Abstract
The physiological and biochemical changes in fruit ripening produce key attributes of fruit quality including color, taste, aroma and texture. These changes are driven by the highly regulated and synchronized activation of a huge number of ripening-associated genes. In tomato (Solanum lycopersicum), a typical climacteric fruit, the MADS-box transcription factor RIN is one of the earliest-acting ripening regulators, required for both ethylene-dependent and ethylene-independent pathways. Although we previously identified several direct RIN targets, many additional targets remain unidentified, likely including key ripening-associated genes. Here, we report the identification of novel RIN targets by transcriptome and chromatin immunoprecipitation (ChIP) analyses. Transcriptome comparisons by microarray of wild-type and rin mutant tomatoes identified 342 positively regulated genes and 473 negatively regulated genes by RIN during ripening. Most of the positively regulated genes contained possible RIN-binding (CArG-box) sequences in their promoters. Subsequently, we selected six genes from the positively regulated genes and a ripening regulator gene, CNR, and assayed their promoters by quantitative ChIP-PCR to examine RIN binding. All of the seven genes, which are involved in cell wall modification, aroma and flavor development, pathogen defense and transcriptional regulation during ripening, are targets of RIN, suggesting that RIN may control multiple diverse ripening processes. In particular, RIN directly regulates the expression of the ripening-associated transcription factors, CNR, TDR4 and a GRAS family gene, providing an important clue to elucidate the complicated transcriptional cascade for fruit ripening.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fruit ripening occurs as the final stage of fruit development in flowering plants and is a critical period for the determination of fruit quality. During ripening, many kinds of fruit drastically change in composition and texture, becoming more attractive for consumption. In general, fully ripened fruits become softened and rich in vitamins, organic acids, sugars, volatiles, dietary fibers and pigments with antioxidant activity (e.g., carotenoids and flavonoids). These physiological and biochemical changes during ripening are mainly brought about by up- or down-regulation of numerous genes in a highly synchronized fashion. Fruit ripening is thus considered to be a well-coordinated genetically programmed phenomenon. In climacteric fruits such as tomato, apple and banana, ripening is mainly controlled by ethylene and ripening-related transcription factors. However, the regulatory mechanism controlling ripening is both extremely complicated and largely unclear.
In tomato, extensive research has focused on the effect of ethylene and on several ripening mutations such as ripening inhibitor (rin), non ripening (nor) and Colorless non-ripening (Cnr). These mutations result in a similar non-ripe fruit phenotype that includes the inhibition of expression of most ripening-related genes, a lack of climacteric ethylene production during ripening and the inability to respond to exogenous ethylene (Giovannoni 2004; Knapp et al. 1989; Lincoln and Fischer 1988; Thompson et al. 1999). These facts suggest that RIN, NOR and CNR lie upstream of ethylene production and regulate fruit ripening by both ethylene-dependent and -independent pathways. RIN, NOR and CNR encode transcription factors (Giovannoni 2004; Manning et al. 2006; Vrebalov et al. 2002). In addition to RIN, NOR and CNR, additional transcription factor genes, including tomato AGAMOUS-LIKE 1 (TAGL1), HD-ZIP HOMEOBOX PROTEIN-1 (LeHB-1), and APETALA2a (SlAP2a), play a crucial role in fruit ripening (Chung et al. 2010; Gimenez et al. 2010; Itkin et al. 2009; Karlova et al. 2011; Lin et al. 2008; Pan et al. 2010; Vrebalov et al. 2009). Despite the discovery of these genes regulating fruit ripening, the transcriptional regulatory pathway for fruit ripening and the direct interactions between the ripening-related transcription factors are still largely unknown.
The rin mutant has been well characterized and frequently used for molecular and physiological studies on fruit ripening in tomato. RIN encodes a MADS-box protein, and the wild-type RIN locus is adjacent to a MADS-box gene, Macrocalyx (MC; also called LeMADS-MC). A deletion stretching over a part of the protein-coding region of RIN and the intergenic region between RIN and MC causes the rin phenotype (Vrebalov et al. 2002). Gene repression and mutant complementation have demonstrated that RIN regulates tomato ripening (Vrebalov et al. 2002), including both ethylene-dependent and ethylene-independent ripening pathways. RIN belongs to the SEPALLATA (SEP) subfamily of MADS-box genes and is expressed in a ripening-specific manner (Ito et al. 2008; Vrebalov et al. 2002). Recently, antisense suppression revealed that a SEP-like gene (FaMADS9) is responsible for fruit ripening of non-climacteric strawberry (Seymour et al. 2011). This finding suggests that SEP family genes play a central role in the transcriptional regulatory pathway of ripening in both climacteric and non-climacteric fruits. Thus, it is important to understand which genes are targets of the SEP family proteins that are involved in fruit development and how they regulate expression of these targets. To understand the genetic mechanism regulating fruit ripening, transcriptome analyses of tomato have been performed (Alba et al. 2005; Fei et al. 2004; Ozaki et al. 2010). These analyses have provided meaningful genetic information that offers useful hints for elucidating RIN regulation of the expression of ripening-induced genes. Nevertheless, the transcriptional cascade downstream from RIN is still ambiguous because the ripening-induced genes identified by these analyses include both direct RIN target genes and non-targets that are regulated by ethylene or other factors. Recently, we have established a method to identify direct RIN target genes by chromatin immunoprecipitation (ChIP) analysis with an anti-RIN antibody. Using this method, we demonstrated that RIN binds to the promoter regions of six genes involved in ethylene synthesis and cell wall modification and also to the promoter of RIN itself (Fujisawa et al. 2011; Ito et al. 2008). However, the number of target genes that were identified in these previous studies is limited, and therefore a large portion of the targets of RIN remains to be identified.
Here, we report the comprehensive identification of ripening-associated genes whose expression is affected by the rin mutation by microarray analysis comparing wild-type and rin mutant tomato fruits. We also describe the identification of seven novel RIN target genes by subsequent ChIP analysis with the anti-RIN antibody. Our results suggest that RIN regulates cell wall modification, volatile production and pathogen defense during ripening, in addition to climacteric ethylene synthesis. These results also suggest that RIN directly regulates the expression of other key ripening transcription factor genes, CNR, TDR4 and a novel GRAS family gene. We discuss the relationship of RIN with CNR and TDR4 in the transcriptional regulatory pathway for fruit ripening.
