Abstract
Binding protein (BiP) is a chaperone protein involved in the folding of secretory proteins in the ER lumen. OsBiP1 is constitutively expressed in various tissues, whereas the expression of OsBiP4 and OsBiP5 (OsBiP4&5) is not detected in any tissue under normal conditions. However, expression of OsBiP4&5 was highly and specifically activated under ER stress conditions induced by DTT treatment, OsBiP1 knockdown, OsBiP1 overexpression, OsIRE1 overexpression, or various exogenous recombinant proteins in transgenic rice. In contrast, OsBiP4&5 did not accumulate in OsIRE1 knockdown transgenic rice even after DTT treatment. When the subcellular localization of OsBiP4&5 was investigated in seed endosperm cells under the ER stress condition, OsBiP4&5 were localized to the ER, but did not participate in ER-derived protein body (PB-I) formation in a different manner to OsBiP1. These results indicate that OsBiP4&5 levels were positively correlated with stress levels in the ER. Taken together, these results suggest that OsBiP4&5 are ER stress-related BiP proteins that are regulated by OsIRE1/OsbZIP50 pathway and that they may have a distinct function from that of OsBiP1 in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Our previous work generated transgenic rice plants that accumulate health-promoting proteins or peptides in their seeds, which are beneficial for human diets (Takagi et al. 2005; Yasuda et al. 2006a; Wakasa et al. 2011a, b). To create these transgenic plants, it was important to maximize the accumulation of desired recombinant proteins in ER-derived protein bodies through the secretory pathway. However, when certain recombinant proteins or peptides accumulate to high levels in rice endosperm tissue, an aberrant grain phenotype with floury and shrunken features has sometimes been observed (Oono et al. 2010). We showed that this decrease in grain quality correlated with an ER stress response (Oono et al. 2010). The ER stress response is a mechanism to maintain ER homeostasis by balancing the folding capacity and the folding demand imposed on the ER. In yeast and mammal cells, the ER stress response is activated to relieve the load of unfolded protein in the ER lumen and to recover homeostasis, thus the ER stress response is also referred to as the “unfolded protein response” (UPR). At least three distinct intracellular signal transduction pathways are activated by the ER stress response (Chakrabarti et al. 2011). It is thought that the ER stress response also regulates the folding and removal of misfolded or unfolded proteins by the ER-associated degradation (ERAD) pathway, the inhibition of translation, and apoptosis (Harding et al. 1999; Bertolotti et al. 2000; Okamura et al. 2000; Oyadomari and Mori 2004; Yamaguchi et al. 2008; Chakrabarti et al. 2011).
Avoidance or alleviation of ER stress in transgenic rice grains is expected to lead to higher accumulation of recombinant proteins without a decrease in seed quality. Therefore, understanding the molecular mechanisms underlying the ER stress response will enable optimum production of beneficial recombinant proteins in rice. However, most studies on ER stress or protein quality control mechanisms have been done in yeast and mammals. To control ER stress in rice, a system to monitor the ER stress level with easily identifiable markers in various tissues (especially seed tissue) is required.
BiP is one of the main ER chaperones and belongs to the heat shock protein 70 (HSP70) family. BiP has a signal peptide at the N terminus and an ER retention signal (i.e., His/Lys-Asp-Glu-Leu) at the C terminus. BiP interacts with nascent immature proteins, which are synthesised on polysomes attached to the ER, and is involved in folding and assembling the newly synthesised polypeptides within the ER lumen. Human and rodents BiP (GRP78) or yeast BiP (KAR2) is encoded by a single gene (Fu et al. 2008; Kimata and Kohno 2010, Rose et al. 1989; Ting and Lee 1988; Wooden et al. 1988). In contrast, at least five OsBiP genes [Os02g0115900 (OsBiP1), Os03g0710500 (OsBiP2), Os05g0367800 (OsBiP3), Os05g0428600 (OsBiP4) and Os08g0197700 (OsBiP5)] have been identified in rice (Hayashi et al. 2012). Their amino acid sequences share 66–94 % identity to each other (Fig. S1). OsBiP1, one of the major rice BiP proteins, is constitutively expressed in plant tissues and is up-regulated in response to ER stress (Oono et al. 2010; Wakasa et al. 2011c; Hayashi et al. 2012). Overexpression and knock down of OsBiP1 in rice endosperm induces a severe ER stress response and leads to a deterioration of grain properties (Yasuda et al. 2009; Wakasa et al. 2011c). Reduction of OsBiP1 also resulted in the induction of OsBiP2, OsBiP3, OsBiP4, and OsBiP5, which were characterised as novel BiP proteins associated with the ER stress response. Furthermore, expression of these novel BiPs was regulated by OsIRE1-mediated mRNA splicing of the OsbZIP50 transcriptional factor (OsIRE1/OsbZIP50) in a manner similar to the signalling pathways IRE1/HAC1 in yeast and IRE1/XBP1 in mammals (Hayashi et al. 2012).
