Abstract
Proteomics analysis has been performed in leaf tissue from field date palm trees showing the brittle leaf disease (BLD) or maladie des feuilles cassantes, the main causal agent of the date palm decline in south Tunisia. To study the evolution of the disease, proteins from healthy and affected leaves taken at three disease stages (S1, S2 and S3) were trichloroacetic acid acetone extracted and subjected to two-dimensional gel electrophoresis (5–8 pH range). Statistical analysis showed that the protein abundance profile is different enough to differentiate the affected leaves from the healthy ones. Fifty-eight variable spots were successfully identified by matrix-assisted laser desorption ionization time of flight, 60 % of which corresponded to chloroplastic ones being involved in the photosynthesis electronic chain and ATP synthesis, metabolic pathways implicated in the balance of the energy, and proteases. Changes in the proteome start at early disease stage (S1), and are greatest at S2. In addition to the degradation of the ribulose-1.5-bisphosphate carboxylase oxygenase in affected leaflets, proteins belonging to the photosynthesis electronic chain and ATP synthesis decreased following the disease, reinforcing the relationship between BLD and manganese deficiency. The manganese-stabilizing proteins 33 kDa, identified in the present work, can be considered as protein biomarkers of the disease, especially at early disease step.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Date palm (Phoenix dactylifera L.) cultures in Tunisian oases suffer from a disease called brittle leaf disease (BLD), known in French as maladie des feuilles cassantes (MFC), which became epidemic ever since 1980 (Triki et al. 2003) and has been reported in eastern Algeria (Saadi et al. 2006). The most characteristic symptom of the disease concerns the plant leaves that become brittle and break easily when flexed and squeezed. The disease evolution has been divided into three stages (1) the first one (S1) is characterized by chlorosis of a few fronds, (2) in the second stage (S2), leaflets become brittle, twisted and frizzled and (3) the symptoms extend in stage 3 (S3) to the entire plant that finally dies (Triki et al. 2003). Symptoms extend to the upper fronds until the whole plant becomes affected and stunted, stops growing, and finally dies (Triki et al. 2003). Although, the MFC epidemiology suggested a biotic etiology, so far no pathogen associated with the disease could be found (Triki et al. 2003). There are two main features of this disease: a manganese (Mn) deficiency in the leaflets of affected palms (Namsi et al. 2007) and the abnormal accumulation of a set of novel RNAs transcribed from the date palm chloroplast genome (Marqués et al. 2008).
Leaflets of affected palms have been shown to contain significantly lower Mn concentrations than those of healthy controls (Namsi et al. 2007). Soils in which MFC develops are Mn deficient and contain about half the manganese content compared to healthy soils (Namsi et al. 2007). Moreover, Mn is the only element for which significant differences are seen in adult leaflets between healthy trees and MFC-affected trees (Namsi et al. 2007). Transcriptomic analysis of MFC-affected leaves showed that a repression of manganese-related genes, photosynthesis, ion transport and protein synthesis occurred in affected leaves, while stress response, metabolism associated, protein synthesis and signal transduction genes were up-regulated by the disease (Saidi et al. 2010).
Manganese deficiency in green plants is known to affect preferentially the activity of the oxygen-evolving system in the photosynthetic apparatus. Higher plants responded to manganese deficiency either by adjusting the number of chloroplasts per cell to the limited Mn supply, or by forming disorganized chloroplasts with low chlorophyll content (Homann 1967). Recently, at the proteomics level, Marqués et al. (2011) reported that Mn-binding PSBO and PSBP proteins, components of the oxygen-evolving complex of photosystem II, were decreased in affected date palm leaves not yet entering into final decline (leaves from stage S2).
The objective of this work is the characterization of the proteome changes that occur in date palm leaves affected by MFC disease from the beginning of the disease (stage S1) to the death of the palm trees (stage S3). In addition, we are interested in finding protein biomarkers of MFC disease that reinforce its relationship to Mn deficiency by studying the function of some identified proteins which may be a key in this disease.
Materials and methods
Plant material
Leaf tissues from field date palm (Phoenix dactylifera L. cv. Deglet Nour) trees (resulting from offshoot propagation) are used in the present work. The healthy and affected palm trees belong to the same cultivar. In the MFC-affected oasis (southern Tunisia; Degache), the following trees were selected: healthy control (HC), trees with mild (S1), moderate (S2), and severe (S3) symptoms. The three stages of MFC symptoms (S1, S2 and S3; Fig. 1) were identified and selected based on the symptoms as described by Triki et al. (2003). Four to six years may elapse between the appearance of the first symptoms (stage S1) and death of the tree (stage S3). HC leaflets were collected from the same oasis as that of the affected palm trees, but at different locations, where MFC symptoms have never been observed.
