Abstract
The biosynthesis of flavonoids, important secondary plant metabolites, has been investigated extensively, but few mutants of genes in this pathway have been identified in rice (Oryza sativa). The rice gold hull and internode (gh) mutants exhibit a reddish-brown pigmentation in the hull and internode and their phenotype has long been used as a morphological marker trait for breeding and genetic study. Here, we characterized that the gh1 mutant was a mutant of the rice chalcone isomerase gene (OsCHI). The result showed that gh1 had a Dasheng retrotransposon inserted in the 5′ UTR of the OsCHI gene, which resulted in the complete loss of OsCHI expression. gh1 exhibited golden pigmentation in hulls and internodes once the panicles were exposed to light. The total flavonoid content in gh1 hulls was increased threefold compared to wild type. Consistent with the gh1 phenotype, OsCHI transcripts were expressed in most tissues of rice and most abundantly in internodes. It was also expressed at high levels in panicles before heading, distributed mainly in lemmas and paleae, but its expression decreased substantially after the panicles emerged from the sheath. OsCHI encodes a protein functionally and structurally conserved to chalcone isomerases in other species. Our findings demonstrated that the OsCHI gene was indispensable for flux of the flavonoid pathway in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flavonoids are a large family of polyphenolic secondary metabolites that are widely distributed in the plant kingdom and are responsible for the coloration of most flowers, fruits and seeds. Besides providing pigmentation, they have a wide variety of biological functions, such as protection against ultraviolet light, modulating polar auxin transport and mediating signal transduction in pollen germination (Koes et al. 2005; Mol et al. 1998). As integral components of our diet, flavonoids have shown antioxidant, anti-proliferative and anti-tumor properties that are beneficial to humans and therefore have attracted increasing attention from researchers. Although there are thousands of different flavonoids in nature, and each plant species has its distinct components and content, those natural products share the same basic structure, modification of which leads to marvelous diversity (Ross and Kasum 2002).
Most flavonoids are biosynthesized from malonyl-CoA and p-coumaroyl-CoA, two precursors derived from carbohydrate metabolic and phenylpropanoid pathways, respectively. The synthesis pathway is initiated by the catalysis of chalcone synthase (CHS), leading to the yellow colored chalcone. In most plant, chalcones are only intermediate to other classes of flavonoids, such as flavanones, dihydroflavonols and ultimately to the anthocyanins, the predominant hydrosoluble pigments in plants. Their production results from the coordinate induction of a series of enzymes, including chalcone isomerase (CHI) and flavonoid hydroxylases (Schijlen et al. 2004).
As one of the best-characterized enzymes in the flavonoid synthesis pathway, CHI was first isolated from soybean (Glycine max) and is ubiquitous in all plants studied (Moustafa and Wong 1967). Earlier studies on this enzyme were focused on the biochemical activity and structure, especially in dicots. Its purification and characterization has been performed for decades in a wide array of species, such as soybean, sweat orange (Citrus sinensis) and petunia (Petunia hybrida) (Bednar and Hadcock 1988; Boland and Wong 1975; Fouche and Dubery 1994; Moustafa and Wong 1967; Van Tunen and Mol 1987). The 3D structure of CHI from alfalfa (Medicago sativa) has been determined, providing insight into aspects of this enzyme’s mode of action, including active site, reaction stereoselectivity and catalytic mechanism (Jez et al. 2000). In recent years, facilitated by molecular genetic approaches great advances have been made in characterizing the genes encoding CHI in various species. Most studies have initiated by investigating genetic loci generating detectable color changes and have revealed a relationship between the CHI gene activity, flavonoid content variation and color change of the petals, fruits or seeds. The Arabidopsis (Arabidopsis thaliana) transparent testa5 (tt5) mutants, named for their reduced seed coat pigmentation, are mutants of the Arabidopsis CHI gene (Shirley et al. 1995). In petunia, two distinct CHI genes denoted as CHI-A and CHI-B are present, and reduction of CHI-A activity causes the accumulation of naringenin chalcone, a yellow pigment, thus changing the pollen color from white to yellow or from blue to green (van Tunen et al. 1988; van Tunen et al. 1990; van Tunen et al. 1991). Similar results were reported in monocots. The natural mutant of CHI in onion (Allium cepa) presents a bleached gold bulb color, which may result from the accumulation of chalcone derivatives (Kim et al. 2004). In barley (Hordeum vulgare), a defective CHI gene causes substantial reduction of flavonoid levels in the primary leaves and accumulation of a novel phenolic compound which is only synthesized in the mutant (Reuber et al. 1997).
Except for a possible role in phenotypic color changes, much remains to be discovered on molecular characterization of the CHI genes, such as expression pattern, subcellular distribution, or other functional aspects of the CHI mutants. Even less information is available on the rice (Oryza sativa) CHI gene OsCHI or the rice flavonoid metabolism, due to the scarcity of mutants of this pathway (Fig. 1). OsCHI was first isolated through sequence homology to the maize (Zea mays) CHI gene, and it was determined that there is only one CHI locus in rice, compared to three in maize (Druka et al. 2003). Later, the biological roles of OsCHI were investigated by complementation in the Arabidopsis tt5 mutant (Shih et al. 2008).