OsbZIP50 is activated by OsIRE1-mediated cytoplasmic splicing of its mRNA. Protein translated from unspliced OsbZIP50 mRNA is not translocated into the nucleus. Under ER stress conditions, unconventional splicing of OsbZIP50 mRNA by OsIRE1 causes a frameshift in the C-terminal region of OsbZIP50, which results in the appearance of a nuclear localization signal in the newly translated OsbZIP50 (Hayashi et al. 2012). The Arabidopsis AtbZIP60 is also activated by AtIRE1-mediated splicing, similar to the activation of rice OsbZIP50 (Deng et al. 2011; Nagashima et al. 2011).
In this study, in order to address the relationship between the OsIRE1/OsbZIP50 signalling and BiP proteins induced under ER stress conditions, we prepared anti-OsBiP4&5 polyclonal antibody, and demonstrated the potential utility of anti-OsBiP4&5 antibody as a marker to evaluate ER stress levels in several rice tissues. Furthermore, based on expression analysis of OsBiP4 and OsBiP5 in some rice tissues under various ER stress situations using the anti-OsBiP4&5 antibody, the functions of ER stress specific-OsBiPs in the ER stress response are discussed.
Materials and methods
Plant materials
Callus, root, and seed tissues from rice (Oryza sativa L. cv. Kitaake or cv. Koshihikari) were used as samples. Seeds were obtained from National Institute of Agrobiological Sciences, Tsukuba, Ibaraki, Japan. Transgenic rice lines included the following: constitutive OsIRE1-knock down (KD), constitutive OsIRE1-overexpression (OE), endosperm-specific OsIRE1-OE, endosperm-specific OsBiP1-KD, endosperm-specific OsBiP1-OE, and various recombinant proteins, including lactostatin peptide derived from β-lactoglobulin (Nagaoka et al. 2001; Wakasa et al. 2011a), novokinin peptide derived from ovalubumin (Onishi et al. 2004; Wakasa et al. 2011b), APL4 peptide derived from human collagen type II (Wakamatsu et al. 2009), modified GLP-1 (mGLP-1) derived from human glucagon-like protein 1 (Yasuda et al. 2006a, b), human β-amyloid (Oono et al. 2010), and human cathelicidin (Sang and Blecha 2008). These proteins were expressed under the control of rice endosperm-specific promoters and were produced by Agrobacterium-mediated transformation. Expression gene cassette constructs are shown in Fig. S2. These transgenic and non-transgenic rice were grown in a closed greenhouse.
RT-PCR analyses of OsBiP1, OsBiP2, OsBiP3, OsBiP4, OsBiP5, OsbZIP50, and 17S ribosomal RNA were performed as described previously (Hayashi et al. 2012).
Antibody preparation
The MH2-GGAPEDGNVDDED-OH peptide derived from OsBiP5 (Os08g0197700) was synthesised and used to raise anti-OsBiP5 polyclonal antibody in a rabbit (Scrum Inc., Tokyo, Japan). This antibody reacts with OsBiP5 and OsBiP4 (Os05g0428600). OsBiP4 is almost identical to OsBiP5 in amino acid sequence (94 % identity, Fig. S1), molecular weight, and expression pattern (Hayashi et al. 2012). Since the amino acid sequence (MH2-GGAPEDGNVDDED-OH) of the antigen peptide was identified in only OsBiP4 and OsBiP5, the antibody was expected to specifically recognize OsBiP4 and OsBiP5. We confirmed that anti-OsBiP4&5 antibody did not react with OsBiP1, OsBiP2, or OsBiP3 proteins (Fig. S3).
Recombinant proteins of full-length OsBiP1 and the upstream region of the putative transmembrane domain of OsbZIP50 were expressed in E. coli, and the purified recombinant proteins were used to raise anti-BiP1 and anti-OsbZIP50 polyclonal antibodies in a rabbit (Scrum Inc.). For the production of rabbit polyclonal antibodies against calnexin (Os04g0402100), lactostatin, novokinin, APL4, mGLP-1, and β-amyloid, specific peptides for these proteins were used to raise polyclonal rabbit antibodies (Scrum Inc.). Anti-cathelicidin polyclonal rabbit antibody was purchased from Abcam plc. (Cambridge, UK).
DTT treatment of rice tissues
Calli were produced from mature rice seeds on N6D medium [4 g/L CHU salt mixture (Wako, Osaka, Japan), 30 g/L sucrose, 2.78 g/L proline, 100 mg/L myo-inositol, 300 mg/L casamino acid, 1 mL/L 1,000× N6-vitamin, 2 mg/L 2,4-D, 0.4 % gelrite, pH 5.8] for 5 weeks at 30 °C (16 h light/8 h dark). DTT was used to induce the ER stress response in calli and in roots of seedlings. Calli were placed on N6D medium containing 2 mM DTT for 0, 0.5, 1, 2, 4, 8, and 24 h at 30 °C (16 h light/8 h dark). Roots of seedlings (1 week after germination) were treated with MS liquid medium [4 g/L MS salt mixture (Wako), 30 g/L sucrose, 1 mL/L 1,000× B5-vitamin, pH 5.8] containing 2 mM DTT under the same time-course as used for calli. After DTT treatment, roots and calli were immediately frozen in liquid N2 before the extraction of total proteins.
