe deficiency. The existence of nutrient-specific signals acting in conjunction with ethylene is logical since this hormone has also been involved in the responses to other nutrient deficiencies, such as K and P deprivation (Jung et al. 2009; Lei et al. 2011). As an example, ACC up-regulated P-acquisition genes in P-deficient plants, but had almost no effect in the P-sufficient ones (Lei et al. 2011). On the other hand, the existence of Fe-related repressive signal(s) moving from leaves to roots is a way for shoots to communicate their Fe status to roots. Evidence for shoot to root communication also exists for the regulation of the uptake of other mineral nutrients, such as N and S, where phloem-transported aminoacids (N) and glutathione (S) appear to regulate their uptake (Liu et al. 2009). Recently, small RNAs have been implicated in the long-distance communication via phloem between shoots and roots following nutrient deprivation (Buhtz et al. 2010).
Without discarding a role for small RNAs and other compounds in the shoot to root communication of Fe deprivation, the results found with the opt3-2 (Figs. 1, 6) and dgl (Fig. 3) mutants suggest a role for a Fe-peptide moving in the phloem in this process. The existence of a Fe-peptide acting as a long-distance repressive signal of Fe responses should not be considered extraordinary. Peptides in plants play crucial roles in biotic and abiotic stress responses (Germain et al. 2006) and in animals, the peptide hepcidin is a key regulator of iron homeostasis (Clark et al. 2011). In plants, it is tempting to speculate that the probable Fe-peptide moving in the phloem could act by interfering with ethylene (and NO) synthesis and signaling (Figs. 7, 8). In this respect, it should be mentioned that the flagellin-derived flg22 peptide regulates the release of an ethylene response factor in the ethylene signaling of Arabidopsis (Bethke et al. 2009). Consequently, it is possible that other peptides (such as a probable Fe-peptide) could also interact with this signaling pathway. In supporting this view, it should be mentioned that all the Fe-related genes inhibited by foliar Fe application in Arabidopsis (Figs. 1, 6) have also been identified as iron-deficiency ethylene-dependent genes (García et al. 2010). Future research should be focused on the identification of this probable Fe-binding peptide (i.e., the unidentified substrate of the OPT3 transporter; Lubkowitz 2011) and, if found, on its possible relationship with ethylene synthesis and signaling.
Abbreviations
- ACC:
-
1-Aminocyclopropane-1-carboxylic acid
- EDDHA:
-
N,N′-ethylenebis[2-(2-hydroxyphenyl)-glycine]
- Ferrozine:
-
3-(2-Pyridyl)-5,6-bis(4-phenyl-sulfonic acid)-1,2,4-triazine
- NA:
-
Nicotianamine
- GSNO:
-
S-nitrosoglutathione
References
Bacaicoa E, Mora V, Zamarreño AM, Fuentes M, Casanova E, García-Mina JM (2011) Auxin: a major player in the shoot-to-root regulation of root Fe-stress physiological responses to Fe deficiency in cucumber plants. Plant Physiol Biochem 49:545–556
Bethke G, Unthan T, Uhrig JF, Pöschl Y, Gust AA, Scheel D, Lee J (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106:8067–8072
Bienfait HF, De Weger LA, Kramer D (1987) Control of development of iron-efficiency reactions in potato as a response to iron deficiency is located in the roots. Plant Physiol 83:244–247
Brumbarova T, Bauer P (2005) Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol 137:1018–1026
Buhtz A, Pieritz J, Springer F, Kehr J (2010) Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol 10:64
Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ (2010) Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis thaliana. Plant Physiol 154:810–819
Clark RJ, Tan CC, Preza GC, Nemeth E, Ganz T, Craik DJ (2011) Understanding the structure/activity relationships of the iron regulatory peptide hepcidin. Chem Biol 18:336–343
Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144:197–205
Enomoto Y, Hodoshima H, Shimada H, Shoji K, Yoshihara T, Goto F (2007) Long-distance signals positively regulate the expression of iron uptake genes in tobacco roots. Planta 227:81–89