Protein extraction and 2-DE electrophoresis
Leaves (10 leaves per plant/one plant per replicate/three replicates) from healthy and affected trees were collected, thoroughly washed with water, dried with filter paper, frozen in liquid nitrogen, and stored at −80 C until protein extraction. Leaves were ground to a fine powder in liquid nitrogen using a mortar and pestle and the powder (0.5 g) was subjected to a TCA–acetone–phenol protein extraction procedure (Wang et al. 2006; Maldonado et al. 2008). The final pellet was dissolved in 40–200 μl of 7 M urea, 2 M thiourea, 4 % CHAPS and 100 mM DTT (Bio-Lyte 3/10, Bio-Rad). Once the pellet was solubilized and the insoluble material eliminated by centrifugation, the protein content was quantified by the method of Bradford (1976), using bovine serum albumin as standard.
For 2-DE, IPG strips (17 cm, 5–8 pH linear gradient; Bio-Rad) were passively rehydrated for 16 h with 400 μg protein in 300 μl of IEF solubilisation buffer [7 M urea; 2 M thiourea; 4 % (w/v) CHAPS; 0.2 % (v/v) IPG buffer 5–8, 100 mM DTT; and 0.01 % (w/v) bromophenol blue]. The strips were loaded onto a Bio-Rad Protean IEF cell system and proteins were electrofocused (Maldonado et al. 2008). Strips were equilibrated and subjected to second-dimension SDS-PAGE, then stained with Colloidal Coomassie Blue G-250 (Maldonado et al. 2008).
Images were digitized with a GS-800 calibrated densitometer (Bio-Rad) and analyzed with the PDQuest™ software (Bio-Rad), using a guided protein spot detection method (Chich et al. 2007). Normalized spot volumes based on total quantity in valid spots were calculated for each 2-DE gel and used for statistical evaluation of differential protein abundance. Experimental M r values were calculated by mobility comparisons with molecular weight standards of low range (Bio-Rad) run in a separate marker lane on the SDS-gel, while pI was determined using a 5–8 linear scale over the total dimension of the IPG strips.
MS analysis and protein identification
Spots were automatically excised (Investigator ProPic, Genomic Solutions), transferred to Multiwell 96 plates and digested with modified porcine trypsin (sequencing grade; Promega) using a ProGest (Genomics Solution) digestion station. The digestion protocol used was that of Schevchenko et al. (1996), with minor variations. Gel plugs were destained by incubation (twice for 30 min) with 200 mM ammonium bicarbonate in 40 % acetonitrile (ACN) at 37 °C, then subjected to three consecutive dehydration/rehydration cycles with pure ACN and 25 mM ammonium bicarbonate in 40 % ACN, respectively, and finally dried at room temperature for 10 min. Trypsin (20 μl) at a final concentration of 12.5 ng/μl in 25 mM ammonium bicarbonate was added to the dry gel pieces and the digestion proceeded at 37 °C overnight.
Peptides were extracted from gel plugs by adding 10 μl of 1 % TFA (15 min incubation), and were then purified by ZipTip μ-C18 (Millipore). Peptides were deposited onto matrix-assisted laser desorption ionization (MALDI) plate using the dry droplet method (ProMS, Genomic Solutions) and the α-cyano hydroxycinnamic acid as a matrix at 5 μg/μl concentration in 70 % ACN, 0.1 % TFA. Samples were analyzed in a 4700 Proteomics Analyzer MALDI-(TOF/TOF) Mass Spectrometer (Applied Biosystems), in the m/z range 800–4,000, with an accelerating voltage of 20 kV, in reflectron mode and with a delayed extraction set to 120 ns. Spectra were calibrated using the trypsin autolysis peaks at m/z = 842.509 and m/z = 2211.104 as internal standards. The three most abundant ions were then subjected to MS/MS analysis, providing information that could be used to determine the peptide sequence.
A combined search (MS plus MS/MS) was performed using GPS Explorer™ software v 3.5 (Applied Biosystems) over NCBInr database using the MASCOT v 1.9 search engine (Matrix Science Ltd., London; http://www.matrixscience.com). The following parameters were allowed: taxonomy restrictions to Viridiplantae, one missed cleavage, 10 ppm mass tolerance in MS and 0.5 Da for MS/MS data, cysteine carbamidomethylation as a fixed modification and methionine oxidation as a variable modification. The confidence in the peptide mass fingerprinting matches (P ≤ 0.05) was based on the MOWSE score and confirmed by the accurate overlapping of the matched peptides with the major peaks of the mass spectrum.