Materials and methods
Microarray
The tomato fruits of a wild-type line (a Kagome Co., LTD breeding line, PK331) were harvested at the mature green (G) and pink coloring (P; 4 days after the breaker) stages. The fruits of a rin mutant (a Kagome Co., LTD breeding line, PK353) were also harvested at the G stage and the same ages as the wild-type P stage, as described previously (Kitagawa et al. 2005), because rin mutant fruit does not normally reach the P stage. Total RNA was extracted and purified with an RNeasy Plus Mini kit (Qiagen, Hilden, Germany) from the wild-type and rin mutant tomato fruits as described previously (Kitagawa et al. 2005). RNA integrity was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Hybridization to Agilent-022270 Tomato Gene Expression Microarray 44 K slides (platform ID: GPL10570; Agilent Technologies) was performed according to the manufacturer’s instructions. Briefly, the first-strand cDNA and Cy3-labeled cRNA were synthesized from 0.2 μg of total RNA using a Quick Amp Labeling Kit (Agilent Technologies). The Cy3-labeled cRNA was hybridized to the microarray slides. The fluorescent signal of Cy3 on each probe of the slides was scanned using an Agilent Technology Microarray Scanner at a resolution of 5 μm. The signal intensities of spots for the probes were monitored using Feature Extraction Software ver. 10.5.1.1 (Agilent Technologies). To compare the results of the microarray, signal intensities were normalized by per chip normalization to the 75th percentile using the GeneSpring version ver. 10.0 software (Agilent Technologies). The raw and normalized microarray data are MIAME compliant and have been deposited in the Gene Expression Omnibus database (GEO) database (DataSet GSE28564) at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/geo/). The probes with data flagged as outliers in non-uniformity or population were excluded from the analyses. The changes in the expression level of tomato genes were evaluated by calculating the fold change ratio (FC) of the signal intensity of the probes in the wild type at the P stage relative to those at the G stage (FCWT) or in the rin mutant fruits at the same age as the wild-type P stage relative to those at the G stage (FCrin). P values for the changes between the G and P stages in respective lines were calculated by two-tailed Welch’s t test using the log2-scaled signal intensities of the probes from three independent experiments. To estimate false discovery rates (FDR), q values were calculated from the p values using the QVALUE program (Storey and Tibshirani 2003) with the default setting. Further, genes positively or negatively regulated by RIN were detected by analyzing the expression change score (ECS, the ratio of FCWT relative to FCrin). The number of differentially expressed genes was counted by means of a BLASTN (Altschul et al. 1997) similarity search of tomato ESTs that were used to design probes on the microarray against the annotation provided by the International Tomato Annotation Group (ITAG) version 2 (ITAG2; http://www.solgenomics.net/genomes/Solanum_lycopersicum/index.pl). For this purpose, we adopted the predicted gene showing the highest similarity (at least ≥100 bp alignment length and ≥95% identity) with each EST. Functional annotation of tomato ITAG2-predicted genes was carried out by similarity search using the BLASTP program (Altschul et al. 1997) with an e-value cutoff <0.01 against the Arabidopsis protein database with gene ontology (GO) information provided by TAIR (TAIR10; http://www.arabidopsis.org/).
In silico motif search
Promoter regions (2 kb of the 5′ upstream region of the start codon) of tomato genes were identified from a draft genome sequence (WGS) of tomato released by the International Tomato Genome Sequencing Consortium (version 2.31: http://www.solgenomics.net/about/tomato_sequencing.pl) using the ITAG2 annotation. The promoters were also identified using the BLASTN program (Altschul et al. 1997) against the WGS version 2.31. The FUZZNUC program included in the EMBOSS package (Rice et al. 2000) was used to search the promoter sequences for possible RIN-binding CArG-box motif (CArG-box) sequences [C(C/T)(A/T)6(A/G)G, C(A/T)8G and C(C/T)(A/T)G(A/T)4(A/G)G] (Fujisawa et al. 2011; Ito et al. 2008), ERF-domain containing protein-binding sequences [the GCC-box sequence (AGCCGCC)] (Ohme-Takagi and Shinshi 1995), EIN3/EIL protein-binding sequences [A(T/C)G(A/T)A(C/T)CT] (Kosugi and Ohashi 2000) and the SQUAMOSA-PROMOTER BINDING PROTEIN (SBP)-box protein-binding sequence (CCGTAC) (Cardon et al. 1997; Liang et al. 2008).
Chromatin immunoprecipitation and enrichment test
Chromatin immunoprecipitation experiments were performed as previously described (Fujisawa et al. 2011; Ito et al. 2008) using ripening tomato fruit at the P stage where the expression of RIN is strongly induced. Briefly, DNA fragments bound by RIN in vivo were recovered by ChIP with anti-RIN antibody and purified. The anti-RIN antibody was raised against a 24-amino-acid peptide (YHRYNYGTLEGTQTSSDSQNNYQE, Cys-labeled at the N-terminus) of RIN. The polyclonal rabbit antibody was purified by ammonium sulfate precipitation, ion exchange chromatography and affinity chromatography. The efficacy and specificity of this antibody were tested by enzyme-linked immunoassay (ELISA; data not shown) and Western blotting analysis in our previous study (Ito et al. 2008). Pools of chromatin DNA treated with pre-immune serum without anti-RIN antibody (PI-treated chromatin DNA) and the total input chromatin DNA without ChIP treatment were used as a template for the negative control and standard, respectively. Using the resulting DNA pools as template, the enrichment levels of CArG-box sites in the promoters of ripening-induced genes were monitored by quantitative ChIP-PCR (qChIP-PCR) using the PowerSYBR Green PCR master mix (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions, as previously described (Fujisawa et al. 2011). Nucleotide sequences of the oligonucleotide primers specific to the respective CArG-box sites used in this study are listed in Supplementary Table S1. The measurements [quantification cycle (Cq) values] for the CArG-box sites were normalized with those for the Actin gene, which is free from RIN-binding and was used for qChIP-PCR (Fujisawa et al. 2011; Ito et al. 2008). The enrichment levels were represented as fold changes relative to the input DNA.
Gene expression analysis
The expression of ripening-associated transcription factors was analyzed by reverse transcription PCR (RT-PCR). The tomato fruits of the wild type were harvested at the G, P and red ripe (R; 7 days after the breaker) stages. The fruits of the rin mutant were also harvested at the G stage and at the same ages as the wild-type P and R fruits, as described previously (Kitagawa et al. 2005). Total RNA was extracted and purified from these wild-type and rin mutant tomato fruits with an RNeasy Plus Mini kit (Qiagen) as described above. Further, total RNA was also extracted and purified from tomato (Ailsa Craig cultivar) flower, leaf, root and lateral bud. Complementary DNA was synthesized from the total RNAs with a PrimeScript II first cDNA strand synthesis kit (Takara Biotech, Otsu, Japan). As PCR template, 2 μl of cDNA synthesis reaction mixture was added to 20 μl of reaction mixture containing 1× reaction buffer with 2 mM Mg2+, 0.2 mM of each dNTP, 0.1 unit of ExTaq DNA polymerase (Takara Biotech) and 0.2 μM of each oligonucleotide primer specific to the RIN (5′-ATGGCATTGTGGTGAGCAAAG-3′ and 5′-GTTGATGGTGCTGCATTTTCG-3′) (Fujisawa et al. 2011), CNR (5′-CAAATGGGAAGGGAAGAGAAGC-3′ and 5′-ATCGACCTGGCAAGAAGGATGT-3′), TDR4 (5′-ACCTTCTCGAAACGTCGATCTG-3′ and 5′-TATCCTCTCCATGCAGGAATCG-3′), or Solyc07g052960 genes (5′-ATAAGGCCATTGAAAGGCAAAC-3′ and 5′-CTCCATGAAGGCACCGATATTC-3′) or a gene encoding the clathrin adaptor complexes medium subunit (CAC; SGN-U314153) as a reference (Exposito-Rodriguez et al. 2008) with PCR conditions of 94°C for 2 min, followed by 25 cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 1 min. The PCR products were analyzed by 2% agarose gel electrophoresis.