García MJ, Lucena C, Romera FJ, Alcántara E, Pérez-Vicente R (2010) Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J Exp Bot 61:3885–3899
García MJ, Suárez V, Romera FJ, Alcántara E, Pérez-Vicente R (2011) A new model involving ethylene, nitric oxide and Fe to explain the regulation of Fe-acquisition genes in Strategy I plants. Plant Physiol Biochem 49:537–544
Germain H, Chevalier E, Matton DP (2006) Plant bioactive peptides: an expanding class of signaling molecules. Can J Bot 84:1–19
Graziano M, Lamattina L (2007) Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant J 52:949–960
Grusak MA, Pezeshgi S (1996) Shoot-to-root signal transmission regulates root Fe(II) reductase activity in the dgl mutant of pea. Plant Physiol 110:329–334
Grusak MA, Welch RM, Kochian LV (1990) Physiological characterization of a single-gene mutant of Pisum sativum exhibiting excess iron accumulation. I. Root iron reduction and iron uptake. Plant Physiol 93:976–981
Han ZH, Han CQ, Xu XF, Wang Q (2005) Relationship between iron deficiency stress and endogenous hormones in iron-efficient versus inefficient apple genotypes. J Plant Nutr 28:1887–1895
Ivanov R, Brumbarova T, Bauer P (2011) Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol Plant 5:27–42
Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21:607–621
Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol 150:257–271
Kneen BE, LaRue T, Welch RM, Weeden NF (1990) Pleiotropic effects of brz. A mutation in Pisum sativum (L.) cv ‘Sparkle’ conditioning decreased nodulation and increased iron uptake and leaf necrosis. Plant Physiol 93:717–722
Landsberg EC (1984) Regulation of iron-stress-response by whole plant activity. J Plant Nutr 7:609–621
Lei M, Zhu C, Liu Y, Karthikeyan AS, Bressan RA, Raghothama KG, Liu D (2011) Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol 189:1084–1095
Li C, Zhu X, Zhang F (2000) Role of shoot in regulation of iron deficiency responses in cucumber and bean plants. J Plant Nutr 23:1809–1818
Ling HQ, Koch G, Baumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher-plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96:7098–7103
Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P (2011) Interaction between the bHLH transcription factor FIT and the ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23:1815–1829
Liu TY, Chang CY, Chiou TJ (2009) The long-distance signaling of mineral macronutrients. Curr Opin Plant Biol 12:312–319
Lubkowitz M (2011) The oligopeptide transporters: a small gene family with a diverse group of substrates and functions? Mol Plant 4:407–415
Lucena C, Waters BM, Romera FJ, García MJ, Morales M, Alcántara E, Pérez-Vicente R (2006) Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J Exp Bot 57:4145–4154
Lucena C, Romera FJ, Rojas CL, García MJ, Alcántara E, Pérez-Vicente R (2007) Bicarbonate blocks the expression of several genes involved in the physiological responses to Fe deficiency of Strategy I plants. Funct Plant Biol 34:1002–1009
Maas FM, van de Wetering DAM, van Beusichem Ml, Bienfait HF (1988) Characterization of phloem iron and its possible role in the regulation of Fe-efficiency reactions. Plant Physiol 87:167–171
Marentes E, Grusak MA (1998) Mass determination of low-molecular-weight proteins in phloem sap using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. J Exp Bot 49:903–911
Marentes E, Stephens BW, Grusak MA (1997) Characterization of a phloem mobile chelator involved in the phloem transport of iron from vegetative tissues to developing seeds of pea. Abstract 9th International Symposium on Iron Nutrition and Interactions in Plants, Stuttgart/Germany, p 74
Meiser J, Lingam S, Bauer P (2011) Post-translational regulation of the Fe deficiency bHLH transcription factor FIT is affected by iron and nitric oxide. Plant Physiol 157:2154–2166
Pich A, Manteuffel R, Hillmer S, Scholz G, Schmidt W (2001) Fe homeostasis in plant cells: Does nicotianamine play multiple roles in the regulation of cytoplasmic Fe concentration? Planta 213:967–976