Protein extraction, SDS-PAGE, and immunoblotting
The total proteins were prepared using extraction buffer (50 mM Tris–HCl pH 6.8, 8 M urea, 4 % SDS, 20 % glycerol, 5 % 2-mercaptoethanol, 0.01 % bromophenol blue). Extraction buffer was added to the roots, calli, and seed powder and then vortexed for more than 1 h at room temperature. The mixture was centrifuged at 12,000g for 20 min at room temperature, and the crude soluble protein sample was decanted into a new tube. Total proteins were subjected to immunoblot analysis using polyclonal rabbit antibody after electrophoresis on 12 % SDS-PAGE.
Confocal immunohistochemical analysis
Immature wild-type and transgenic rice seeds were collected at 15 days after flowering (DAF) and used for immunocytochemical analysis using the anti-OsBiP1, anti-OsBiP4&5 and anti-calnexin rabbit polyclonal antibody as described by Yasuda et al. (2006b). Briefly, 200 μm of section sample was treated with 3.7 % formaldehyde in phosphate buffer saline (PBS) for 1 h. Then, the sample was incubated with cell wall digestion solution (1 % cellulose and 0.1 % pectolyase in PBS) at 30 °C for 10 min. Primary antibodies (anti-OsBiP1, anti-OsBiP4&5 and anti-calnexin antibodies) were used at a 1:300 dilution. Alexa488-conjugated goat anti-rabbit IgG (Invitrogen) was used at a 1:500 dilution as the secondary antibody. Rhodamine B was used to stain ER-derived protein bodies (PB-I). The samples were observed through a confocal laser scanning microscope (FLUOVIEW; Olympus).
Results
Expression of OsBiP2, OsBiP3, OsBiP4, and OsBiP5 is regulated by the ER stress signalling pathway through OsIRE1-mediated splicing of OsbZIP50 mRNA (Hayashi et al. 2012). As shown in Fig. 1, expression of these OsBiPs was exclusively detected in ER-stressed cells, such as roots treated with DTT (Fig. 1a) and OsBiP1-KD seeds (Fig. 1b). Furthermore, the spliced form of OsbZIP50 was detected only in ER-stressed cells (Fig. 1). On the other hand, expression and splicing of OsbZIP50 mRNA were strongly suppressed in OsIRE1-KD rice, even during stress induction with DTT treatment (Fig. 1a).
Expression of OsBiP1, OsBiP2, OsBiP3, OsBiP4, OsBiP5, and OsbZIP50 in ER-stressed cells. a RT-PCR analysis of DTT-treated root mRNA. Wt, wild type; OsIRE1-KD, OsIRE1 knock down transgenic plant; −, no-DTT treatment; +, DTT treatment; ubZIP50, unspliced OsbZIP50; sbZIP50, spliced OsbZIP50; hybrid, annealing products of sbZIP50 and ubZIP50. b RT-PCR analysis of OsBiP1-KD seed mRNA. DAF days after flowering, OsBiP1-KD seed-specific OsBiP1 knock down transgenic plant
We prepared antibodies against these proteins, and an antibody that specifically recognized OsBiP4 and OsBiP5 was successfully produced (anti-OsBiP4&5 antibody). We used this antibody as a marker to investigate if the accumulation levels of OsBiP4 and OsBiP5 were correlated with stress levels in the ER lumen of various rice tissues.
Calli and roots were treated with 2 mM DTT as an ER stress-inducing agent, and proteins were extracted and then subjected to immunoblot analysis using the anti-OsBiP4&5 antibody (Fig. 2). OsBiP4&5 proteins were clearly detected at 4 h after DTT treatment for both tissues. OsbZIP50 protein was detected at 1 h after DTT treatment and approximately 3 h before the appearance of OsBiP4&5 (Fig. 2), suggesting that the induced OsbZIP50 mRNA was smoothly spliced by OsIRE1 in these experiments. Furthermore, it is notable that the production of OsBiP4&5 is dependent on ER stress, and levels increased markedly in response to longer treatments with DTT. In contrast, OsBiP1 showed constitutive expression in both roots and calli, and levels increased in response to DTT (Fig. 2), however, changes in the OsBiP4&5 levels in callus and root were more pronounced than that of OsBiP1 after DTT treatment.