Quantitative RT-PCR analyses were performed basically as previously described (Fujisawa et al. 2011). Briefly, 1 μl of cDNA synthesis reaction mixture was applied as a template for analysis using PowerSYBR Green PCR master mix (Applied Biosystems) according to the manufacturer’s instructions. Copy numbers of the objective transcripts were calculated from measurements of the quantification cycle (Cq) using standard curves generated from a series of diluted PCR products for the respective genes. The data were normalized with that of the CAC gene as a reference (Exposito-Rodriguez et al. 2008).
Results
Identification by microarray analysis of tomato genes that are positively or negatively regulated by RIN
To identify genes that are regulated by RIN during ripening, we comprehensively monitored the expression of genes in wild-type and rin mutant tomatoes by microarray analysis with RNAs isolated from wild-type fruits that were harvested at the pre-ripening (mature green, G) and ripening (pink coloring, P) stages and from rin mutant fruits that were harvested at the G stage and the same age as the wild-type P fruit. Note that the rin tomatoes harvested at the same age as the wild-type P fruit expressed the mutated RIN gene and did not normally reach the pink stage. First, we analyzed changes in the expression level of tomato genes using the fold change ratio of signal intensity of the probes on the microarray (FCWT for the wild type and FCrin for the rin mutant; for more information, see “Materials and methods”) to detect differentially expressed genes during ripening. Of the 42,745 probes for which we obtained valid signal data, 1,399 and 2,965 probes showed substantial up-regulation (FCWT > 5 and p < 0.05) and down-regulation (FCWT < 0.2 and p < 0.05) with ripening in the wild-type fruit, respectively. On the other hand, 285 and 1,160 probes showed up-regulation (FCrin > 5 and p < 0.05) and down-regulation (FCrin < 0.2 and p < 0.05), respectively, in the rin mutant fruit at the same age as the wild-type P stage relative to the fruit at the G stage. FDR for the p values (<0.05) were calculated as q values, resulting in 4.1% for the wild type and 12.9% for the rin mutant.
Next, we identified genes whose expression was significantly affected by the rin mutation using the ECS, which was defined as the ratio of FCWT to FCrin. As a result, 841 of the 1,399 up-regulated probes apparently showed RIN-dependent up-regulation (ECS > 5), whereas 811 of the 2,965 down-regulated probes showed RIN-dependent down-regulation (ECS < 0.2). A similarity search showed that these 841 and 811 probes were derived from at least 342 and 473 ITAG2-predicted genes in the tomato genome annotation, respectively (Supplementary Tables S2 and S3). This result indicates that our screening could detect 342 genes positively and 473 genes negatively regulated by RIN. The positively regulated gene set included not only the RIN target genes that we have identified, namely 1-Aminocyclopropane-1-carboxylic acid (ACC) synthase 2 (LeACS2), LeACS4, Polygalacturonase (PG), β-Galactosidase 4 (TBG4), Endo-(1,4)-β-mannanase 4 (LeMAN4), α-Expansin 1 (LeEXP1) and RIN itself, but also probable non-targets such as ACC oxidase 1 (LeACO1), Invertase (INV) and Phytoene synthase 1 (PSY1) (Fujisawa et al. 2011; Ito et al. 2008).
Functional analysis of the genes positively or negatively regulated by RIN based on GO
To provide an overview of the expected functions of the genes whose expression is affected by RIN, we classified these genes into functional categories by GO, based on their similarities to their Arabidopsis homologs. We found that the positively and negatively regulated gene sets contained ≥10 genes that were classified into 12 biological processes each, 15 molecular functions each and 12 and 22 cellular components, respectively (Table 1). Some of these categories likely reflect ripening phenomena. For example, the numbers of positively regulated genes were also relatively larger than those of negatively regulated genes in the category related to response to stress: “defense response to fungus, incompatible interaction (GO:0009817)” (Table 1). These categories included ethylene-inducible genes encoding pathogenesis-related (PR) proteins and NP24, which is a putative osmotin protein (Van Kan et al. 1995) (Supplementary Table S2). The positive regulation of these genes might be induced mainly by the increase in climacteric ethylene in the ripening fruit.
In contrast to the examples shown above, the numbers of negatively regulated genes were larger than those of positively regulated genes in several categories involved in photosynthesis, including photosynthesis in the biological process, chlorophyll binding in the molecular function category and chloroplast or light-harvesting complex in the cellular component category (Supplementary Table S2). This may reflect chlorophyll degradation and the transition from chloroplast to chromoplast during the onset of ripening.
Intriguingly, the category associated with transcription factors (GO:0003700) included ten or more genes that were positively and negatively regulated by RIN (Table 1), which may explain in part the changes in the expression patterns of vast numbers of genes at the onset of ripening. These categories included transcription factor genes: a ripening repressor, SlAP2a and a ripening-associated MADS-box gene, TDR4 (Supplementary Table S2). In contrast, other well-known ripening regulator genes, NOR (Solyc10g006880) and CNR (Solyc02g077920), were up-regulated significantly (p < 0.05) at the ripening stage in both the wild-type and the rin mutant fruits [for NOR (probe name A_96_P193259), FCWT = 9.43, FCrin = 4.56 and ECS = 2.07; for CNR (probe name A_96_P079454), FCWT = 4.77, FCrin = 1.71 and ECS = 2.80], suggesting that the expression of NOR and CNR is controlled by both RIN-dependent and -independent mechanisms.
Identification of binding sequences of RIN or ethylene responsive factors in promoter regions of the genes positively regulated by RIN
Because RIN is a positive ripening regulator with transcriptional activation activity (Ito et al. 2008), we focused on identifying genes positively regulated by RIN in a direct manner for further analysis. To identify the direct RIN target genes, we searched for possible RIN-binding sequences [CArG-box sequences: C(C/T)(A/T)6(A/G)G, C(A/T)8G and C(C/T)(A/T)G(A/T)4(A/G)G] (Fujisawa et al. 2011; Ito et al. 2008) in the entire tomato genome (provided by the International Tomato Genome Sequencing Project, version 2.31; 782 Mb). In addition, we also searched for the binding sequences of the ethylene signaling factors, EIN3/EIL [A(C/T)G(A/T)A(C/T)CT] and ERF1 (GCC box; AGCCGCC), to confirm that the identified genes are potentially regulated by either RIN or ethylene, or both. These searches found that CArG-box sequences appeared at an extremely high frequency (in each strand, one CArG-box site per 0.7 kb), which is much higher than their frequency of appearance in ethylene-signaling factor-binding sites (in each strand, one EIN3/EIL binding or GCC box site per 6.3 kb).