Proposed scheme of the major branch pathways of flavonoid biosynthesis in rice. The enzymes are: OsANS1 anthocyanidin synthase, OsCGT C-glucosyltransferase, OsCHS1 chalcone synthase, OsDFR dihydroflavonol-4-reductase, OsF3H flavanone hydroxylase, OsF3′H flavonoid-3′-hydroxylase, OsFNSI-1 flavone synthase I, OsMaT-2 flavonoid malonyltransferase, RF5 flavonol 3-O-glucosyltransferase, ROMT-9 flavonoid 3′-O-methyltransferase, RUGT-5 and RUGT-10 UDP-glycosyltransferase. The mutant identified for OsDFR is shown in italic between brackets
Most of the researched flavonoid pathway mutants are focused on flowers and seeds, such as the seed color mutants in Arabidopsis and maize or floral color changing mutants of carnation (Dianthus caryophyllus), snapdragon (Antirrhinum majus) and petunia plants (Grotewold 2006; Koes et al. 2005; Lepiniec et al. 2006). The coloration of seed coat in rice has also been genetically deciphered in the past several years (Furukawa et al. 2007; Sweeney et al. 2006). Rice seeds are enveloped by hulls which provide protection against damage from UV irradiation and may also aid in seed dispersal. Hulls include lemmas and palea, which are suggested to be counterparts of sepals in eudicots (Ambrose et al. 2000). Except for studies on golden hull and internode2 (gh2) and inhibitor for brown furrows (ibf), limited work has been done to determine the mechanism of pigmentation in rice hulls or their counterparts in other species, such as lemmas and paleae in maize or sepals in Arabidopsis (Cui et al. 2007; Zhang et al. 2006). Here we analyzed another natural rice hull color mutant golden hull and internode1 (gh1), and found that it was a mutant of the OsCHI gene. Our findings support an essential role for OsCHI in the flavonoid pathway of rice hulls and indicate the possible role flavonoid metabolism might take in the coloration of rice hulls.
Materials and methods
Plant materials
The original rice (Oryza sativa) gh1 mutant was kindly provided by Dr. G. S. Khush at the International Rice Research Institute (Manila, Philippines). The gh1 gene was transferred to Zhefu802 (ssp. indica) by ten rounds of backcrosses with Zhefu802 (Zeng et al. 2003). We obtained the isogenic line with the gh1 gene, while Zhefu802 served as the wild type. The gh1 mutant line was further crossed with a japonica rice variety, Chunjiang06, to construct the F2 mapping population. All plant materials were grown in paddy fields in Beijing in summer unless otherwise stated.
Map-based cloning and complementation tests
Markers used for fine mapping gh1 are provided in Suppl. Table S1. The gh1 locus was located to a narrow ~50-kb region between the two STS markers S5 and S6. A Dasheng retrotransposon insertion was identified in the Os03g0819600 (OsCHI) gene. We created a complementation construct pCGH by inserting an 8-kb genomic DNA fragment containing the entire OsCHI coding region (946 bp), along with the 3,865-bp upstream sequence and the 3,175-bp (947–4,122 bp after ATG) downstream sequence, to the binary vector pCAMBIA1300 (CAMBIA, Australia). A control plasmid, pCGHC, containing only a partial downstream (1,158–4,122 bp after ATG) region was also constructed. The two binary plasmids were introduced into Agrobacterium tumefaciens by electroporation and transformed into rice calli from scutellum for complementation testing according to a published method (Lee et al. 1999).
Genomic DNA southern blotting
Genomic DNA was extracted by standard phenol–chloroform technique. Fifteen micrograms of genomic DNA was digested overnight with Bgl II or Hind III and separated by electrophoresis in 0.8% agarose gels. Completion of digestion was checked with ethidium bromide staining. Genomic DNA was blotted onto a nitrocellulose (Amersham), and hybridized overnight with 32P-labeled gene-specific probes. A 1.3-kb fragment covering the 5′ UTR and the entire OsCHI coding region (155 bp before ATG to 1,130 bp after ATG) was amplified by PCR from the wild-type (Zhefu802) genomic DNA and used to generate probes. Primer sequences are listed in Suppl. Table S2.
Phenotypic characterization
Mutant and wild-type plants, seeds, internodes and panicles at proper developmental stages were observed and photographed. For histological studies, spikelets from wild-type and gh1 at the heading stage were fixed in FAA (formalin/acetic acid/alcohol) overnight at 4°C. After dehydration in a graded ethanol series, the samples were embedded in Technovit 7100 resin (Hereaus Kulzer). Five micron transverse sections were cut using a microtome (Leica RM2145) and photographed.
Gene expression analysis
Total RNA was extracted from various tissues using TRIpure reagent (BioTeke). For RT-PCR, 25 ng of cDNA template were amplified using corresponding gene specific primer pairs. Ubiquitin was amplified as the control. Quantitative RT-PCR was performed on a cycler apparatus (Bio-Rad) using the SYBR Green PCR Master Mix (TIANGEN) according to the manufacturer’s instructions. Amplification was conducted in 96-well optical reaction plates with the following protocol: 94°C for 4 min, 40 cycles of 94°C for 15 s, 58°C for 15 s, and 72°C for 15 s. Expression levels of target gene were quantified using the BioRad CFX96 real-time PCR detection system (Bio-Rad) by a relative quantification method (DDCT). The statistical significance was analyzed by Student’s t test. Data were presented as mean values of at least two biological repeats with SE. Ubiquitin was chosen as a control to normalize all data. For the β-glucuronidase (GUS) histochemical assay, a 2792-bp genomic fragment upstream of the OsCHI gene translation start codon was PCR amplified and cloned into the vector pCAMBIA1301. The resulting plasmid was transformed into rice, and the resulting transgenic plants were analyzed by GUS staining assay as previously described (Zhang et al. 2011). For in situ hybridization, the OsCHI sense and antisense probes were synthesized with T7 RNA polymerase using the digoxigenin RNA labeling kit (Roche). Tissue fixation and RNA in situ hybridization were performed as previously described (Hong et al. 2010). Primer sequences are listed in Suppl. Table S2.