Statistical analysis
For statistical analysis, treatment and cluster analysis of protein abundance values, the web-based software NIA array analysis tool was utilized (Sharov et al. 2005) available at http://lgsun.grc.nia.nih.gov/anova/index.html. This software tool selects statistically valid protein spots based on analysis of variance (ANOVA). After uploading the data table (Supplementary Table 1) and indication of biological replications, the data were statistically analyzed using the following settings: error model max (average, actual), 0.01 proportions of highest variance values to be removed before variance averaging, 10 degrees of freedom for the Bayesian error model, 0.05 FDR threshold, and zero permutations.
The entire data set was analyzed by principal component analysis (PCA) using the following settings: covariance matrix type, four principal components, twofold change threshold for clusters, and 0.5 correlation threshold for clusters. PCA results were represented as a biplot, with consistent proteins in those experimental situations located in the same area of the graph. To perform this statistical cluster analysis, a software tool designed for the analysis of biological gene chip data was used (Sharov et al. 2005), which has also been successfully employed in the analysis of protein abundance data (Brumbarova et al. 2008; Castillejo et al. 2010a, b; Sghaier-Hammami et al. 2012).
The identified proteins were subjected to heat mapping using Euclidean distance (Eisen et al. 1998), with average linkage algorithm of the Genesis software package allowing a definition of some expression groups, the presence or absence of some of them being a characteristic of a particular group (Sturn et al. 2002).
Results
Leaves collected from MFC-affected trees were verified as having symptoms, and the three stages of MFC symptoms (S1, S2 and S3) were selected as described by Triki et al. (2003). Total proteins were extracted from HC and affected leaflets from the first symptoms of the disease (stage S1) to the death of the tree (stage S3, Fig. 1).
Protein extraction and 2-DE analysis
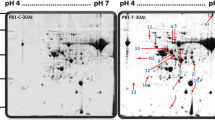
Proteins from HC and affected leaves at each disease stage (three biological replicates) were extracted based on TCA–acetone–phenol methods. The different protein samples (400 μg) were separated by 2-DE (Fig. 2) with IEF carried out in the 5–8 pH range. In all, 364 spots could be resolved in the master gel with most of them (297 spots) being variable between samples (affected leaves and HC). The total number of resolved spots increased in affected leaves from the different stages (S1, S2 and S3) compared to the HC (Table 1), and were the highest at S2. The number of proteins that increased in abundance was greater than the number of proteins that decreased in abundance. MFC-affected leaves at S2 presented higher number of proteins that increased in abundance (227 spots, Table 1) than the ones affected at S1 and S3 (142 and 77 spots, respectively).
Real 2-DE proteome map of healthy control (HC) and affected leaves taken from different disease stages (S1, S2 and S3). The relative M r is given on the left, while the pI is given at the top. Five hundred μg of proteins was separated in the first dimension on an immobilized, linear, 5–8 pH gradient and in the second dimension on a 12 % polyacrylamide–SDS gel
Statistical and expression cluster analyses
The whole protein data set was analyzed by PCA. The first three principal components accounted for 99.99 % of the biological variability of each spot dataset (Table 2). MFC-affected leaves from the three stages (S1, S2 and S3) and the HC are clearly separated (Fig. 3a). We found that the experimental conditions could be divided into three large clusters in a dendrogram, namely, cluster 5, cluster 6 and cluster 7 (Fig. 3b). This clustering indicated that affected leaves from stages S1 and S3 had a similar protein profile (clustering closest together; cluster 5). MFC-affected leaves from stages S2 were further away from the HC than those from S1 and S3.
Statistical protein expression cluster analysis of healthy control (HC) and affected leaves taken from different disease stages (S1, S2 and S3), using the ANOVA-based NIA array analysis tool (Sharov et al. 2005). a Two-dimensional plot showing separation of samples plotted in the first and the second component space by principal component analysis (PCA). b Dendrogram showing hierarchical clustering of samples. The expression clusters are numbered from 1 to 7
Protein identification and differential expression analysis
Out of the 364 resolved spots, 297 were considered as being variable between samples after multivariate statistical analysis. One hundred and thirteen consistent proteins, present in three biological replicates, thus preventing the assignment of normalized volume values to missing spots for multivariate analysis (Valledor and Jorrín 2011), were subjected to MALDI-(TOF/TOF) mass spectrometry analysis, resulting in 58 confidence hits (Table 3). The major group of identified proteins corresponded to chloroplastic ones (60 %), these being involved in the photosynthesis electronic chain and ATP synthesis (31 %), carbohydrate metabolism and Calvin cycle [40 %, with 7 large ribulose-1.5-bisphosphate carboxylase/oxygenase large subunit (RubisCO)], and protein fate (14 %). Twelve percent of proteins were stress-related ones. Actin and porin have also been identified (Table 3).