Next, we analyzed the promoter sequence (2-kb upstream of the start codon) of each of the 342 genes positively regulated by RIN to find any possible binding sequences for RIN or the ethylene-signaling factors. The results showed that 218 (64%) of the genes contained one or more CArG-box sequences in their promoters but no ethylene-signaling factor-binding sequences (Supplementary Table S4), suggesting these genes are candidates for RIN targets independently of the ethylene pathway. The results also showed that 111 (32%) of the genes contained both CArG-box and ethylene-signaling factor-binding sequences (Supplementary Table S4), suggesting these genes are candidates for RIN targets that are affected by the ethylene pathway. The remaining 13 genes (4%) did not contain any CArG-box sequences (Supplementary Table S4), suggesting these genes are potentially non-targets of RIN and likely regulated by other ripening regulators or indirectly by RIN. No significant difference in the proportion of genes with a CArG-box in their promoters was found between the up-regulated genes (96%) and all tomato genes (33,663 of the 35,802 genes: 94%) (Supplementary Table S4). By contrast, the proportion of positively regulated genes with one or more ethylene-signaling factor-binding sequence (113 genes, 33%) was higher than that of all the tomato genes (8,537 of the 35,802 genes, 24.3%), consistent with the increase in the mRNA level of ethylene-inducible genes during ripening (Supplementary Table S4).
Binding of RIN to CArG-box sequences in the promoters of ripening-induced genes
By using ChIP, we previously identified a subset of RIN targets, which include well-known ripening-associated genes for ethylene synthesis and cell wall modification (Fujisawa et al. 2011). To identify additional targets from the genes positively regulated by RIN, we examined RIN binding to the promoters of putative target genes by a qChIP-PCR enrichment test, which consisted of immunoprecipitating chromatin using the anti-RIN antibody and quantitative PCR analysis to determine whether the promoter sequences were enriched in the precipitate. Based on the presence of CArG-box sequences in their promoters and their ripening-related functions as described below, we selected the following putative target genes: Cel2 (Solyc09g010210: FCWT = 256.9, ECS = 1,081.8), LeXYL1 (Solyc10g047030: FCWT = 66.0, ECS = 16.6), TomloxC (Solyc01g006540: FCWT = 227.2, ECS = 24.9), NP24 (Solyc08g080640: FCWT = 121.7, ECS = 54.7), TDR4 (Solyc06g069430: FCWT = 20.0, ECS = 9.7) and a GRAS gene, Solyc07g052960 (FCWT = 683.5, ECS = 100.6) (Supplementary Table S2).
Cel2 and LeXYL1, both of which encode enzymes involved in cell wall modification, were highly expressed in the ripening fruit but not in the rin mutant in our microarray analysis (Supplementary Table S2), consistent with previous reports (Gonzalez-Bosch et al. 1996; Itai et al. 2003; Lashbrook et al. 1994), suggesting that these genes may play a role in fruit softening during ripening.
TomloxC encodes a lipoxygenase involved in fatty-acid-derived volatile synthesis in tomato fruit (Chen et al. 2004). The expression of TomloxC is likely up-regulated during fruit ripening by both ethylene and developmental factors including RIN, CNR and NOR (Griffiths et al. 1999; Kovacs et al. 2009), but is not induced by wounding (Heitz et al. 1997). Our microarray data also showed that the up-regulation of TomloxC strongly depended on RIN (Supplementary Table S2).
NP24 encodes a putative osmotin protein belonging to the pathogenesis-related group 5 (PR-5) protein family (Grenier et al. 1999). In general, osmotin family genes are induced in response to osmotic stress and wounding and by phytohormones such as abscisic acid, ethylene, methyl jasmonate and salicylic acid (Kononowicz et al. 1992; Larosa et al. 1992; Raghothama et al. 1997; Rodrigo et al. 1991; Singh et al. 1989; Xu et al. 1994). NP24, especially isoform I, also accumulates during ripening (Pressey 1997), consistent with our microarray finding that NP24 mRNA levels increased in a RIN-dependent manner (Supplementary Table S2). These observations suggest the possibility that RIN transcriptionally regulates NP24, although we could not exclude the possibility that this increase may be induced by ethylene as in the case of other osmotin family genes.
TDR4 has been identified as a ripening-induced gene and shows similarity to the Arabidopsis gene FRUITFULL (FUL) (Busi et al. 2003; Litt and Irish 2003; Pnueli et al. 1991). Consistent with this prospect, our microarray data showed that TDR4 was among the genes whose expression was significantly affected by RIN (ECS > 5) (Supplementary Table S2). Further, the involvement of RIN, CNR and TDR4 in the same regulatory network has been proposed based on their expression profiles in the rin and Cnr mutants (Eriksson et al. 2004; Seymour et al. 2008). Thus, we also selected CNR as a putative target in spite of the lower ECS than five as described above.
The Solyc07g052960 gene (cDNA clone LEFL2034M18; GenBank Accession No. AK327648) encoded a predicted protein composed of 429 amino acid residues, which showed significant similarity to GRAS family proteins such as the grape (Vitis vinifera) hypothetical protein (RefSeq Accession No. XP_002275420, 73% amino acid identity), Ricinus communis putative DELLA protein DWARF8 (GenBank Accession No. EEF46646, 73% identity), poplar (Populus trichocarpa) GRAS family transcription factor (GRAS13; GenBank Accession No. EEE75706, 71% identity) and A. thaliana scarecrow-like protein 32 (AtSCL32, also named AtGRAS-18; GenBank Accession No. AEE78610.1, 47% identity). We preferentially selected the Solyc07g052960 gene as a putative target to be subjected to the qChIP-PCR test, because the Solyc07g052960 gene showed an extensively high degree of RIN-dependence: the highest FCWT value (683.5) and the highest ECS value (100.6) among the genes of the category associated with the transcription factors (GO:0003700) (Supplementary Table S2).
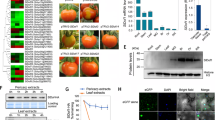
The promoters of each of these genes contained one to four CArG-box sequences (Fig. 1; Table 2). We thus evaluated enrichment of these CArG-box sequences in immunoprecipitated (IPed) DNA that was recovered from ripening fruits at the pink stage with the anti-RIN antibody. Note that the three CArG-box sites in XYL1-a, -b and 07g052960-c were excluded from this test because their flanking DNA sequences were not suitable for designing adequate primers for qChIP-PCR. As shown in Fig. 1, the ChIP treatment highly enriched the CArG-box sites of Cel2-a (16.3-fold relative to the input), TomloxC-a and -b (25.5- and 27.6-fold, respectively), NP24-c and -d (10.9- and 11.4-fold, respectively), CNR-a, -b and -c (22.0-, 16.0- and 11.9-fold, respectively) and 07g052960-b (18.1-fold). The ChIP treatment moderately enriched NP24-b and -d (8.0- and 8.5-fold, respectively), XYL1-c (6.2-fold), TomloxC-c (6.0-fold), TDR4-a and -b (5.0- and 7.1-fold, respectively), and 07g052960-a (9.1-fold), but gave a slightly low-level enrichment of 07g052960-d (3.0-fold). As a negative control, we observed no enrichment of an intron sequence within the DnaJ-like protein gene (Accession No. AF124139), which contains no CArG-box sequences (Fig. 1). Also, ChIP assays with the pre-immune serum (PI) instead of the anti-RIN antibody resulted in no enrichment (0.5- to 2.2-fold) of any of the sequences examined (Fig. 1). The enrichment level of NP24-a (1.9-fold) was close to that of the negative control, indicating that there was no binding of RIN to this site (Fig. 1). These observations indicated that RIN binds in vivo to all the gene promoters examined in this study.