CHI activity assay
The recombinant OsCHI protein was obtained by cloning the OsCHI ORF into the expression vector pET-30a (Novagen), which was transformed into Escherichia coli strain BL21 (DE3) (DingGuo). The soluble histidine (His)-tagged fusion protein was purified using Ni–NTA affinity column (Qiagen) following the instruction manual. The primer sequences are listed in Suppl. Table S2. The CHI activity was performed at 25°C by monitoring absorption loss at 390 nm due to naringenin chalcone isomeration on a Beckman DU-800 spectrophotometer. The reaction proceeded in a 1 ml mixture of 50 mM Tris–HCl buffer (pH 7.6) containing 1% ethanol and 100 μM naringenin chalcone in the presence or absence of purified OsCHI (~50 μg). Naringenin chalcone was synthesized from naringenin (Sigma) as described previously (Moustafa and Wong 1967).
Subcellular localization
To determine its exact subcellular location, the OsCHI ORF was fused in-frame with GFP transcribed from a 35S promoter. The resulting plasmid was transfected or co-transfected with mRFP/mCherry-tagged markers for ER into rice protoplasts as previously described by Zhou et al. (2009). The transfected cells were observed with a confocal laser scanning microscope (Leica TCS SP5). Primer sequences are listed in Suppl. Table S2.
Pigment analysis
Anthocyanin analysis, total flavonoid content measurement, and HPLC separation of methanol extracts were performed according to Li et al. (2006). Lignin content was quantified according to Li et al. (2003). Briefly, dry hulls were ground into powder and incubated in corresponding extraction solutions. Absorbance values were obtained using a Beckman DU-800 spectrophotometer. HPLC analysis was performed using an Agilent 1100 system (Agilent) with a Zorbax 300 SB-C18 reverse-phase analytical column (250 × 4.5 mm, particle size 4 μm). A photodiode array detector (Agilent) was used to record online spectra, from 190 to 400 nm, of the eluates with the peak monitored at 360 nm.
Phylogenetic analysis
The amino acid sequences of CHI proteins were retrieved through database search using the amino acid sequences of alfalfa CHI (M91079). The multiple sequence alignments were performed by Clustal_X 1.83 (Thompson et al. 1997) with parameters of Weight matrix: Gonnet; Gap opening penalty: 10.0 and Gap extension penalty: 0.10. Phylogenetic trees were constructed with the aligned CHI protein sequences using MEGA 3.1 software (Kumar et al. 2004) based on the neighbor-joining method with parameters of Poisson correction model, pairwise deletion, and bootstrap (1,000 replicates; random seed).
Results
Morphological characterization of gh1
Compared to the wild-type variety Zhefu802 (an indica cultivar), gh1 exhibited an obvious golden pigmentation in the panicle and internode at maturity, while coloration of the hull was the most significant phenotype (Fig 2a, b). As shown in Fig. 2c, gh1 panicles before heading were not significantly different from wild-type panicles at the same stage. Once the panicles grew out of the sheaths, golden pigmentation of hulls occurred conspicuously in gh1 panicles, exhibiting a marked difference from the green wild-type panicles (Fig. 2d). Visual inspection of cross sections of wild-type and gh1 spikelets at the heading stages revealed that the pigments in gh1 hulls were deposited mainly in the epidermal layer and the underlying parenchyma cells (Fig. 2e, f). When the mutants were grown in the greenhouse in winter, the golden coloration of hulls was less pronounced (Suppl. Fig. S1). Except for the golden coloration in the specific tissues mentioned above, we did not observe any abnormality in vegetative growth, heading date or panicle branching patterns, as well as any obvious alteration regarding fertility, seed weight or seed number (per panicle) in the gh1 plants (Suppl. Fig. S2).