Induction of ER stress by DTT treatment in callus and root. Calli (left panels) and roots (right panels) of seedlings were treated with 2 mM DTT for 0, 0.5, 1, 2, 4, 8, and 24 h. SDS-PAGE and immunoblot analyses using anti-OsBiP4&5, anti-OsbZIP50, and anti-OsBiP1 antibodies are shown. −, no-DTT treatment; +, 2 mM DTT treatment; sbZIP50, spliced OsbZIP50
Next, we examined the accumulation of OsBiP4&5 in the roots of constitutive OsIRE1-KD transgenic rice subjected to DTT treatment and in the roots of OsIRE1-OE transgenic rice without DTT treatment (Fig. 3). Overexpression of IRE1 induces an ER stress response in animal and plant tissues (Hayashi et al. 2012; Yoshida et al. 2001), therefore, we examined whether OsbZIP50 was translocated to the nuclei of OsIRE1-OE transgenic rice seed cells (Fig. S4). In roots of the OsIRE1-KD line, OsBiP4&5 and OsbZIP50 were not detected even under stress-induction treatments with DTT. In contrast, roots of the OsIRE1-OE line accumulated OsBiP4&5 constitutively, even without DTT treatment. OsbZIP50 proteins in the OsIRE1-OE line were detected as two bands, representing inactive (unspliced) and active (spliced) forms of OsbZIP50, respectively. In non-stressed cells, the results suggested that OsBiP4&5 is primarily regulated by the OsIRE1/OsbZIP50 signalling pathway because OsBiP4&5 accumulation depended on the accumulation of the active (spliced) form of OsbZIP50. However, the amount of active OsbZIP50 that accumulated in OsIRE1-OE roots was much lower than that in roots treated with DTT, whereas BiP4&5 levels were much higher in OsIRE1-OE roots. This lack of correlation between OsBiP4&5 levels and the active form of OsbZIP50 may be explained by the fact that DTT treatment induces only a short period of ER stress; this is in contrast to the constitutive activation of the ER stress signalling pathway caused by overexpression of OsIRE1, which results in greater accumulation of OsBiP4&5.
The relationship between OsBiP4&5 accumulation and OsbZIP50 in OsIRE1-KD and OsIRE1-OE lines. Root tissues of wild-type and independent transgenic rice lines with or without DTT treatment are used as samples. SDS-PAGE and immunoblot analyses using anti-OsBiP4&5, anti-OsbZIP50, and anti-OsBiP1 antibodies are shown. Wt, wild type; KD, OsIRE1-KD; OE, OsIRE1-OE; −, no-DTT treatment; +, DTT treatment for 8 h; ubZIP50, unspliced OsbZIP50; sbZIP50, spliced OsbZIP50
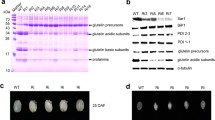
Next, we investigated the relationship between OsBiP4&5 levels and the ER stress response in seed tissue. As DTT treatments cannot be applied to seed tissue, rice seeds harvested from transgenic plants in which ER stress was induced by the overexpression or suppression of OsBiP1 or high accumulation of exogenous recombinant proteins were used for these experiments. OsBiP1, the major BiP protein in rice, is constitutively expressed in various rice tissues. Transgenic rice seeds in which OsBiP1 is suppressed (OsBiP1-KD) exhibited severe ER stress (Wakasa et al. 2011c). On the other hand, very high expression of OsBiP1 (OsBiP1-OE) also induces the ER stress response in transgenic rice seeds (Yasuda et al. 2009; Wakasa et al. 2011c). OsBiP1-KD and OsBiP1-OE transgenic rice seeds display a severely abnormal phenotype with floury and shrunken features (Yasuda et al. 2009; Wakasa et al. 2011c). Furthermore, both transgenic lines suppressed the accumulation of seed storage proteins compared with non-transgenic rice seed. Thus, these abnormal grain phenotypes may be useful as an indicator of ER stress. We used OsBiP1-KD, OsBiP1-OE, and OsIRE1-OE transgenic seeds for the evaluation of the OsBiP4&5 antibody in seed tissues. As shown in Fig. 4a, wild-type seed displayed a normal phenotype, whereas aberrant phenotypes such as floury and shrunken features were observed in the transgenic seeds. Immuno-blot analysis shows that inactive OsbZIP50, active OsbZIP50, and OsBiP4&5 proteins were detected in the transgenic rice seeds, but these proteins were not detected in wild-type seed (Fig. 4b).