In 46 % of total identified proteins, the same protein has been identified from different spots, and we can speculate that they were the products of isogenes or post-translational modifications (PTMs), variants of specific gene products differing in pI and/or Mr. Thus, the following proteins are examples of those with more than two spots: ATP synthase CF1 beta chain (found in 4 spots), oxygen-evolving system (OEC) of photosystem II 33 kDa subunit or manganese-stabilizing proteins (MSP-33 kDa; found in 6 spots), RubisCO (4), enolase (3), ATP-dependent Clp protease (6). For those spots with significantly lower experimental values than theoretical Mr, protein degradation can be proposed (RubisCO; spots 6102, 6106, 6101, 7311 and 8310), and for those with higher experimental M r (putative glutamine synthetase cytosolic isozyme; spots 2509 and 7401), PTMs such as ubiquitination or aggregation processes, but these must be experimentally validated.
The different expression dynamics of each identified protein could be distinguished in the heat map allowing a definition of three expression groups (Fig. 4). The first one showed high protein abundance throughout the HC (Fig. 4, in red) and a low one for the affected leaves from three stages (in green), including proteins of the photosynthesis electronic chain and ATP synthesis. The second group showed an opposite trend to the first one, and the third group showed high expression protein abundance only at the S2 stage. The second and third groups resemble the proteases and proteins from Calvin cycle and carbohydrate metabolism (Fig. 4).
Hierarchical clustering of the differentially regulated proteins identified in this study. A heat map representation shows the protein value according to the level of normalized experiments which are indicated from −3 (minimum positive expression; green color) to 3 (maximum positive expression; red color); black indicates zero expression. Leaves were sampled from both healthy control (HC) and affected leaves taken from different disease stages (S1, S2 and S3)
Proteins belonging to the photosynthesis electronic chain and ATP synthesis (ATP synthase; spots 3502, 4603, 3601, 2604 and 2203, and MSP-33 kDa subunit proteins; spots 2213, 2205, 2211, 4206 and 3204) showed a decrease in affected leaves from three disease stages (Figs. 4, 5). On the contrary, ATP-dependent Clp protease proteins (spots: 3801, 3802, 4802, 5801, 4803 and 4801) increased in abundance in affected leaflets, these being more abundant at S3 stage (spots: 3801, 3802, 4802 and 5801). For those RubisCO proteins (spots: 6613, 5209, 6102, 6106, 6101, 7311 and 8310) identified in the present study, 87.5 % of which present lower (0.5 fold) experimental values than theoretical Mr, protein degradation processes can be proposed. RubisCO proteins increased in the affected leaflets (Figs. 4, 5). MFC led to the decrease in MSP-33 kDa subunit proteins (spots 2205, 2211, 2213, 3204, 4201 and 4206) and ATP synthase beta subunit (spots 2203, 2604, 3502, 3601 and 4603) in date palm leaves from three characteristic stages. Chloroplastic (phosphoglycerate kinase, 3505; sedoheptulose-1.7-bisphosphatase, 1513 and d-ribulose-5-phosphate-3-epimerase, 5106), and cytosolic (enolase, 3710 and 3711; triosephosphate isomerase, 5208 and 5206; and glyceraldehyde 3-phosphate dehydrogenase, 7520, 7608, 7609; Figs. 4, 5) metabolic enzymes increased in the affected leaflets. Two chloroplastic proteins have been identified as stress-related proteins in our work in response to MFC; the heat shock 70 kDa (Hsp; spot 3712) and chaperonin 60 subunit beta 1 (4713) which increased in the affected leaves taken from stage S2 and S3, respectively (Figs. 4, 5). Other stress-related proteins identified as peroxiredoxin (spots 2005 and 3002) increased only at the S2 stage. Protein (spot 2607) identified as actin increased at stages S1 and S3; however, it was not detected in affected palm leaf from stage S2. Finally, we have observed an increase in mitochondrial outer membrane protein porin of 34 kDa from affected leaflets at S2, and this was not observed at S3.
Discussion
In palm trees with early MFC symptoms (stage S1), some fronds show chlorosis and have a dull, olive green color. At this disease stage, we cannot discriminate exactly the HC from the affected tree visually (for that reason HC was taken from a location where MFC symptoms have never been detected), however, at S2 and S3 stages, the presence of the MFC disease is clearly seen. Healthy and affected leaves result from offshoot propagation also named vegetative propagation. The offshoots develop from axillary buds on the trunk of the mother plant and consequently conserve the same genetic information. Therefore, all the protein variations observed may result only from the disease’s progression.
The different protein samples were separated by 2-DE with IEF carried out in the 5–8 pH range, taking into consideration that the use of the 3–10 pH range in date palm leaves tissues (Gómez-Vidal et al. 2009), resulted in obtaining of protein spots concentrated in 5–8 pH region. Similar results were also observed for somatic and zygotic date palm embryos Sghaier-Hammami et al. (2009a, b, 2010). 2-DE protein profile of the HC seems to be poorer in protein spots than MFC-affected leaves at S1, S2 and S3 stages, which may be due to the fact that it is easier to extract proteins from leaves affected by MFC, compared to the fibrous HC counterparts. Moreover, MFC-affected leaves at S2 presented higher number of proteins that increased in abundance than the ones affected at S1 and S3. Indeed, it is during this stage that several metabolic and phenotypic changes take place in the plant and leaves (Saadi et al. 2006).