CArG-box sites in the ripening-induced gene promoters and their enrichment in ChIP-DNA. a Position of the CArG-box sites (indicated by the thin open rectangles) found in the region 2 kb upstream of the ripening-induced genes. A pair of primers specific to each site is indicated by pairs of filled arrowheads. When two or more sites are analyzed in the same promoter, they are distinguished by the lower-case letters (a–e) above them. The position of the SBP-binding site found in the TDR4 promoter is indicated by a thin filled rectangle. b ChIP enrichment test of the CArG-box sites. Bars represent the relative DNA amounts of CArG-box sequences in the IPed DNA recovered using either anti-RIN antibody or pre-immune serum (PI) to those in the total input chromatin DNA. Data are the means from two independently prepared IPed DNAs. Error bars indicate the standard deviation of each mean
Expression patterns and specificity of the transcription factors targeted by RIN
Among the RIN targets, ripening-associated transcription factors are keys to elucidating the ripening regulatory mechanism. Thus, to analyze the expression patterns of the CNR, TDR4 and Solyc07g052960 genes in greater detail, we used quantitative RT-PCR (qRT-PCR) to measure mRNA levels in wild-type and rin mutant tomatoes at different stages of development (or different ages in the case of the rin mutant). The mRNA level of each gene was represented as the copy number per copy of the clathrin adaptor complexes medium subunit gene (CAC), which has been identified as a suitable expression control for tomato fruit (Exposito-Rodriguez et al. 2008).
In the wild-type fruits, RIN mRNA was accumulated to substantial levels at the P and R stages but not at the G stage; in the rin mutant fruits, the wild-type RIN mRNA was not accumulated at any stages (Fig. 2), as previously reported (Fujisawa et al. 2011). The mRNA levels of CNR and TDR4 were elevated in the wild-type fruits at the P and R stages compared with that at the G stage (Fig. 2). In the rin mutant fruits, the increases of CNR and TDR4 diminished substantially at all ages examined (Fig. 2). These results of CNR and TDR4 are consistent with our microarray analysis described above and previous reports (Busi et al. 2003; Eriksson et al. 2004; Manning et al. 2006). The mRNA level of Solyc07g052960 gene increased substantially in the wild-type fruits at the P and R stages compared with the level at the G stage (Fig. 2). In the rin mutant fruits, the expression of Solyc07g052960 gene was highly inhibited at all ages examined (Fig. 2).
Gene expression analyses of transcription factors that are direct targets of RIN. mRNA accumulations of the transcription factors in the wild type (filled bars) and rin mutant (open bars) tomato fruits at the mature green (G), pink-coloring (P) and red ripe (R) stages analyzed by qRT-PCR. In the case of the rin mutant, the fruits harvested at the G stage and the same ages as the wild-type P and R fruits were used. Data are the means and standard deviation (error bars) of two biological replicates. CAC was used as a reference for normalization of the measurements among the samples
To examine whether these transcription factors were expressed in other tissues (flower, root, leaf or lateral bud) or were specific to fruit, we monitored their expression by RT-PCR. The results showed that a detectable level of CNR transcript was observed in all the above-ground parts analyzed of the plants (Fig. 3). The TDR4 transcript was detected in all tissues examined (Fig. 3), in agreement with a previous report (Busi et al. 2003). In contrast to these genes, the Solyc07g052960 gene transcript was not detected in any tissues other than fruit, similar to the expression of RIN (Fig. 3) (Ito et al. 2008).
Discussion
Global identification of tomato genes positively and negatively regulated by RIN
During tomato ripening, RIN plays a central role as a ripening regulator. The identification of direct target genes of RIN will therefore provide important clues to understanding the complicated transcriptional cascade regulating ripening. To achieve this, we first screened for RIN target genes by performing microarray analyses of ripening fruits of wild-type and rin mutants based on their ECS values, which represent the degree of dependence of gene expression on RIN. As a result, we identified at least 342 genes positively regulated and 473 genes negatively regulated by RIN, corresponding to 2.0 and 2.7% of the 17,307 ITAG2 predicted genes analyzed by microarray. An earlier paper reported that a number of ripening-related mRNAs were identified whose accumulation is affected by the rin mutation (Picton et al. 1993). Through the microarray analysis, we have achieved the identification of numerous rin-affected genes, including the previously identified genes by Picton et al. (1993) encoding such as uridine diphosphate (UDP) glucuronosyl transferase (Solyc10g085230), short-chain alcohol dehydrogenase (Solyc10g080900), E4 (Solyc03g111720), E8 (Solyc09g089580) and phytoene synthase (Solyc03g031860) (Supplementary Table S2). Thus, a comparative expression analysis using microarray with ECS value is an effective approach to identify mutation-affected genes.
These positively or negatively regulated genes were classified into categories associated with fruit-ripening phenomena, including ethylene production, softening and chlorophyll degradation. As described above, the high ECS gene set contains 13 RIN targets including RIN itself; these targets were identified in our previous and present studies (Fig. 4; Supplementary Table S2). However, the set also included LeACO1 and PSY1, which were previously excluded from the subset of direct targets of RIN (Fujisawa et al. 2011) and 13 positively regulated genes lacking a CArG box in the promoters (Supplementary Table S4). The presence of these probable non-targets, which may be induced by RIN in an indirect manner or by other ripening regulators (e.g., NOR, TAGL1, SlAP2a or LeHB-1), indicates that the use of the ECS value is effective for initial screening of RIN target genes but not sufficient to identify direct targets. Screening for the presence/absence of CArG-box sequences in the promoter of genes was also not sufficient to identify RIN targets due to the high frequency of CArG-box sites in the tomato genome (Supplementary Table S4). The previously reported observation that not all CArG-box sites are bound by RIN (Fujisawa et al. 2011; Ito et al. 2008) makes it difficult to perceive actual target sites. To solve this, it is necessary to elucidate how RIN recognizes and binds specifically to actual target sites during ripening.
A schematic representation of the proposed model for the transcriptional regulation of fruit ripening involving RIN, CNR and TDR4. Arrows indicate the direction of the transcriptional regulatory pathway. The autoregulation of RIN was proposed in our previous study (Fujisawa et al. 2011). A broken line means that RIN may interact with TDR4 to form a functional complex. The RIN target genes identified in this study are underlined
To identify actual targets of RIN from the set of high ECS candidate genes identified by microarray, we conducted qChIP-PCR with the anti-RIN antibody to examine the in vivo binding of RIN to the promoters of the Cel2, LeXYL1, TomloxC, NP24, TDR4 and Solyc07g052960 genes. This analysis proved that all of these genes are direct targets of RIN, suggesting that RIN target genes are enriched in the high ECS gene set. All of the 14 targets that were identified in our previous and current studies (Fig. 4) contain one or more CArG-box sequences in their promoter sequences. In addition, three targets (PG, LeXYL1 and NP24) are included in the subset of 111 genes whose promoters also contain ethylene-signaling factor-binding sequences (Supplementary Table S2), suggesting that their expressions may be affected by both RIN and ethylene-signaling factors. The remaining ten targets except for CNR are included in the subset of 218 genes whose promoters contain no ethylene-signaling factor-binding sequences (Supplementary Tables S2 and S4), supporting the idea that RIN is the main regulator of the transcription of these genes. This could be applied in part to CNR, whose promoter contains three CArG boxes but lacks any ethylene signaling factor-binding sequences. However, we observed in this study that the ECS of CNR was relatively lower than those of the other targets and that CNR was expressed nonspecifically to the ripening fruit. These observations suggest that additional regulatory factor(s), which is independent of RIN and ethylene, contributes to the expression of CNR during ripening in parallel with RIN. More comprehensive ChIP analyses with massively parallel DNA sequencing in combination with our results would lead to the effective elucidation of RIN target genes, as was done with the Arabidopsis floral MADS-box proteins SEP3 (Kaufmann et al. 2009) and AP1 (Kaufmann et al. 2010).