Ramírez L, Simontacchi M, Murgia I, Zabaleta E, Lamattina L (2011) Nitric oxide, nitrosyl iron complexes, ferritin and frataxin: a well equipped team to preserve plant iron homeostasis. Plant Sci 181:582–592
Rodriguez-Castrillón JA, Moldovan M, García JI, Lucena JJ, García-Tomé ML, Hernández-Apaolaza L (2008) Isotope pattern deconvolution as a tool to study iron metabolism in plants. Anal Bioanal Chem 390:579–590
Rogers EE, Guerinot ML (2002) FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14:1787–1799
Romera FJ, Alcántara E, De la Guardia MD (1992) Role of roots and shoots in the regulation of the Fe efficiency responses in sunflower and cucumber. Physiol Plant 85:141–146
Romera FJ, García MJ, Alcántara E, Pérez-Vicente R (2011) Latest findings about the interplay of auxin, ethylene and nitric oxide in the regulation of Fe deficiency responses by Strategy I plants. Plant Signal Behav 6:167–170
Roschzttardtz H, Séguéla-Arnaud M, Briat JF, Vert G, Curie C (2011) The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. Plant Cell 23:2725–2737
Santi S, Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183:1072–1084
Santi S, Cesco S, Varanini Z, Pinton R (2005) Two plasma membrane H+-ATPase genes are differentially expressed in iron-deficient cucumber plants. Plant Physiol Biochem 43:287–292
Schmidke I, Krüger C, Frömmichen R, Scholz G, Stephan UW (1999) Phloem loading and transport characteristics of iron in interaction with plant-endogenous ligands in castor bean seedlings. Physiol Plant 106:82–89
Scholz G, Schlesier G, Seifert K (1985) Effect of nicotianamine on iron uptake by the tomato mutant ‘chloronerva’. Physiol Plant 63:99–104
Schuler M, Rellán-Álvarez R, Fink-Straube C, Abadía J, Bauer P (2012) Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24:2380–2400
Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G (2006) Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta 223:291–305
Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146:589–601
Stephan UW, Scholz G (1993) Nicotianamine: mediator of transport of iron and heavy metals in the phloem? Physiol Plant 88:522–529
Venkatraju K, Marschner H (1981) Inhibition of iron-stress reactions in sunflower by bicarbonate. Z Pflanzenernähr Bodenkd 144:339–355
Walker EL, Connolly EL (2008) Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol 11:530–535
Waters BM, Lucena C, Romera FJ, Jester GG, Wynn AN, Rojas CL, Alcántara E, Pérez-Vicente R (2007) Ethylene involvement in the regulation of the H+-ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiol Biochem 45:293–301
Wu T, Zhang HT, Wang Y, Jia WS, Xu XF, Zhang XZ, Han ZH (2012) Induction of root Fe(III) reductase activity and proton extrusion by iron deficiency is mediated by auxin-based systemic signalling in Malus xiaojinensis. J Exp Bot 63:859–870
Yuan YX, Wu HL, Wang N, Li J, Zhao WN, Du J, Wang DW, Ling HQ (2008) FIT interacts with AtbHLH038 and AtbHLH039 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18:385–397
Acknowledgments
We thank Dr Michael Grusak (USDA/ARS, Houston, Texas, USA), Dr Ross Welch (USDA/ARS, Ithaca, New York, USA), and Dr Petra Bauer (Saarland University, Saarbrücken, Germany) for kindly providing seeds of the Sparkle [dgl,dgl] mutant, the brz mutant, and the chloronerva mutant. We also thank the Arabidopsis Biological Resource Center (Ohio State University, USA) for providing seeds of the frd3-3 mutant. This work was supported by the European Regional Development Fund from the European Union, the “Ministerio de Educación y Ciencia” (Projects AGL2007-64372 and AGL2010-17121), and the “Junta de Andalucía” (Research Groups AGR115 and BIO159, and Project AGR-3849). Funding to MGS and GS was provided by a Grant from the USA National Science Foundation Plant Genome Program (Award #0820769).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
García, M.J., Romera, F.J., Stacey, M.G. et al. Shoot to root communication is necessary to control the expression of iron-acquisition genes in Strategy I plants. Planta 237, 65–75 (2013). https://doi.org/10.1007/s00425-012-1757-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1757-0