Pigmented hulls and internodes of the gh1 mutant. a Seeds and internodes of wild-type plants. b Seeds and internodes of gh1 plants displaying golden pigmentation. c Panicles before heading (enveloped by sheath) of the wild-type and gh1 plants. d Panicles after heading (out of sheath). e Transverse sections of wild-type hulls after panicle heading. f Transverse sections of gh1 hulls after panicle heading exhibited significant pigmentation in the silicified cells and parenchyma cells. Scale bars 10 μm
gh1 is a mutant of the OsCHI gene
We analyzed individual plants in the F1 and F2 generation from a cross between gh1 and Chunjiang06 (a japonica cultivar) during the heading stage. In the F1 progenies, all plants showed the wild-type phenotype, indicating that it is a recessive trait. In the F2 population, the segregation ratio of the wild-type and gh1 plants was close to 3:1. These results suggested that the gh1 phenotype was controlled by one recessive gene. To determine the molecular basis of the phenotype, map-based cloning was used to isolate the GH1 gene. We generated a large F2 mapping population between gh1 and Chunjiang06, of which 980 segregants showing the gh1 mutant phenotype were used for fine mapping. gh1 was previously located to 0 cM on the short arm of chromosome 5 according to linkage analysis (Zeng et al. 2003), but through map-based cloning we relocated it to the near-telomere region on chromosome 3. Sequence-tagged site (STS) markers (Suppl. Table S1) were applied to narrow its genetic location to an approximately 50-kb region between two STS markers, S5 and S6 (Suppl. Fig. S3a). Using the databases of the National Center for Biotechnology Information (NCBI), 27 putative genes were identified in this region. Most of those genes are unknown proteins, except four genes which are annotated with putative functions. Sequencing of these four genes suggested that three of them had the same sequences in gh1 and wild-type genomes except the gene Os03g0819600, sequencing of Os03g0819600 in gh1 genome suggested that it had the same open reading frame (ORF) sequence as the wild-type, but when amplifying its 5′ untranslated region (5′ UTR), we obtained a product from the gh1 genome which was much longer than the wild-type counterpart (Suppl. Fig. S3c). Sequencing the long PCR product revealed that the head and tail sequences of this product matched the 5′ UTR sequence of Os03g0819600, and this long sequence was caused by an 7.3-kb insertion in a site 12 bp upstream of ATG (Fig. 3a). The full-length sequence of the insert indicated that it was a Dasheng retrotransposon (Fig. 3b). The Dasheng retrotransposon in the gh1 locus had the Dasheng typical 439-bp long terminal repeat (LTR) and a 5-bp target site duplication in its insertion site, as well as tandem repeats of an 89-bp unit (Jiang et al. 2002). Besides, no Os03g0819600 transcripts were detected in gh1 leaves in contrast to the substantial expression in wild-type leaves (Suppl. Fig. S4), indicating the total knockout of Os03g0819600 in gh1. Os03g0819600 encodes a putative rice chalcone isomerase and has been named as OsCHI before. Therefore we regarded OsCHI as the candidate GH1 gene.
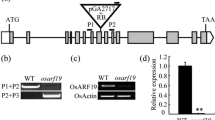
OsCHI is the GH1 gene. a A schematic representation of the OsCHI gene. Black boxes indicate exons of the ORF and white boxes represent the 5′-UTR and 3′-UTR. The arrow indicates the location of the Dasheng retrotransposon insertion. b Diagram of the Dasheng transposon in the gh1 locus. Black arrows represent the 89-bp tandem repeat. LTR long terminal repeat, PBS primer binding site, PPT polypurine tract, TSD target site duplication. c Transgenic lines of pCGH demonstrated complete complementation of the gh1 phenotype and wild-type level of OsCHI expression. Ubiquitin (UBQ) was used as a control
The insertion was verified through Southern blot analysis using the probe of the region covering the 5′ UTR and the entire coding region of OsCHI. No Bgl II or Hind III sites exist in the genomic sequence of OsCHI, and southern blot analysis of the wild-type genomic DNA after digestion with Bgl II and Hind III showed single bands; meanwhile in the gh1 genomic DNA after digestion with Bgl II and Hind III, the detected DNA fragments were longer than those in the wild-type, suggesting that the gh1 allele of OsCHI contained an insertion (Suppl. Fig. S5).
Complementation experiments further confirmed OsCHI as the candidate gene. The plasmid pCGH, containing the entire open reading frame (ORF) of OsCHI with regions 3,865-bp upstream and 3,175-bp downstream of the coding sequence, and the control vector pCGHC containing a partial coding region of the ORF were introduced into calli from gh1 mutants (Suppl. Fig. S3b). Dozens of independent transgenic lines were obtained from the two constructs. All the nine lines of pCGH demonstrated complete complementation of the gh1 phenotype, while no lines of the control vector rescued the gh1 mutant (Fig. 3c). In addition, these pCGH lines had wild-type level of OsCHI expression. Consequently, we determined OsCHI to be the GH1 gene controlling the gh1 phenotype in rice, and the insertion of the retrotransposon Dasheng in the 5′ UTR of OsCHI led to this phenotype. For clarity, OsCHI instead of GH1 was used to refer to the gene responsible for the gh1 phenotype in the following text.
OsCHI is a functional chalcone isomerase localized to the ER
OsCHI encodes a putative rice chalcone isomerase, and homology searches revealed that it is the only rice gene manifesting homology with the alfalfa CHI encoding gene (Fig. 4a). In addition, most of the residues composing the active site cleft of the alfalfa CHI were found to be conserved in OsCHI (Jez et al. 2000). Consequently, we tested whether OsCHI could exhibit activity similar to that of the other chalcone isomerases. The OsCHI protein was expressed in E. coli, purified and its enzymatic activity assessed in vitro. The results indicated that OsCHI was capable of catalyzing the isomerization of naringenin chalcone into naringenin (Suppl. Fig. S6a).