Role of RIN in determining fruit qualities such as tomato-fruit softening, aroma and flavor development and pathogen defense and stress response, taste and pigmentation during ripening
By comparative transcriptome analysis using the rin mutant fruit, we here reveal that the expression levels of numerous genes involved in ripening processes changes are actually affected by the rin mutation, indicating that RIN regulates directly or indirectly these gene expressions as described below.
We previously identified PG, TBG4, LeEXP1 and LeMAN4, which are involved in cell wall modification during fruit ripening, as direct targets of RIN (Fujisawa et al. 2011). In addition to these genes, we reveal here that two other cell-wall modification enzyme genes, Cel2 and LeXYL1, are also direct targets of RIN. It is of particular interest that many genes involved in cell wall modification activity are targets of RIN. Moreover, the positively regulated gene set includes other cell-wall modifying genes encoding Cel5, glucan endo-1,3-β-d-glucosidase (tomQ′b) and xyloglucan endotransglycosylase 4 (XET4), and genes similar to Arabidopsis genes encoding such as glycosyl hydrolase superfamily proteins, pectin lyase-like superfamily proteins, pectinacetylesterase family protein and expansin-like proteins (Supplementary Table S2). These gene activities may be also required for the RIN-dependent fruit softening during ripening. Previous studies showed that the suppression of several gene expressions for cell wall modification enzymes, such as PG, PME and TBG4, results in a limited effect on the inhibition of fruit softening (reviewed by Giovannoni 2004), in contrast to the rin mutation, which results in complete inhibition of softening. These facts suggest that fruit softening is achieved by the cooperation of many genes involved in cell wall modification, with RIN playing a crucial role in the transcriptional regulation of these genes.
We also reveal that TomloxC, which is involved in aroma and flavor generation (Chen et al. 2004), is a direct target of RIN. Griffiths et al. (1999) concluded that, during ripening, a developmental pathway initiates TomloxC expression and an ethylene-dependent pathway enhances TomloxC mRNA levels once its expression has been initiated. Our findings suggest that RIN is a necessary component of the developmental pathway of ripening that initiates TomloxC expression. In plants, lipoxygenases participate in the metabolic pathway that forms volatile C6 aldehydes and alcohols, such as n-hexanal, (Z)-3-hexenal, (E)-2-hexenal and (Z)-3-hexenol, which are components of fruit quality (Alexander and Grierson 2002; Chen et al. 2004; Ortiz-Serrano and Gil 2010). The suppression of TomloxC expression caused a reduction in the accumulation of these volatiles in ripening tomatoes, indicating that TomloxC plays a key role in the volatile production (Chen et al. 2004). Thus, RIN may contribute to the aroma and flavor development in ripening tomato fruit through direct transcriptional regulation of TomloxC. Although lipoxygenases generally act as key enzymes in jasmonate synthesis, it remains unclear whether TomloxC participates in jasmonate synthesis during ripening, because endogenous jasmonate concentrations are not associated with TomloxC transcript accumulation (Fan et al. 1998). Moreover, the positively regulated gene set includes two genes (Solyc08g066220 and Solyc08g066240, Supplementary Table S2) that encode proteins significantly similar to the tomato aromatic amino acid decarboxylases (AADC1A, AADC1B and AADC2; 65–66% amino acid identities), belonging to pyridoxal phosphate (PLP)-dependent transferase superfamily. AADCs are known to participate in synthesis of other flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde in tomato fruit (Tieman et al. 2006). Therefore, RIN might contribute to the aroma and flavor development also via the RIN-dependent transcriptional regulation of the two genes although further analyses for their roles in the volatile production are required.
Our analyses showed that RIN could bind preferentially to the NP24 promoter at the two CArG-box sites, indicating that NP24 is also a direct target of RIN. This osmotin homolog is induced by ethylene, possibly via ERFs that bind to two GCC boxes in its promoter (Hongxing et al. 2005; Raghothama et al. 1997; Zhang et al. 2004). The high level of ethylene accumulation during ripening has thus been expected to lead to the induction of NP24 in the ripening fruit. In addition, the binding of RIN to the NP24 promoter revealed in this study suggests the participation of RIN in the transcriptional regulation of NP24 during ripening. Such bimodal regulation is also observed for other ripening-related genes, such as PG and LeACS2, that are regulated by ethylene and RIN (Fujisawa et al. 2011). Thus, RIN regulation of NP24 expression may contribute to ripening, although the role of NP24 in the ripening tomato is unknown so far. A possible role of NP24 in the ripening tomato is expected to be pathogen defense, due to its antifungal activity as a β-1,3-glucanase (Grenier et al. 1999). Further, the positively regulated gene set includes biotic or abiotic stress-inducible genes encoding such as chitinase family proteins, PR proteins, peroxidase family proteins and glutathione S-transferases (Supplementary Table S2).
The positively regulated gene set also includes genes involved in carotenoid and flavonoid synthesis pathways (Supplementary Table S2). For carotenoid synthesis, the set includes the genes encoding phytoene synthases (PSY1 and PSY2), 15-cis-ζ-carotene isomerase (Z-ISO) and carotenoid isomerase (CRTISO). Both Z-ISO and CRTISO are required to convert colorless 15-cis-phytoene to the red-colored all-trans-lycopene. Moreover, the set also includes four genes encoding 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase (ISPE), 1-d-deoxyxylulose 5-phosphate synthase (DXS), geranylgeranyl pyrophosphate synthase 2 (GGPS2) and hydroxy methylglutaryl CoA reductase 2 (HMGR2). In isoprenoid synthesis, ISPE, DXS and GGPS2 belong to the 1-deoxy-d-xylulose-phosphate/2-C-methylerythritol 5-phosphate (DOXP/MEP) pathway in plastids, whereas HMGR2 belongs to the mevalonate (MVA) pathway in cytosol. DOXP/MEP pathway, and possibly also MVA pathway, lies upstream of the carotenoid pathway (Lichtenthaler 2007). For flavonoid synthesis, the positively regulated gene set includes the gene encoding a protein similar to Arabidopsis chalcone synthase (CHS). CHS catalyzes the condensation of 4-coumaroyl CoA and three malonyl CoA to produce naringenin chalcone, which is the first step of flavonoid synthesis pathway (Winkel-Shirley 2001). Further, the set also includes the genes encoding phenylalanine ammonia lyase 1 (PAL1) and cinnamate-4-hydroxylase (C4H, also named CYP73A5), involved in the core reactions of the general phenylpropanoid pathway upstream of the flavonoid pathway (Winkel-Shirley 2001). Thus RIN controls the pigment accumulation during ripening via up-regulation of the expression of the genes involved in the carotenoid and flavonoid biosynthesis pathways.