OsBiP4&5 accumulation in seed tissues under ER stress conditions. Rice seeds from wild-type, OsBiP1-KD, OsBiP1-OE, and OsIRE1-OE transgenic lines are used as samples. SDS-PAGE and immunoblot analyses using anti-OsBiP4&5, anti-OsbZIP50, and anti-OsBiP1 antibodies are shown. The positions of OsBiP1, glutelin precursor, glutelin acidic or basic, globulin, and prolamin on the SDS-PAGE gel are indicated on the right side of the panel. ubZIP50, unspliced OsbZIP50; sbZIP50, spliced OsbZIP50
The accumulation of OsBiP4&5 was investigated in transgenic rice seeds expressing various recombinant proteins. We generated a number of transgenic rice lines expressing different recombinant proteins or bioactive peptides to provide greater nutritional qualities and human health-promoting functions to wild-type rice. We investigated the relationship between the seed phenotype and OsBiP4&5 levels in transgenic rice expressing lactostatin (Wakasa et al. 2011a), novokinin (Onishi et al. 2004; Wakasa et al. 2011b), APL4 (one of the analogue peptides of the type II collagen-reactive T cell epitope) (Wakamatsu et al. 2009), mGLP-1 (Yasuda et al. 2006a, b), β-amyloid (Oono et al. 2010), and cathelicidin (Sang and Blecha 2008). Lactostatin, novokinin, and APL4 were expressed as fusion proteins with the seed storage protein glutelin, whereas mGLP-1, β-amyloid, and cathelicidin were linked to the ER retention signal peptide (Lys-Asp-Glu-Leu) at their C termini and were directly expressed as secretory proteins. When lactostatin and APL4 were produced as fusion proteins in transgenic rice seeds, the resulting phenotypes were not significantly different to those of the wild-type seeds. Transgenic seeds accumulating novokinin displayed a slightly chalky phenotype. However, rice seeds accumulating mGLP-1, β-amyloid, and cathelicidin resulted in severely abnormal phenotypes (Fig. 5a), suggesting that they were under strong ER stress conditions compared with the wild-type and other transgenic rice seeds. Importantly, OsBiP4&5 accumulation levels were closely correlated with the observed deterioration of seed phenotypes. A very faint level of OsBiP4&5 was detected in transgenic rice seeds expressing lactostatin or APL4 (Fig. 5b), which exhibited normal phenotypes. A weak level of BiP4&5 was detected in transgenic rice seed expressing novokinin. However, remarkably high levels of OsBiP4&5 were detected in transgenic seeds expressing mGLP-1, β-amyloid, or cathelicidin, which exhibited severely abnormal phenotypes (Fig. 5a, b). These results indicate that ER stress and severely aberrant seed phenotypes are strongly correlated with OsBiP4&5 accumulation levels induced by ER stress in seeds. Notably, the highest level of OsBiP1 was observed in transgenic rice expressing mGLP-1; relatively higher levels of OsBiP1 accumulation compared to wild-type rice seed were observed in these transgenic rice seeds. However, the severe seed phenotype observed in transgenic lines expressing mGLP-1, β-amyloid, or cathelicidin could not be caused by the accumulation of OsBiP1, because OsBiP1 levels were not significantly different among the lines expressing lactostatin, novokinin, APL4, β-amyloid, and cathelicidin, but the grain phenotypes were quite different. The relationship between seed quality and concentration of OsBiP4&5 was further examined for transgenic rice accumulating mGLP-1 (Fig. 5c). The results indicate that increases in OsBiP4&5 levels were dependent on the level of mGLP-1 accumulation, leading to the severe grain phenotypes with floury and shrunken features.
OsBiP4&5 accumulation in transgenic rice seeds expressing various recombinant proteins. a Phenotype of wild-type and transgenic seeds accumulating various recombinant proteins. b Immunoblot analysis of seeds in a. Wt wild type, lactostatin pentapeptide derived from bovine milk β-lactoglobulin with hypocholesterolemic activity, novokinin a new potent anti-hypertensive peptide based on the sequence of novokinin (2–7) derived from ovalbumin, APL4 one of the altered peptide ligands that controls type II collagen-reactive T cells, GLP-1 a 30-amino acid peptide hormone involved in insulin stimulation, β-amyloid dimers of mature 42-amino acid β-amyloid peptide (Aβ1–42), cathelicidin antimicrobial peptide derived from human. c Phenotypes of transgenic rice seeds accumulating mGLP-1 at various levels, and immunoblot analysis of these seeds
Finally, to examine the function of ER stress-associated BiP, such as OsBiP4 and OsBiP5, the subcellular localization of OsBiP4&5 in IRE1-OE endosperm cells was investigated by indirect histochemical analysis using confocal microscopy (Fig. 6). ER-derived protein body-I (PB-I) was stained by rhodamine, and OsBiP1 and OsBiP4&5 were detected using anti-BiP1 and anti-BiP4&5 primary antibodies and Alexa 488-conjugated anti-rabbit IgG secondary antibody. Furthermore, calnexin protein was used as a marker protein localized to the ER (Gupta and Tuteja 2012; Takahashi et al. 2012) (Fig. 6m–o).
Indirect immunohistochemical analysis of OsBiP1 and OsBiP4&5 in rice cells. Left panels show localization of OsBiPs (green), middle panels show localization of PB-I (red), and right panels show the merged images. a–c Localization of OsBiP1 in wild-type seeds. d–f Localization of OsBiP1 in OsIRE1-OE transgenic rice seeds. g–i Localization of OsBiP4&5 in wild-type seeds. j–l Localization of OsBiP4&5 in OsIRE1-OE transgenic rice seeds. m–o Localization of calnexin, an ER marker protein, in the wild type
OsBiP1 was localized predominantly to the ER and periphery of PB-I in wild-type seeds as described in a previous report (Yasuda et al. 2009), and similar localization patterns were observed in transgenic OsIRE1-OE seeds (Fig. 6a–f). Although OsBiP4&5 levels were very low in wild-type seeds (Fig. 6g), they were primarily detected in transgenic OsIRE1-OE seeds, namely ER-stressed seeds (Fig. 6j). As the localization of OsBiP4&5 displayed a very similar fluorescent pattern to that of calnexin (Fig. 6m), OsBiP4&5 was considered to be localized to the ER, with very little localization to the periphery of PB-I (Fig. 6j).