Comparatively, a 2-DE protein map (with a 3–10 pH range) from similar proteomics studies realized on affected leaves at S2 stage (Marqués et al. 2011) showed that the leaflets of MFC-affected and HC palms do not contain striking qualitative differences either in the number or in the amount of the most abundant protein species detected by colloidal Coomassie blue staining (Marqués et al. 2011). This could be explained by the use of different extraction methods and different ranges of pH. In a proteomics study, the protein extraction protocol greatly affects the quality of the extract and every modification in sample preparation generates variability for both protein identification and quantitation (Maldonado et al. 2008; Zhang et al. 2011; Sghaier-Hammami et al. 2012). It is important to emphasize that the TCA–acetone–phenol method was previously successfully used for date palm tissues such as leaves Gómez-Vidal et al. (2009) and embryos (Sghaier-Hammami et al. 2009a, b, 2010).
Nowadays, the employment of statistics analyses is being widely used, these being the basis of system biology because of their capacity to interrelate different variables and they can sometimes be helpful in selecting candidate spots (Kreutz and Timmers 2008). PCA is a strong approach and is used to obtain an overview of the main variation and any interrelations between spots in the protein patterns (Valledor and Jorrín 2011). Heat map analysis was considered as the most accurate procedure for proteomics data (Meunier et al. 2007). Following the use of the PCA and heat map analyses, it is clear that the remaining spots were different enough to differentiate MFC-affected leaves from the healthy ones, and that the greatest change in the 2-DE protein profile occurred during the S2 disease stage.
The proteomic analysis carried out in the present work has shown that among the 58 successfully identified proteins, 54 have not been reported previously in similar proteomics studies of the MFC-date palm (Marqués et al. 2011). Half of the total identified proteins, may result from a product of isogenes or post-translational modifications (PTMs), variants of specific gene products differing in pI and/or Mr. The presence of protein isoforms with different expression patterns has been reported previously in date palm (Sghaier-Hammami et al. 2009a, b, 2010; Marqués et al. 2011) and in other species like Arabidopsis (Sghaier-Hammami et al. 2012).
Differential identified proteins belonged to different groups including those of photosynthesis and Calvin cycle, carbohydrate metabolism, amino acid biosynthesis, protein fate (synthesis and turnover), stress-related proteins and cytosqueleton. Seven RubisCO protein species have been identified in the present work, six of which seem to be a product of protein degradation processes. It has been demonstrated that RubisCO can be degraded in intact pea chloroplasts (Mitsuhashi et al. 1992). During senescence, RubisCO accounts for about 90 % of the degraded proteins (Miller and Huffaker 1985). Under abiotic stress, a rapid net degradation of RubisCO fragments of the large subunit may become detectable on immunoblots. However, during natural senescence, often only the bands representing the intact large and small subunits (but no fragments) are visible on stained gels or immunoblots (Feller et al. 2008). A clear degradation of RubisCO and other chloroplast proteins can be observed during initiated leaf senescence as well as during or after abiotic stress phases (Crafts-Brandner and Egli 1987; Crafts-Brandner et al. 1998; Demirevska-Kepova et al. 2004; Feller et al. 2008). The observed leaf chlorosis during the S2 disease stage may be in relation with the RubisCO degradation. The degradation of RubisCO reinforces the hypothesis that MFC-affected date palm leaves are caused by an abiotic stress (Mn deficiency).
The increase in RubisCO proteins, as well as the increase in ATP-dependent chloroplastic proteases is observed in MFC-affected leaflets. Chloroplast proteases participate in chloroplast biogenesis during adaptation to changing environmental conditions through degradation of certain proteins, and maintenance of protein quality through degradation of unassembled or damaged proteins (Adam et al. 2006). Roberts et al. (2003) reported the role of serine protease activity in senescing wheat leaves for which RubisCO was a target protein. The ATP-dependent chloroplast proteases identified in our study are classified as serine endopeptidases (http://www.genome.jp/dbget-bin/www_bget?ec:3.4.21.92), and may enhance photoinhibition (http://www.uniprot.org/uniprot/Q9SXJ7). The increase in ATP-dependent clp proteases may also play an important role for the photosystem complexes, since the formation and maintenance of a functional thylakoid electron transport chain require various protease activities, operating in parallel, as well as in series (Dominic et al. 2011). Our previous results (Saidi et al. 2012) showed that MFC led to a decrease in photosynthetic activity and the diminution of chlorophyll content by enzymatic degradation via chlorophyllase. Consequently, we can consider RubisCO and ATP-dependent clp proteases as MFC proteins biomarkers.