Sequence comparisons of OsCHI and CHIs in other species. a Primary structure of OsCHI and full-length protein alignment of CHIs from alfalfa, Arabidopsis, maize, snapdragon, and sorghum. Sequences were aligned using the CLUSTAL W program. Conserved amino acid residues are highlighted in black shade. The residues of the CHI active site cleft are marked with arrows. b A neighbor-joining tree built on the full-length protein sequences of CHI proteins in higher plants using MEGA 3.1. Bootstrap values are percentage of 1,000 replicates. The black dot shows the position of OsCHI. The scale bar is an indicator of genetic distance based on branch length. A physcomitrella (Physcomitrella patens) protein showing similarity to OsCHI was used as the outgroup. Accession numbers of the sequences used to build the tree are as follows: alfalfa (M91079), Arabidopsis (At3g55120), barley (AK374952), chrysanthemum (Chrysanthemum × morifolium, EF094934), cotton (Gossypium hirsutum EF187439), grape (Vitis vinifera, FJ468358), oil palm (Elaeis oleifera, FJ940770), maize (EU964438), onion (AY541034), pear (Pyrus communis, EF446163), physcomitrella (PHYPADRAFT_233922), poplar (Populus trichocarpa EF146619), rice (Os03g0819600), snapdragon (M68326), sorghum (Sb01g003330), tobacco (Nicotiana tabacum, AB213651), wheat (AK330782)
A BLASTn research of rice genome sequence databases using the OsCHI protein sequence as the template detected no other rice proteins with significant homology. Amino acid sequence comparison of CHIs from a variety of angiosperms revealed high homology between OsCHI and other CHIs. Phylogenetic analysis estimated that OsCHI belonged to a monocot clade comprised of the grass species maize CHI (EU964438), sorghum (Sorghum bicolor) CHI (Sb01g003330), barley CHI (AK374952) and wheat (Triticum aestivum) CHI (AK330782) (Fig. 4b).
To determine the subcellular localization of OsCHI, a green fluorescent protein (GFP) sequence was fused to the ORF of OsCHI driven by the 35S promoter and transformed into rice protoplasts. Unlike the ubiquitous distribution in the control cells expressing GFP alone, the CHI-GFP chimeric proteins displayed a filamentous pattern and were distributed abundantly around the nucleus although they were also found throughout the cell (Suppl. Fig. S6b, c). Furthermore, the distribution pattern of CHI-GFP was identical to that of the co-transformed red fluorescent protein (RFP)-tagged endoplasmic reticulum (ER) marker (Suppl. Fig. S6d). These findings indicate that OsCHI is an ER-localized chalcone isomerase.
The overall conclusion from the above characterization is that OsCHI is a single copy gene in rice encoding a chalcone isomerase, which is functionally and structurally conserved to chalcone isomerases in other species.
Expression pattern of OsCHI
Quantitative RT-PCR (qRT-PCR) was performed using RNA from the wild-type plant to draw an expression profile of OsCHI. The results showed OsCHI to be expressed in most of the tissues analyzed, including leaf, internode, leaf sheath and panicles at later development stages. Strong expression of OsCHI was detected in the internodes and leaf sheaths, as well as in leaves and panicles before heading. After heading OsCHI transcripts in the panicles decreased conspicuously. However, no expression of OsCHI expression above background was found in the panicles before or after heading of gh1 (Fig. 5a).
Tissue expression profile of the OsCHI gene. a qRT-PCR analysis of OsCHI expression. R root, In internode, Sh sheath, L leaf, S seedling, YP young panicles, BP booting panicles (panicles enveloped by the sheath), HP heading panicles (panicles stretching out of the sheath). b–d OsCHI expression pattern revealed by GUS staining: b transverse section of internode; c spikelet; d transverse section of spikelet. e Wild-type spikelet hybridized with OsCHI antisense probe. f Control hybridization with OsCHI sense probe. Scale bars, 100 μm
To localize the temporal and spatial patterns of OsCHI expression, we generated transgenic rice plants expressing the GUS reporter gene fused with the OsCHI promoter. GUS staining of transgenic plants demonstrated that OsCHI was preferentially expressed in the spikelets, especially those at the booting stages (the stages when panicles are enveloped by the leaf sheathes) (Fig. 5d). GUS expression was also visualized in internodes and leaves, with high levels in the vascular bundles, epidermis and mesophyll cells (Fig. 5b, c).
Given the marked phenotype in gh1 spikelets, we performed a more detailed characterization of OsCHI transcripts in panicles by inspecting the in situ expression of the OsCHI gene on sections of wild-type spikelets. The signal was obviously distributed in lemmas and paleae, especially in the epidermal layer and the underlying parenchyma cells (Fig. 5e). The signals were not visualized in spikelets probed with a sense strand OsCHI RNA as a negative control (Fig. 5f). In summary, OsCHI appear to be ubiquitously expressed while with preferential expression in internodes, leaves and panicles. The abundance of OsCHI transcripts in the panicles decrease substantially as the panicles stretch out of the sheath.