Discussion
We previously reported the expression of novel ER stress responsive BiP genes (OsBiP2–OsBiP5) in roots and cultured cells treated with the ER stress agents DTT and tunicamycin (Hayashi et al. 2012). The present study identified a positive correlation between ER stress levels and the accumulation of two BiP proteins, OsBiP4 and OsBiP5, in callus, roots (Figs. 2, 3) and maturing rice seed (Figs. 4, 5) by immunoblot analysis with an anti-OsBiP4&5 antibody. Expression of the OsBiP4&5 genes was very sensitive to various ER stress conditions but was hard to detect under normal conditions. This result is in marked contrast with the finding that OsBiP1 was constitutively expressed in various tissues under normal conditions, although expression was upregulated by ER stress. ER stress-specific induction of OsBiP4&5 is under the control of the OsIRE1/OsbZIP50 signalling pathway; thus, OsBiP4&5 expression is strongly suppressed not only in OsIRE1-KD (Fig. 3) but also in OsbZIP50-KD (Hayashi et al. 2012). By contrast, OsBiP1 is mainly regulated by the other signal transduction pathway via OsbZIP39 (Takahashi et al. 2012).
It is notable that the phenotypic deterioration in rice grains, caused by the production of foreign recombinant proteins, was more correlated with high levels of OsBiP4&5 accumulation than with OsBiP1 levels. However, the levels of OsBiP1, OsBiP4&5, OsbZIP50 and OsIRE1 were somewhat contradictory, although the OsBiP4&5 level was regulated by OsIRE1/OsbZIP50 signalling. As shown in Fig. 3, the level of OsBiP4&5 in OsIRE1-OE roots (constitutive ER stress) was higher than that in roots treated with DTT (transient induction of ER stress), whereas the amount of active OsbZIP50 in OsIRE1-OE roots was much lower than that in roots treated with DTT. Furthermore, the OsBiP4&5 level in OsBiP1-KD seeds was much higher than that in OsIRE1-OE seeds, although the level of active OsbZIP50 remained low (Fig. 4). In the case of ER stress induced by DTT treatment or in OsIRE1-OE, differences in ER stress levels, treatment times and ER stress signalling pathways may explain why there was only a weak relationship between OsBiP50 and OsBiP4&5 levels. The high abundance of OsBiP4&5 in OsBiP1-KD seeds might be because OsBiP4&5 must complement the low amount of OsBiP1. OsBiP1-KD probably triggers severe ER stress in the absence of the BiP chaperone (Wakasa et al. 2011c). However, the present data cannot address why OsbZIP50 is less abundant in cells lacking OsBiP1 in spite of its high abundance in OsBiP4&5. Another explanation might be that since OsbZIP50 is a transcriptional factor, it is not as stable as BiP proteins, considering that OsBiP1 stably accumulates even in mature rice seed. In the stable transgenic plants, such as OsIRE1-OE and OsBiP1-KD, OsBiP4&5 may accumulate in stressed tissues, whereas the OsbZIP50 level may be strictly regulated by the stress level and undergo prompt turnover. Thus, the difference in stability between OsbZIP50 and OsBiP4&5 might also explain the non-parallel relationship of these accumulation levels. Further work is required to understand the correlation between OsBiP1, OsBiP4&5 and OsbZIP50 in response to ER stress in rice.
We demonstrated that differences in the intracellular localization of OsBiP1 and OsBiP4&5 were observed in wild type and OsIRE1-OE endosperm cells (Fig. 5). OsBiP1 localised at the periphery of PB-Is and the ER, whereas OsBiP4&5 was predominantly distributed within the ER. These results indicate that OsBiP genes may show different functions under different types of ER or environmental stress, and during developmental processes. Indeed, differential expression of BiP genes has been reported in Arabidopsis (Iwata et al. 2008). AtBiP3 shows a similar expression pattern to OsBiP4&5 (Chen and Brandizzi 2012; Deng et al. 2011; Nagashima et al. 2011; Hayashi et al. 2012), suggesting that OsBiP4&5 may be a counterpart of AtBiP3. Plant BiP is encoded by multiple genes (five in rice and three in Arabidopsis), thus, plant BiP genes are thought to have become functionally differentiated throughout evolution. By contrast, mammalian BiP (GRP78) and yeast BiP (KAR2) are encoded by a single gene (Ting and Lee 1988; Wooden et al. 1988; Rose et al. 1989; Fu et al. 2008; Kimata and Kohno 2010). GRP78 plays a critical role as a chaperone during protein quality control, irrespective of ER stress conditions. Knock out of GRP78 in mice results in early embryonic lethality (Luo et al. 2006). Deletion of the KAR2 gene in yeast also causes a recessive lethal mutation (Rose et al. 1989). Thus, difference in gene copy number between plants and animals may explain the difference in responses to various stresses. Since plants cannot move, they have developed/evolved multiple defence mechanisms and protein quality control systems to protect themselves against stresses, including ER stress.