MFC led to the decrease in MSP-33 kDa subunit proteins and ATP synthase beta subunit in affected leaflets from three characteristic stages. Mn is a key element in photosynthetic water reduction and oxygen evolution. It is also a constituent of respiratory enzymes (Adriano 2001). Mn deficiency in green plants is known to affect preferentially the activity of the OEC (Homann 1967). In plants, the integrity and optimal function of the OEC depend on the presence of extrinsic MSP-33 kDa subunit proteins, which play a central role in maintaining the integrity and activity of the manganese cluster (Seidler 1996). The decrease in the ATP synthase, MSP-33 kDa subunit proteins and the luminal OEC proteins (PSBO2, PSB, PSBP and PSBO1) in the study of Marqués et al. (2011) reinforce the hypothesis of the relation between the MFC and Mn deficiency. We can consider the decrease in the MSP-33 kDa subunit proteins and ATP synthase proteins as biomarkers of the Mn deficiency in MFC-affected date palm leaves.
Both the chloroplastic and cytosolic metabolic enzyme increased in the affected leaflets. Similarly, it has been demonstrated that chloroplastic transketolase, triosephosphate isomerase and cytoplasmic malate dehydrogenase increased in diseased leaflets (Marqués et al. 2011).
The typical stress response of a plant is the induction of the stress-related proteins including the chloroplastic Hsp70 kDa and chaperonin 60 subunit beta 1 proteins. Also, our transcriptomic analysis showed that a cDNA fragment related to Hsp70 was up-regulated in affected leaves (Saidi et al. 2010). Hsp may play a role in cross-talk with other mechanisms and function synergistically with other components to decrease cell damage (Hartl 1996; Bukau and Horwich 1998). The Chaperonin 60 subunit beta 1 showed itself to be implicated in cell death and systemic acquired resistance (http://www.uniprot.org/uniprot/P21240) and to play a role in acclimating photosynthesis to heat stress, possibly by protecting RubisCO activase from thermal denaturation (Salvucci 2008). Other stress-related proteins identified such as peroxiredoxin increased only at the S2 stage. Plant 2-Cys peroxiredoxins are post-translationally targeted to chloroplasts, where they protect the photosynthetic membrane against photooxidative damage (Baier and Dietz 1997, 1999). At a transcriptomic level, our group has found that manganese deficiency in the BLD affected date palm induces an oxidative stress especially at S2 stage (Saidi et al. 2012).
Of interest is the increase of actin, which has recently been related to programmed cell death (PCD). Recent live cell imaging techniques have revealed dynamic features and significant roles of cytoskeletons during defense responses and PCD (Higaki et al. 2011). One unexpected result is that we did not detect this protein in the affected palm leaf from stage S2; however, it increased at stages S1 and S3.
Finally, we have observed an increase in mitochondrial outer membrane protein porin of 34 kDa from affected leaflets at S2, and this was not observed at S3. Similarly, this protein was also seen to be increased at S2 in the previous study by Marqués et al. (2011). The porin proteins form a channel through the mitochondrial outer membrane that allows the diffusion of small hydrophilic molecules (http://www.uniprot.org/uniprot/Q6K548). The absence of porin at S3 may be explained by the fact that symptoms extend to the entire plant that finally dies.
In conclusion, the present proteomics work permits the dissection of the MFC disease from the early to the last disease steps (death of the palm tree). Changes in the 2-DE protein profile start at early disease stage (S1), in which we have especially observed a decrease in MSP-33 kDa subunit proteins. From the S2 stage, when leaflets became chlorotic, we have observed degradation of the RubisCO proteins. This is not only specific to Mn deficiency, but confirms that MFC is an abiotic stress. From this stage, MSP-33 kDa subunit proteins continue to decrease together with ATP synthase. One unexpected result is the increase of the actin at the S3 stage, in which leaflets break when flexed and squeezed, but it may be a signal of the PCD. At this stage, MSP-33 kDa subunit proteins also decrease and we have observed an extensive increase of the ATP-dependent clp proteases. All the observed protein changes, related to MFC disease, may also be related to the other stress response. However, the decrease in the MSP-33 kDa subunit proteins is closely related to the Mn deficiency. Therefore, MSP-33 kDa subunit proteins can be considered as a MFC biomarker, specifically at an early MFC disease stage, at which symptoms may not reflect the disease’s intensity. As perspective to this work, it would be of interest to seek an antibody against this protein, and with an ELISA test or immunoblot, we could predict the presence of the MFC disease early. Then, the discrimination of the disease could be avoided (by eliminating the diseased tree), since its progression is apparently transmitted from an affected tree to neighboring trees.