The effect of OsCHI mutation on the expression of flavonoid structural genes
A number of genes have been shown to be involved in the flavonoid pathway in rice, and several of them are expressed in panicles, such as OsCGT (a C-glucosyltransferase gene), OsCHS1 (a chalcone isomerase gene), OsDFR (a dihydroflavonol-4-reductase gene), OsF3H (a flavanone hydroxylase gene), OsF3′H (a flavonoid-3′-hydroxylase gene), OsFNSI-1 (a flavone synthase I gene), OsMaT-2 (a flavonoid malonyltransferase gene), RF5 (a flavonol 3-O-glucosyltransferase gene), ROMT-9 (a flavonoid 3′-O-methyltransferase gene), RUGT-5 (a UDP-glycosyltransferase gene) and RUGT-10 (a UDP-glycosyltransferase gene) (Brazier-Hicks et al. 2009; Hong et al. 2007; Kim et al. 2006a, b, 2009; Ko et al. 2006; Lee et al. 2008; Shih et al. 2008). We wondered whether the expression of flavonoid structural genes in panicles was affected by OsCHI mutation. The expression patterns of these genes in panicles before or after heading were compared in wild-type or gh1 plants by qRT-PCR. Largely, these genes displayed differential expression levels in wild-type panicles before or after heading (Suppl. Fig. S8). Most of the genes tested, except for OsMaT-2 and RF5, showed similar expression patterns but generally slightly lower expression levels in gh1 panicles, compared to that of the wild type. However, the gh1 plants had remarkable accumulations of OsMaT-2 mRNA in panicles before heading as well as RF5 and RUGT-5 transcripts in panicles after heading (Fig. 6). In conclusion, a sizeable proportion of flavonoid structural genes exhibit different expression levels after the panicles stretch out of the sheath in wild type, and most of them have not changed expression patterns significantly in gh1 mutants. However, the expression of three genes which are involved in metabolite modifications, namely OsMaT-2, RUGT-5, and RF5, is greatly stimulated in gh1 panicles.
gh1 accumulates substantial amounts of flavonoids
Since OsCHI is the only rice gene encoding the chalcone isomerase in the flavonoid pathway, we checked whether the flavonoid metabolism was affected in gh1. The total contents of flavonoids and anthocyanins were assayed in wild-type and gh1 hulls at maturity. gh1 accumulated higher levels of total flavonoids in hulls, about threefold more than did the wild type, but the anthocyanin content in hulls was reduced by 30% relative to that in wild-type control plants (Fig. 7). The high-performance liquid chromatography (HPLC) profile also showed that gh1 hulls had accumulated higher levels of some pigments than did the wild-type hulls, although we were not able to isolate and identify these compounds due to technical limitations (Suppl. S9a–c). Lignin content was also measured, while no significant difference was detected in the total lignin content of gh1 and wild-type plants, both with lignin composing about one-third of the cell wall.
Discussion
The color trait of gh mutants has long been used as a selection marker in rice breeding. In a previous study we cloned the GH2 gene and found that it encoded a cinnamyl-alcohol dehydrogenase engaged in synthesizing monolignols to provide precursors for the rice lignin biosynthesis pathway. In this study, we characterize that another gh mutant gh1 is caused by the disruption of the OsCHI gene which encodes an enzyme involved in the flavonoid pathway. Because the monolignol biosynthesis pathway and flavonoid biosynthesis pathway are two major downstream pathways of the phenylpropanoid pathway and use the same precursors, we wondered whether the downregulation of OsCHI would promote the flux to the lignin biosynthesis pathway. However, lignin content measurement demonstrates that there is no significant difference in total lignin content between the gh1 hulls and wild-type ones. Therefore the similar pigmentation phenotype in gh1 and gh2 might result from malfunction of different metabolism pathways.
OsCHI has been annotated as a putative chalcone isomerase. Through in vitro enzymatic assay, we determine it could catalyze the isomeration of naringenin chalcone. Analysis on high-resolution 3D structure of CHI has demonstrated that the active site cleft of this enzyme in alfalfa consists of Arg 36, Gly 37, Leu 38, Phe 47, Thr 48, Ile 50, Tyr 106, Lys 109, Val 110, Asn 113, Thr 190 and Met 191 (Jez et al. 2000). In the current study, most of the residues were found to be conserved in OsCHI, but Leu 38, Thr 190 and Met 191 were replaced by Val 38, Ser 190 and Ile 191, respectively, in the rice enzyme (Fig. 4a). Interestingly, these three residues were also not found to be conserved in CHIs of other grass species including maize, barley and sorghum. Thr190 and Met191 are two residues suggested to be related to substrate preference, and the combination of these two residues could only be seen in legume CHIs (Jez et al. 2000). In CHIs from nonlegumes one or both residues is/are usually replaced, for example, by Ser 190 and Ile 191 in OsCHI. Based on observations that OsCHI could accelerate naringenin chalcone isomerization and that it has typical CHI sequence and active sites, together with the report that the OsCHI gene could complement the Arabidopsis CHI mutant tt5, we conclude that OsCHI encodes a functional rice chalcone isomerase.
CHI is the enzyme catalyzing the second step in the flavonoid biosynthesis pathway. In dicots, mutation of the CHI gene would disrupt the flavonoid pathway and result in phenotypic color changes (Forkmann and Dangelmayr 1980; Shirley et al. 1995; van Tunen et al. 1991). Similarly, in barley defective CHI is demonstrated to block flavone biosynthesis in leaves (Reuber et al. 1997). Although CHI catalyzes a key step for the flux in this pathway, mutants about the CHI gene have not yet been reported in rice or maize. Although the maize CHI1 gene can complement the Arabidopsis tt5 phenotype, cells of the maize inbred Black Mexican Sweet (BMS) lacking CHI1 mRNA as well as CHI1 activity can accumulate high levels of flavonoids. Considering the spontaneous isomeration of chalcones to flavavones in maize cells, it has been suggested that CHI activity may be unnecessary for maize anthocyanin production (Dong et al. 2001; Grotewold et al. 1998). In our study, we characterized that gh1 is a rice CHI mutant. The gh1 plant showed golden pigments deposited in its hulls and internodes at maturity. The only apparent reason for the accumulation of pigments in the hulls was the absence of CHI. Thus the work presented here provided strong evidence that CHI is necessary for the flux through the flavonoid pathway in rice, and this differences between the CHI mutants in rice, barley and maize suggest that the necessity of CHI for the flavonoid metabolism varies between species.