The results of the present study indicate that the level of OsBiP4&5 is regulated by the OsbZIP50/OsIRE1 signalling pathway, which is responsible for the major ER stress response in various rice tissues. OsBiP4&5 accumulation is a valuable marker for evaluating ER stress levels, which can be easily measured by immunoblot analysis using the anti-OsBiP4&5 antibody. Monitoring ER stress levels using this identifiable marker will be useful for basic research investigating the molecular mechanisms underlying the ER stress response, and for the production of transgenic rice seeds that accumulate recombinant proteins.
Further studies on the function of OsBiP4 and OsBiP5 in rice cells under ER stress conditions are required. On-going work aims to produce constitutive or seed-specific OsBiP4&5-KD and OsBiP4&5-OE lines and to examine the function of these transgenic rice lines.
Abbreviations
- ER:
-
Endoplasmic reticulum
- ERAD:
-
ER associated degradation
- HSP:
-
Heat shock protein
- KD:
-
Knockdown
- OE:
-
Overexpression
- PB:
-
Protein body
- UPR:
-
Unfolded protein response
References
Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2:326–332
Chakrabarti A, Chen AW, Varner JD (2011) A review of the mammalian unfolded protein response. Biotechnol Bioeng 108:2777–2793
Chen Y, Brandizzi F (2012) AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J 69:266–277
Deng Y, Humbert S, Liu J-X, Srivastava R, Rothstein SJ, Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108:7247–7252
Fu Y, Wey S, Wang M, Ye R, Liao C-P, Roy-Burman P, Lee AS (2008) Pten null prostate tumorigenesis and AKT activation are blocked by targeted knockout of ER chaperone GRP78/BiP in prostate epithelium. Proc Natl Acd Sci USA 105:19443–19448
Gupta D, Tuteja N (2012) Chaperone and foldases in endoplasmic reticulum stress signalling in plants. Plant Signal Behav 6:232–236
Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274
Hayashi S, Wakasa Y, Takahashi H, Kawakatsu T, Takaiwa F (2012) Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the ER stress response of rice. Plant J 69:946–956
Iwata Y, Fedoroff NV, Koizumi N (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20:3107–3121
Kawakatsu T, Hirose S, Yasuda H, Takaiwa F (2010) Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol 154:1842–1854
Kimata Y, Kohno K (2010) Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr Opin Cell Biol 23:135–142
Luo S, Mao C, Lee B, Lee AS (2006) GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol 26:5688–5697
Nagaoka S, Futamura Y, Miwa K, Awano T, Yamauchi K, Kanamaru Y, Kojima K, Kuwata T (2001) Identification of novel hypocholesterolemic peptides derived from bovine milk β-lactoglobulin. Biochem Biophys Res Commun 281:11–17
Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Scientific Rep 1: Article number 29
Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K (2000) Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279:445–450
Onishi K, Matoba N, Yamada Y, Doyama N, Utsumi S, Yoshikawa M (2004) Optimal designing of β-conglycinin to genetically incorporate RPLKPW, a potent anti-hypertensive peptide. Peptides 25:37–43
Oono Y, Wakasa Y, Hirose S, Yang L, Sakuta C, Takaiwa F (2010) Analysis of ER stress in developing rice endosperm accumulating beta-amyloid peptide. Plant Biotechnol J 8:691–718
Oyadomari S, Mori M (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 11:381–389
Rose MD, Misra LM, Vogel JP (1989) KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1211–1221
Sang Y, Blecha F (2008) Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Anim Health Res Rev 9:227–235
Takagi H, Saito S, Yang L, Nagasaka S, Nishizawa N, Takaiwa F (2005) Oral immunotherapy against a pollen allergy using a seed-based peptide vaccine. Plant Biotechnol J 3:521–533
Takahashi H, Kawakatsu T, Wakasa Y, Hayashi S, Takaiwa F (2012) A rice transmembrane bZIP transcriptional factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol 53:144–153
Ting J, Lee AS (1988) The human gene encoding the 78000 dalton glucose regulated protein and its pseudogene: structure, conservation and regulation. DNA 7:275–286
Wakamatsu E, Matsumoto I, Yoshiga Y, Hayashi T, Goto D, Ito S, Sumida T (2009) Altered peptide ligands regulate type II collagen-induced arthritis in mice. Mod Rheumatol 19:366–371
Wakasa Y, Tamakoshi C, Ohno T, Hirose S, Goto T, Nagaoka S, Takaiwa F (2011a) The hypocholesterolemic activity of transgenic rice seed accumulating lactostatin, a bioactive peptide derived from bovine milk β-lactoglobulin. J Agric Food Chem 59:3845–3850