Abbreviations
- HC:
-
Healthy control
- IEF:
-
Isoelectric focusing
- MALDI:
-
Matrix-assisted laser desorption ionization
- MFC:
-
Maladie des feullles cassantes (brittle leaf disease)
- MSP:
-
Manganese-stabilizing proteins
- TOF:
-
Time of flight
- RubisCO:
-
Ribulose-1.5-bisphosphate carboxylase/oxygenase large subunit
References
Adam Z, Rudella A, van Wijk KJ (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9:234–240
Adriano DC (2001) Manganese. Trace elements in terrestrial environments. Springer, Berlin, pp 547–585
Baier M, Dietz KJ (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12:179–190
Baier M, Dietz KJ (1999) Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol 119:1407–1414
Bradford M (1976) A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Brumbarova T, Matros A, Mock H-P, Bauer P (2008) A proteomic study showing differential regulation of stress, redox regulation and peroxidase proteins by iron supply and the transcription factor FER. Plant J 54:321–334
Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366
Castillejo MA, Curto M, Fondevilla S, Rubiales D, Jorríın JV (2010a) Two-dimensional electrophoresis based proteomic analysis of the pea (Pisum sativum) in response to Mycosphaerella pinodes. J Agric Food Chem 58:12822–12832
Castillejo MA, Kirchev H, Jorrín JV (2010b) Differences in the Triticale (X Triticosecale Wittmack) flag leaf 2-DE protein profile between varieties and nitrogen fertilization levels. J Agric Food Chem 58:5698–5707
Chich JF, David O, Villers F, Schaeffer B, Lutomski D, Huet S (2007) Statistics for proteomics: experimental design and 2-DE differential analysis. J Chromatogr B 849:261–272
Crafts-Brandner SJ, Egli DB (1987) Sink removal and leaf senescence in soybean: cultivar effects. Plant Physiol 85:662–666
Crafts-Brandner SJ, Hölzer R, Feller U (1998) Influence of nitrogen deficiency on senescence and the amounts of RNA and proteins in wheat leaves. Physiol Plant 102:192–200
Demirevska-Kepova K, Simova-Stoilova L, Stoyanova Z, Hölzer R, Feller U (2004) Biochemical changes in barley plants after excessive supply of copper and manganese. Environ Exp Bot 52:253–266
Dominic BP, Olinares PDB, Kim J, van Wijk KJ (2011) The Clp protease system; a central component of the chloroplast protease network. Biochim Biophys Acta 1807:999–1011
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Feller U, Anders I, Mae T (2008) Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J Exp Bot 59:1615–1624
Gómez-Vidal S, Salinas J, Tena M, Lopez-Llorca LV (2009) Proteomic analysis of date palm (Phoenix dactylifera L.) responses to endophytic colonization by entomopathogenic fungi. Electrophoresis 30:2996–3005
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–580
Higaki T, Kurusu T, Hasezawa S, Kuchitsu K (2011) Dynamic intracellular reorganization of cytoskeletons and the vacuole in defense responses and hypersensitive cell death in plants. J Plant Res doi. doi:10.1007/s10265-10011-10408-z
Homann PH (1967) Studies on the manganese of the chloroplast. Plant Physiol 42:997–1007
Kreutz C, Timmers J (2008) Systems biology: experimental design. FEBS J 276:923–942
Maldonado AM, Echevarría-Zomeño S, Jean-Baptiste S, Hernández M, Jorrín-Novo JV (2008) Evaluation of three different protocols of protein extraction for Arabidopsis thaliana leaf proteome analysis by two-dimensional electrophoresis. J Proteomics 71:461–472
Marqués J, Fadda ZG, Duran-Vila N, Flores R, Bové JM, Daròs JA (2008) A set of novel RNAs transcribed from the chloroplast genome accumulates in date palm leaflets affected by brittle leaf disease. Phytopathology 98:337–344
Marqués J, Duran-Vila N, Daròs JA (2011) The Mn-binding proteins of the photosystem II oxygen-evolving complex are decreased in date palms affected by brittle leaf disease. Plant Physiol Biochem 49:388–394
Meunier B, Dumas E, Piec I, Béchet D, Hébraud M, Hocquette JF (2007) Assessment of hierarchical clustering methodologies for proteomic data mining. J Proteome Res 6:358–366
Miller B, Huffaker LRC (1985) Differential induction of endoproteinases during senescence of attached and detached barley leaves. Plant Physiol 78:442–446
Mitsuhashi W, Crafts-Brandne SJ, Feller U (1992) Ribulose-1.5-bisphosphate carboxylase/oxygenase degradation in isolated pea chloroplasts incubated in the light or in the dark. J Plant Physiol 139:653–658