Most mutants defective in CHI show a color change in pigment of some floral organs. For example, in petunia CHI mutants have their pollens changed from white to yellow or from blue to green, and the seed coats of Arabidopsis tt5 mutant are nearly transparent compared to the brown color of the wild-type (Shirley et al. 1995; van Tunen et al. 1991). In gh1 mutants, the hulls have a darker pigmentation, changing from the usual yellow color in wild-type plants to a golden color. The detailed mechanism about the pigmentation is not known yet, but OsCHI disruption might play a central role. OsCHI transcripts are localized substantially in the epidermal layer and the underlying parenchyma cells of the hulls where conspicuous pigmentation is also observed. The parallel between the distribution pattern of OsCHI transcripts in wild-type plants and the golden pigment deposition pattern in gh1 plants, indicates that like CHIs in other species, OsCHI is also engaged in floral organ pigmentation.
In conclusion, this study provides to our knowledge the first characterization of a rice chalcone isomerase mutant. The hulls of this mutant exhibited golden pigmentation, and this characteristic phenotype has long been used as a selection marker in rice breeding. In addition to the role in seed production, the gh1 hull coloration might provide other ecological or physiological advantages to the plants such as ultraviolet protection, facilitation of seed dispersal, defense against pathogens and other possible benefits. Therefore, further analysis of gh1 and identifying the causal pigments will contribute to a comprehensive knowledge of these beneficial properties and an enhanced understanding of the flavonoid pathway.
Abbreviations
- ANS:
-
Anthocyanidin synthase
- CGT:
-
C-glycosyltransferase
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol 4-reductase
- ER:
-
Endoplasmic reticulum
- F3H:
-
Flavanone 3-hydroxylase
- F3′H:
-
Flavonoid 3′-hydroxylase
- FGT:
-
Flavonoid 3-O-glucosyltransferase
- FNS:
-
Flavone synthase
- GFP:
-
Green fluorescent protein
- gh :
-
golden hull and internode
- GUS:
-
β-Glucuronidase
- HPLC:
-
High-performance liquid chromatography
- ibf :
-
inhibitor for brown furrows
- RFP:
-
Red fluorescent protein
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- STS:
-
Sequence-tagged Site
- tt :
-
transparent testa
- UGT:
-
UDP-glycosyltransferase
References
Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5:569–579
Bednar RA, Hadcock JR (1988) Purification and characterization of chalcone isomerase from soybeans. J Biol Chem 263:9582–9588
Boland MJ, Wong E (1975) Purification and kinetic properties of chalcone-flavanone isomerase from soya bean. Eur J Biochem 50:383–389
Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284:17926–17934
Cui JJ, Fan SZ, Shao T, Huang ZJ, Zheng DL, Tang D, Li M, Qian Q, Cheng ZK (2007) Characterization and fine mapping of the ibf mutant in rice. J Integr Plant Biol 49:678–685
Dong X, Braun EL, Grotewold E (2001) Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol 127:46–57
Druka A, Kudrna D, Rostoks N, Brueggeman R, von Wettstein D, Kleinhofs A (2003) Chalcone isomerase gene from rice (Oryza sativa) and barley (Hordeum vulgare): physical, genetic and mutation mapping. Gene 302:171–178
Forkmann G, Dangelmayr B (1980) Genetic control of chalcone isomerase activity in flowers of Dianthus caryophyllus. Biochem Genet 18:519–527
Fouche SD, Dubery IA (1994) Chalcone Isomerase from Citrus Sinensis: purification and characterization. Phytochemistry 37:127–132
Furukawa T, Maekawa M, Oki T, Suda I, Iida S, Shimada H, Takamure I, Kadowaki K (2007) The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J 49:91–102
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J, Maddock S, Clair GS, Bowen B (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10:721–740
Hong BS, Kim JH, Kim NY, Kim BG, Chong Y, Ahn JH (2007) Characterization of uridine-diphosphate dependent flavonoid glucosyltransferase from Oryza sativa. J Biochem Mol Biol 40:870–874
Hong LL, Qian Q, Zhu KM, Tang D, Huang ZJ, Gao L, Li M, Gu MH, Cheng ZK (2010) ELE restrains empty glumes from developing into lemmas. J Genet Genomics 37:101–115
Jez JM, Bowman ME, Dixon RA, Noel JP (2000) Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat Struct Biol 7:786–791
Jiang N, Bao Z, Temnykh S, Cheng Z, Jiang J, Wing RA, McCouch SR, Wessler SR (2002) Dasheng: a recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics 161:1293–1305
Kim S, Jones R, Yoo KS, Pike LM (2004) Gold color in onions (Allium cepa): a natural mutation of the chalcone isomerase gene resulting in a premature stop codon. Mol Genet Genomics 272:411–419
Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH (2006a) Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67:387–394