Wakasa Y, Zhao H, Hirose S, Yamauchi D, Yamada Y, Yang L, Ohinata K, Yoshikawa M, Takaiwa F (2011b) Anti-hypertensive activity of transgenic rice seed containing an 18-repeat novokinin peptide localized in the nucleolus of endosperm cells. Plant Biotechnol J 9:729–735
Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, Hayashi S, Yang L, Takaiwa F (2011c) Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J 65:675–689
Wooden SK, Kapur RP, Lee AS (1988) The organization of the rat GRP78 gene and A23187-induced expression of fusion gene products targeted intracellulary. Exp Cell Res 178:84–92
Yamaguchi Y, Larkin D, Lala-Lemus R, Ramos-Castaneda J, Liu M, Arvan P (2008) Endoplasmic reticulum (ER) chaperone regulation and survival of cells compensating for deficiency in the ER stress response kinase, PERK. J Biol Chem 283:17020–17029
Yasuda H, Tada Y, Hayashi Y, Jomori T, Takaiwa F (2006a) Expression of the small peptide GLP-1 in transgenic plants. Transgenic Res 14:677–684
Yasuda H, Hayashi Y, Jomori T, Takaiwa F (2006b) The correlation between expression and localization of a foreign gene product in rice endosperm. Plant Cell Physiol 47:756–763
Yasuda H, Hirose S, Kawakatsu T, Wakasa Y, Takaiwa F (2009) Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant Cell Physiol 50:1532–1543
Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891
Acknowledgments
The authors thank Ms. Y. Ikemoto, K. Miyashita, Y. Suzuki, M. Utsuno, and Y. Yajima for technical assistance. This research was supported by the research grant, “Genomics and Agricultural Innovation, GMC0003”, from the Ministry of Agriculture, Forestry and Fisheries of Japan to F. Takaiwa.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2012_1714_MOESM2_ESM.tif
Supplementary Fig. S2 Binary vector constructs used to produce transgenic rice lines in this study. a Constitutive suppression of OsIRE1. b Constitutive expression of OsIRE1. c Seed-specific expression of OsIRE1. d Seed-specific suppression of OsBiP1. e Seed-specific expression of OsBiP1. f Seed-specific expression of lactostatin as a fusion protein with seed storage protein GluA1. g Seed-specific expression of novokinin as a fusion protein with seed storage protein GluA1. h Seed-specific expression of APL4 as a fusion protein with seed storage protein GluA1. i Seed-specific expression of GLP-1 with the ER retention signal. j Seed-specific expression of β-amyloid with the ER retention signal. k Seed-specific expression of cathelicidin with the ER retention signal. Ubi P, ubiquitin gene promoter; GluB1 P, seed-specific glutelin B1 promoter; Nos T, nopaline synthase terminator; GluB1 T, glutelin B1 terminator; RSIS, RNA silencing inducible sequence (Kawakatsu et al. 2010; Wakasa et al. 2011c); sp, signal peptide; KDEL, ER retention signal (TIFF 222 kb)
425_2012_1714_MOESM3_ESM.tif
Supplementary Fig. S3 Anti-OsBiP4&5 antibody does not react with OsBiP1, OsBiP2, or OsBiP3 proteins. 6 × His tag-fused OsBiP1, OsBiP2, OsBiP3 and OsBiP5 were produced by the E. coli expression system. Because amino acid sequence identity is high between OsBiP4 and OsBiP5, the OsBiP5 recombinant protein was used as a representative protein for both proteins. Expression of recombinant protein by E. coli was performed by using the pCold shock vector II (Takara) and BL21-codon plus RIL E. coli strain (Stratagene). An immunoblot obtained with the anti-His-tag antibody is shown as a loading control (TIFF 192 kb)
425_2012_1714_MOESM4_ESM.tif
Supplementary Fig. S4 Indirect immunohistochemical analysis of OsbZIP50 in seeds. a SDS-PAGE and immunoblot analysis in wild-type and seed-specific OsIRE1-OE transgenic rice seed. b Localization of OsbZIP50 in wild-type seed. Left panel shows the merged image of rhodamine (red, position of PB-I) and DAPI (light blue, position of nuclei) stain. Right panel shows the merged image of rhodamine (red) and Alexa 488 (green) OsbZIP50 signal. c Localization of OsbZIP50 in seeds of three independent OsIRE1-OE transgenic lines. The three panels show the merged images of rhodamine (red) and Alexa 488 (green) OsbZIP50 signals in three independent transgenic rice seeds. Arrows indicate nuclei (TIFF 1.73 mb)
Rights and permissions
About this article
Cite this article
Wakasa, Y., Hayashi, S. & Takaiwa, F. Expression of OsBiP4 and OsBiP5 is highly correlated with the endoplasmic reticulum stress response in rice. Planta 236, 1519–1527 (2012). https://doi.org/10.1007/s00425-012-1714-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1714-y