Namsi A, Montarone M, Serra P, Ben Mahamoud O, Takrouni ML, Zouba A, Khoualdia O, Bové JM, Duran-Vila N (2007) Manganese and brittle leaf disease of date palm trees. J Plant Pathol 89:49–60
Roberts IN, Murray PF, Caputo CP, Passeron S, Barneix AJ (2003) Purification and characterization of a subtilisin-like serine protease induced during the senescence of wheat leaves. Physiol Plantarum 118:483–490
Saadi I, Namsi A, Ben Mahamoud O, Takrouni ML, Zouba A, Bove JM, Duran–Vila N (2006) First report of ‘maladie des feuilles cassantes’ (brittle leaf disease) of date palm in Algeria. Plant Pathol 55:572
Saidi MN, Ladouce N, Hadhri R, Grima–Pettenati J, Drira N, Gargouri–Bouzid R (2010) Identification and characterization of differentially expressed ESTs in date palm leaves affected by brittle leaf disease. Plant Sci 179:325–332
Saidi MN, Jbir R, Ghorbel I, Namsi A, Drira N, Gargouri–Bouzid R (2012) Brittle leaf disease induces an oxidative stress and decreases the expression of manganese–related genes in date palm (Phoenix dactylifera L.). Plant Physiol Biochem 50:1–7
Salvucci ME (2008) Association of Rubisco activase with chaperonin-60b: a possible mechanism for protecting photosynthesis during heat stress. J Exp Bot 59:1923–1933
Schevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858
Seidler A (1996) The extrinsic polypeptides of photosystem II. Biochim Biophys Acta 1277:35–60
Sghaier-Hammami B, Valledor L, Noureddine D, Jorrín Novo JV (2009a) Proteomic analysis of the development and germination of date palm (Phoenix dactylifera L.) zygotic embryos. Proteomics 9:2543–2554
Sghaier-Hammami B, Drira N, Jorrín Novo JV (2009b) Comparative 2-DE proteomic analysis of date palm (Phoenix dactylifera L.) somatic and zygotic embryos. J Proteomics 73:161–177
Sghaier-Hammami B, Jorrín-Novo JV, Gargouri-Bouzid R, Drira N (2010) Abscisic acid and sucrose increase the protein content in date palm somatic embryos, causing changes in 2-DE profile. Phytochemistry 71:1223–1236
Sghaier-Hammami B, Redondo-López I, Maldonado-Alconada AM, Echevarría-Zomeño S, Jorrín-Novo JV (2012) A proteomic approach analysing the Arabidopsis thaliana response to virulent and avirulent Pseudomonas syringae strains. Acta Physiol Plant 34:905–922
Sharov AA, Dudekula DB, Ko MS (2005) A web-based tool for PC and significance analysis of microarray data. Bioinformatics 21:2548–2549
Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18:207–208
Triki MA, Zouba A, Khoualdia O, Ben Mahamoud O, Takrouni ML, Garnier M, Bové JM, Montarone M, Poupet A, Flores R, Darós JA, Fadda ZGN, Moreno P, Duran-Vila N (2003) Maladie des feuilles cassantes” or brittle leaf disease of date palms in Tunisia: biotic or abiotic disease? J Plant Pathol 85:71–79
Valledor L, Jorrín JV (2011) Back to the basics: maximizing the information obtained by quantitative two dimensional gel electrophoresis analyses by an appropriate experimental design and statistical analyses. J Proteomics 74:1–18
Wang W, Vignani R, Scali M, Cresti M (2006) A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27:2782–2786
Zhang YM, Zhao J, Xiang Y, Bian X, Zuo Q, Shen Q, Gai J, Xing H (2011) Proteomics study of changes in soybean lines resistant and sensitive to Phytophthora sojae. Proteome Sci 9:52
Acknowledgments
This work is supported by ‘‘le Ministère de l’Enseignement Supérieur, de la Recherche Scientifique et de la Technologie’’ of Tunisia. We gratefully acknowledge support from Dr. Lola Gutierrez Blàzquez (Proteomics Service, University of Madrid) for mass spectrometry facilities and technical assistance. Authors thank Dr. Lyudmila Simova from Institute of Plant Physiology, Bulgarian Academy of Sciences for critical review of the manuscript, and Dr. Anne-Lise Haenni from Institut Jacque Monod Paris for her kind help in correcting the English.
Author information
Authors and Affiliations
Corresponding author
Additional information
B. Sghaier-Hammami and M. N. Saidi contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sghaier-Hammami, B., Saidi, M.N., Castillejo, M.Á. et al. Proteomics analysis of date palm leaves affected at three characteristic stages of brittle leaf disease. Planta 236, 1599–1613 (2012). https://doi.org/10.1007/s00425-012-1713-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1713-z