Kim JH, Shin KH, Ko JH, Ahn JH (2006b) Glucosylation of flavonols by Escherichia coli expressing glucosyltransferase from rice (Oryza sativa). J Biosci Bioeng 102:135–137
Kim DH, Kim SK, Kim JH, Kim BG, Ahn JH (2009) Molecular characterization of flavonoid malonyltransferase from Oryza sativa. Plant Physiol Biochem 47:991–997
Ko JH, Kim BG, Hur HG, Lim Y, Ahn JH (2006) Molecular cloning, expression and characterization of a glycosyltransferase from rice. Plant Cell Rep 25:741–746
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lee S, Jeon JS, Jung KH, An G (1999) Binary vectors for efficient transformation of rice. J Plant Biol 42:310–316
Lee YJ, Kim JH, Kim BG, Lim Y, Ahn JH (2008) Characterization of flavone synthase I from rice. BMB Rep 41:68–71
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Li YH, Qian Q, Zhou YH, Yan MX, Sun L, Zhang M, Fu ZM, Wang YH, Han B, Pang XM, Chen MS, Li JY (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15:2020–2031
Li F, Jin Z, Qu W, Zhao D, Ma F (2006) Cloning of a cDNA encoding the Saussurea medusa chalcone isomerase and its expression in transgenic tobacco. Plant Physiol Biochem 44:455–461
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3:212–217
Moustafa E, Wong E (1967) Purification and properties of chalcone-flavanone isomerase from soya bean seed. Phytochemistry 6:625–632
Reuber S, Jende-Strid B, Wray V, Weissenbock G (1997) Accumulation of the chalcone isosalipurposide in primary leaves of barley flavonoid mutants indicates a defective chalcone isomerase. Physiol Plant 101:827–832
Ross JA, Kasum CM (2002) Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu Rev Nutr 22:19–34
Schijlen EGW, de Vos CHR, van Tunen AJ, Bovy AG (2004) Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65:2631–2648
Shih CH, Chu H, Tang LK, Sakamoto W, Maekawa M, Chu IK, Wang M, Lo C (2008) Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228:1043–1054
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18:283–294
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
van Tunen AJ, Mol JN (1987) A novel purification procedure for chalcone flavanone isomerase from Petunia hybrida and the use of its antibodies to characterize the Po mutation. Arch Biochem Biophys 257:85–91
van Tunen AJ, Koes RE, Spelt CE, van der Krol AR, Stuitje AR, Mol JN (1988) Cloning of the two chalcone flavanone isomerase genes from Petunia hybrida: coordinate, light-regulated and differential expression of flavonoid genes. EMBO J 7:1257–1263
van Tunen AJ, Mur LA, Brouns GS, Rienstra JD, Koes RE, Mol JN (1990) Pollen- and anther-specific chi promoters from petunia: tandem promoter regulation of the chiA gene. Plant Cell 2:393–401
van Tunen AJ, Mur LA, Recourt K, Gerats AG, Mol JN (1991) Regulation and manipulation of flavonoid gene expression in anthers of petunia: the molecular basis of the Po mutation. Plant Cell 3:39–48
Zeng DL, Qian Q, Dong GJ, Zhu XD, Dong FG, Teng S, Guo LB, Cao LY, Cheng SH, Xiong ZM (2003) Development of isogenic lines of morphological markers in indica rice. Acta Botanica Sinica 45:1116–1120
Zhang KW, Qian Q, Huang ZJ, Wang YQ, Li M, Hong LL, Zeng DL, Gu MH, Chu CC, Cheng ZK (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140:972–983
Zhang BC, Liu XL, Qian Q, Liu LF, Dong GJ, Xiong GY, Zeng DL, Zhou YH (2011) Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc Natl Acad Sci USA 108:5110–5115
Zhou YH, Li SB, Qian Q, Zeng DL, Zhang M, Guo LB, Liu XL, Zhang BC, Deng LW, Luo GZ, Wang XJ, Li JY (2009) BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J 57:446–462
Acknowledgments
This work was supported by grants from the Ministry of Sciences and Technology of China (2011ZX08001-006 and 2011ZX08009-003), and the National Natural Science Foundation of China (30900885).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2012_1598_MOESM1_ESM.pdf
Suppl. Fig. S1 The golden coloration of gh1 hulls was less pronounced when grown in the greenhouse. Suppl. Fig. S2 The wild-type and gh1 plants. Suppl. Fig. S3 Isolation of the GH1 gene. Suppl. Fig. S4 RT-PCR analysis showing no detectable OsCHI transcripts in gh1 leaves. Suppl. Fig. S5 Southern blotting verifying the presence of an insertion in the OsCHI gene of rice. Suppl. Fig. S6 Molecular characterization of the OsCHI protein. Suppl. Fig. S7 Differential expression of flavonoid pathway genes in panicles before or after heading. Suppl. Fig. S8 Some compounds accumulated in gh1 hulls. Suppl. Table S1 List of the PCR-based molecular markers developed in this study. Suppl. Table S2 The primers for OsCHI molecular characterization. (PDF 728 kb)
Rights and permissions
About this article
Cite this article
Hong, L., Qian, Q., Tang, D. et al. A mutation in the rice chalcone isomerase gene causes the golden hull and internode 1 phenotype. Planta 236, 141–151 (2012). https://doi.org/10.1007/s00425-012-1598-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1598-x