The positively regulated gene set includes also INV and a gene encoding a protein similar to Arabidopsis sucrose synthase (SUS), involved in sucrose metabolism (Nguyen-Quoc and Foyer 2001). The accumulation levels of reducing sugars (fructose and glucose) are increased during ripening (Gautier et al. 2008), suggesting that RIN participates in controlling taste by transcriptional regulation of these genes.
On the other hand, we could not find genes obviously involved in the organic acid synthesis, chlorophyll degradation and the respiratory climacteric in the positively regulated gene set. In summary, RIN plays a pivotal role in determining fruit quality of tomato such as softening, aroma and flavor development, pathogen defense and stress response, taste and pigmentation. In this study, we identify at least seven genes involved in the fruit ripening as direct RIN targets, but many more RIN-dependently expressed genes remain to be identified as targets of RIN or not. Further identification of direct RIN targets will bring a better understanding of the ripening regulatory mechanism.
RIN targets ripening-associated transcription factor genes
So far, a number of transcription factors involved in fruit ripening have been identified, but little is known about their interactions in vivo. In this study, we demonstrate that RIN directly binds to promoters of the ripening-associated transcription factor genes, CNR and TDR4, suggesting that RIN directly regulates the expression of these transcription factors. The direct regulation of CNR by RIN is likely consistent with the phenotypic similarity between rin and Cnr mutants, the fruits of which fail to ripen. On the other hand, the epigenetic modification site (286 bp) of the Cnr mutant allele lies farther upstream (>2 kb) (Manning et al. 2006) in the CNR promoter than the CNR-a, -b and -c sites, and no CArG-box sequences are affected by the epigenetic modifications. In addition, CNR is expressed in tissues where RIN expression is not evident (Fig. 2). These facts imply that other RIN-independent factor(s) also regulate CNR expression; thus the increased level of CNR expression during ripening requires RIN and the additional factor(s) as described above.
TDR4 is expected to be a target of CNR much as in the case of Arabidopsis FUL, whose promoter is bound by an Arabidopsis SBP-like protein, SPL3 (Yamaguchi et al. 2009). In actuality, the Cnr mutation inhibits TDR4 expression during ripening (Eriksson et al. 2004; Manning et al. 2006; Seymour et al. 2008, 2002). We found a sequence, CCGTAC, conserved among SBP-binding sites (Cardon et al. 1997; Liang et al. 2008) at −1,829 to −1,824 bp upstream of the start codon in the TDR4 promoter (Fig. 1), supporting the interaction of CNR with the TDR4 promoter. On the other hand, our results that TDR4 expression is reduced in rin mutant fruit and that RIN binds to the TDR4 promoter suggest that RIN increases TDR4 expression during ripening. These observations indicate that TDR4 expression is likely to be regulated by both RIN and CNR, as previously proposed by Seymour et al. (2002). Although the role of TDR4 in fruit ripening remains unclear, TDR4 (alternatively named TM4) may interact with RIN to form a functional complex (Fig. 4), as previously revealed by yeast two-hybrid assays (Leseberg et al. 2008). Taking these results together, we propose a hypothetical model of the interaction between RIN and two other transcription factors, CNR4 and TDR4, for the regulation of fruit ripening, as shown in Fig. 4. This model could explain how CNR and TDR4 specifically exert their effect on ripening.
Our results also revealed that the ripening-specific Solyc07g052960 gene, which is identical to the previously reported TC118434 (Fei et al. 2004), is a target of RIN. We could not detect ERF-domain-containing protein-binding sequences (the GCC box; Ohme-Takagi and Shinshi 1995) or the EIN3/EIL protein-binding sequences (Kosugi and Ohashi 2000) in the Solyc07g052960 gene promoter [Supplementary Table S2 and the data for the region at least 5 kb upstream of the start codon (not shown)], implying that RIN contributes directly to regulation of the Solyc07g052960 gene but ethylene-signaling transcription factors may not.
A similarity search indicates that the Solyc07g052960 gene belongs to the GRAS gene family. GRAS family members are transcription factors involved in a diverse range of processes, such as root development, shoot maintenance, axillary meristem development, phytochrome signaling and gibberellin signaling (Bolle 2004; Hirsch and Oldroyd 2009). In tomato, the LATERAL SUPPRESSOR (LS) gene, which is required for axillary meristem formation (Greb et al. 2003), and 17 putative genes, some of which are involved in the response to biotic and abiotic stress (Mayrose et al. 2006), have been identified as GRAS family genes. Intriguingly, the Solyc07g052960 gene sequence shows low conservation with these known tomato GRAS genes at the amino acid level (~28% identity). Although further analysis such as suppression or overexpression will be required to clarify the Solyc07g052960 gene function, we expect that the Solyc07g052960 gene plays a role in fruit ripening due to its ripening-specific expression and direct transcriptional regulation by RIN.
Besides CNR, TDR4 and Solyc07g052960 gene, RIN induces the expression of at least 22 genes encoding proteins with transcription factor activity in the positively regulated gene set (Table 1 and Supplementary Table S2), including SlAP2a, which acts as a repressor for ethylene synthesis during ripening (Chung et al. 2010; Karlova et al. 2011). Further analysis to identify direct targets of RIN from these transcription factors will lead to unveiling the transcriptional network for fruit ripening.
References
Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17:2954–2965
Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53:2039–2055
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Busi MV, Bustamante C, D’Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E (2003) MADS-box genes expressed during tomato seed and fruit development. Plant Mol Biol 52:801–815
Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J 12:367–377
Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136:2641–2651
Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64:936–947
Eriksson EM, Bovy A, Manning K, Harrison L, Andrews J, De Silva J, Tucker GA, Seymour GB (2004) Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol 136:4184–4197
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Fan XT, Mattheis JP, Fellman JK (1998) A role for jasmonates in climacteric fruit ripening. Planta 204:444–449
Fei Z, Tang X, Alba RM, White JA, Ronning CM, Martin GB, Tanksley SD, Giovannoni JJ (2004) Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J 40:47–59
Fujisawa M, Nakano T, Ito Y (2011) Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol 11:26
Gautier H, Diakou-Verdin V, Benard C, Reich M, Buret M, Bourgaud F, Poessel JL, Caris-Veyrat C, Genard M (2008) How does tomato quality (sugar, acid, and nutritional quality) vary with ripening stage, temperature, and irradiance? J Agric Food Chem 56:1241–1250
Gimenez E, Pineda B, Capel J, Anton MT, Atares A, Perez-Martin F, Garcia-Sogo B, Angosto T, Moreno V, Lozano R (2010) Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS One 5:e14427
Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell 16(Suppl):S170–S180
Gonzalez-Bosch C, Brummell DA, Bennett AB (1996) Differential expression of two endo-1, 4-[beta]-Glucanase genes in pericarp and locules of wild-type and mutant tomato fruit. Plant Physiol 111:1313–1319
Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17:1175–1187
Grenier J, Potvin C, Trudel J, Asselin A (1999) Some thaumatin-like proteins hydrolyse polymeric beta-1,3-glucans. Plant J 19:473–480
Griffiths A, Barry C, Alpuche-Solis AG, Grierson D (1999) Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J Exp Bot 50:793–798
Heitz T, Bergey DR, Ryan CA (1997) A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol 114:1085–1093
Hirsch S, Oldroyd GE (2009) GRAS-domain transcription factors that regulate plant development. Plant Signal Behav 4:698–700
Hongxing Z, Benzhong Z, Bianyun Y, Yanling H, Daqi F, Wentao X, Yunbo L (2005) Cloning and DNA-binding properties of ethylene response factor, LeERF1 and LeERF2, in tomato. Biotechnol Lett 27:423–428
Itai A, Ishihara K, Bewley JD (2003) Characterization of expression, and cloning, of beta-d-xylosidase and alpha-l-arabinofuranosidase in developing and ripening tomato (Lycopersicon esculentum Mill.) fruit. J Exp Bot 54:2615–2622
Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60:1081–1095
Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55:212–223
Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23:923–941
Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7:e1000090
Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueno F, Krajewski P, Meyerowitz EM, Angenent GC, Riechmann JL (2010) Orchestration of floral initiation by APETALA1. Science 328:85–89
Kitagawa M, Ito H, Shiina T, Nakamura N, Inakuma T, Kasumi T, Ishiguro Y, Yabe K, Ito Y (2005) Characterization of tomato fruit ripening and analysis of gene expression in F-1 hybrids of the ripening inhibitor (rin) mutant. Physiol Plantarum 123:331–338
Knapp J, Moureau P, Schuch W, Grierson D (1989) Organization and expression of polygalacturonase and other ripening related genes in Ailsa Craig Neverripe and Ripening inhibitor tomato mutants. Plant Mol Biol 12:105–116
Kononowicz AK, Nelson DE, Singh NK, Hasegawa PM, Bressan RA (1992) Regulation of the osmotin gene promoter. Plant Cell 4:513–524
Kosugi S, Ohashi Y (2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res 28:960–967
Kovacs K, Fray RG, Tikunov Y, Graham N, Bradley G, Seymour GB, Bovy AG, Grierson D (2009) Effect of tomato pleiotropic ripening mutations on flavour volatile biosynthesis. Phytochemistry 70:1003–1008
Larosa PC, Chen Z, Nelson DE, Singh NK, Hasegawa PM, Bressan RA (1992) Osmotin gene expression is posttranscriptionally regulated. Plant Physiol 100:409–415
Lashbrook CC, Gonzalez-Bosch C, Bennett AB (1994) Two divergent endo-beta-1, 4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell 6:1485–1493
Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L (2008) Interaction study of MADS-domain proteins in tomato. J Exp Bot 59:2253–2265
Liang X, Nazarenus TJ, Stone JM (2008) Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry 47:3645–3653
Lichtenthaler HK (2007) Biosynthesis, accumulation and emission of carotenoids, alpha-tocopherol, plastoquinone, and isoprene in leaves under high photosynthetic irradiance. Photosynth Res 92:163–179
Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55:301–310
Lincoln JE, Fischer RL (1988) Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol 88:370–374
Litt A, Irish VF (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165:821–833
Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952
Mayrose M, Ekengren SK, Melech-Bonfil S, Martin GB, Sessa G (2006) A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol Plant Pathol 7:593–604
Nguyen-Quoc B, Foyer CH (2001) A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot 52:881–889
Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173–182
Ortiz-Serrano P, Gil JV (2010) Quantitative comparison of free and bound volatiles of two commercial tomato cultivars (Solanum lycopersicum L.) during ripening. J Agric Food Chem 58:1106–1114
Ozaki S, Ogata Y, Suda K, Kurabayashi A, Suzuki T, Yamamoto N, Iijima Y, Tsugane T, Fujii T, Konishi C, Inai S, Bunsupa S, Yamazaki M, Shibata D, Aoki K (2010) Coexpression analysis of tomato genes and experimental verification of coordinated expression of genes found in a functionally enriched coexpression module. DNA Res 17:105–116
Pan IL, McQuinn R, Giovannoni JJ, Irish VF (2010) Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J Exp Bot 61:1795–1806
Picton S, Gray J, Barton S, AbuBakar U, Lowe A, Grierson D (1993) cDNA cloning and characterisation of novel ripening-related mRNAs with altered patterns of accumulation in the ripening inhibitor (rin) tomato ripening mutant. Plant Mol Biol 23:193–207
Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E (1991) The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J 1:255–266
Pressey R (1997) Two isoforms of NP24: a thaumatin-like protein in tomato fruit. Phytochemistry 44:1241–1245
Raghothama KG, Maggio A, Narasimhan ML, Kononowicz AK, Wang G, D’Urzo MP, Hasegawa PM, Bressan RA (1997) Tissue-specific activation of the osmotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol Biol 34:393–402
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277
Rodrigo I, Vera P, Frank R, Conejero V (1991) Identification of the viroid-induced tomato pathogenesis-related (PR) protein P23 as the thaumatin-like tomato protein NP24 associated with osmotic stress. Plant Mol Biol 16:931–934
Seymour GB, Manning K, Eriksson EM, Popovich AH, King GJ (2002) Genetic identification and genomic organization of factors affecting fruit texture. J Exp Bot 53:2065–2071
Seymour G, Poole M, Manning K, King GJ (2008) Genetics and epigenetics of fruit development and ripening. Curr Opin Plant Biol 11:58–63
Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K (2011) A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria x ananassa Duch.) fruit, a non-climacteric tissue. J Exp Bot 62:1179–1188
Singh NK, Nelson DE, Kuhn D, Hasegawa PM, Bressan RA (1989) Molecular cloning of osmotin and regulation of its expression by ABA and adaptation to low water potential. Plant Physiol 90:1096–1101
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100:9440–9445
Thompson AJ, Tor M, Barry CS, Vrebalov J, Orfila C, Jarvis MC, Giovannoni JJ, Grierson D, Seymour GB (1999) Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol 120:383–390
Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ (2006) Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA 103:8287–8292
Van Kan JA, Cozijnsen T, Danhash N, De Wit PJ (1995) Induction of tomato stress protein mRNAs by ethephon, 2, 6-dichloroisonicotinic acid and salicylate. Plant Mol Biol 27:1205–1213
Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296:343–346
Vrebalov J, Pan IL, Arroyo AJ, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, Irish VF (2009) Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21:3041–3062
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Xu Y, Chang P, Liu D, Narasimhan ML, Raghothama KG, Hasegawa PM, Bressan RA (1994) Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6:1077–1085
Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17:268–278
Zhang H, Huang Z, Xie B, Chen Q, Tian X, Zhang X, Lu X, Huang D, Huang R (2004) The ethylene-, jasmonate-, abscisic acid- and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220:262–270
Acknowledgments
The authors express their sincere thanks to Drs. M. Kitagawa and J. Kimbara of the Research Institute of Kagome Co., Ltd (Tochigi, Japan) for the provision of tomato fruits. This work was supported in part by the Program for Promotion of Basic and Applied Researches for Innovations in the Bio-oriented Technology Research Advancement Institution (BRAIN) of Japan to Y.I.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fujisawa, M., Shima, Y., Higuchi, N. et al. Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235, 1107–1122 (2012). https://doi.org/10.1007/s00425-011-1561-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1561-2