Abstract
cDNAs and genes encoding a hydroxycinnamoyl-CoA:hydroxyphenyllactate hydroxycinnamoyltransferase (CbRAS; rosmarinic acid synthase) and a hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase (CbHST) were isolated from Coleus blumei Benth. (syn. Solenostemon scutellarioides (L.) Codd; Lamiaceae). The proteins were expressed in E. coli and the substrate specificity of both enzymes was tested. CbRAS accepted several CoA-activated phenylpropenoic acids as donor substrates and d-(hydroxy)phenyllactates as acceptors resulting in ester formation while shikimate and quinate were not accepted. Unexpectedly, amino acids (d-phenylalanine, d-tyrosine, d-DOPA) also yielded products, showing that RAS can putatively catalyze amide formation. CbHST was able to transfer cinnamic, 4-coumaric, caffeic, ferulic as well as sinapic acid from CoA to shikimate but not to quinate or acceptor substrates utilized by CbRAS. In addition, 3-hydroxyanthranilate, 3-hydroxybenzoate and 2,3-dihydroxybenzoate were used as acceptor substrates. The reaction product with 3-aminobenzoate putatively is an amide. For both enzymes, structural requirements for donor and acceptor substrates were deduced. The acceptance of unusual acceptor substrates by CbRAS and CbHST resulted in the formation of novel compounds. The rather relaxed substrate as well as reaction specificity of both hydroxycinnamoyltransferases opens up possibilities for the evolution of novel enzymes forming novel secondary metabolites in plants and for the in vitro formation of new compounds with putatively interesting biological activities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hydroxycinnamic acid esters and amides such as hydroxycinnamoylshikimate, hydroxycinnamoylquinate, hydroxycinnamoyltyramine, hydroxycinnamoylmalate and rosmarinate and their derivatives are widely found in the plant kingdom (Fig. 1; Petersen et al. 2009). It is supposed that most of these compounds are active as plant defense compounds against pathogens and herbivores and as UV protectants. Hydroxycinnamoylshikimate, which in most plants is not accumulated to high amounts, has recently been assigned an important role in the biosynthesis of monolignols (Hoffmann et al. 2004; Wagner et al. 2007).
Different enzyme classes can be involved in the biosyntheses of these compounds, namely the transfer of a hydroxycinnamoyl moiety to an acceptor substrate. The family of BAHD acyltransferases was first described by St. Pierre and De Luca (2000). Serine carboxypeptidase-like acyltransferases transfer hydroxycinnamoyl moieties from hydroxycinnamoyl glucose esters to the acceptor substrate (Steffens 2000; Milkowski and Strack 2004; Stehle et al. 2008a, b). Enzymes of this family are involved, e.g. in the formation of sinapoylmalate in Brassicaceae species (Lehfeldt et al. 2000) or acylated triterpene saponins in Avena sativa (Mugford et al. 2009). A third enzyme family is the family of acyl-CoA N-acyltransferases, represented by tyramine N-hydroxycinnamoyltransferase (THT), serotonin N-hydroxycinnamoyltransferase (SHT) or caffeoyl-CoA:putrescine N-caffeoyltransferase (PCT) (Facchini et al. 2002; Kang et al. 2006). Hydroxycinnamoyl moieties are transferred to aromatic amines (e.g. tyramine, tryptamine) or to polyamines (e.g. putrescine, spermine, spermidine) resulting in neutral or basic amides. The hydroxycinnamoyl amides formed by THT-like enzymes are widely distributed in the plant kingdom (Martin-Tanguy et al. 1978). They can be integrated into the cell wall and might play an important role in plant defense. Further functions in growth and development have been reported (Facchini et al. 2002). It should be pointed out here that N-acylation can also be catalyzed by acyltransferases of the BAHD superfamily, e.g. in the formation of hydroxycinnamoylagmatine in barley or of avenanthramides in oat (Burhenne et al. 2003; Yang et al. 2004). Also the formation of phenylpropanoid polyamine conjugates in Arabidopsis thaliana is catalyzed by a BAHD-like acyltransferase (Fellenberg et al. 2009; Grienenberger et al. 2009; Luo et al. 2009).
The superfamily of BAHD acyltransferases has been named after the first four enzymes identified for this family (St. Pierre and De Luca 2000; D’Auria 2006). Members of this family have been detected in fungi and plants. They are active as monomers and have a molecular mass of 48–55 kDa. The superfamily is characterized by two mostly conserved sequence motifs, the HxxxD(G) motif containing the catalytically active histidine residue and the DFGWG motif close to the C terminus. Phylogenetic analyses reveal four to six clades (St. Pierre and De Luca 2000; D’Auria 2006; Yu et al. 2009). Hydroxycinnamoyltransferases (HCT) are found in several clades. Besides hydroxycinnamoyl-CoA thioesters acetyl-, malonyl-, tigloyl-, benzoyl- or β-phenylalanyl-CoA can be utilized by BAHD acyltransferases for transfer to a wide variety of acceptor substrates as esters or amides. The number of putative BAHD acyltransferase genes in plant genomes has been determined at 64 and 119 in A. thaliana and Oryza sativa, respectively, and 61 in A. thaliana and 94 in Populus trichocarpa (D’Auria 2006; Yu et al. 2009). The lower number reported by Yu et al. (2009) in A. thaliana was explained by erroneous or duplicate annotations. The presence of pseudogenes has been suspected. The crystal structure of vinorine synthase involved in the biosynthesis of the monoterpene indole alkaloid ajmaline in Rauvolfia species was the first solved protein structure (Ma et al. 2005). Other known structures are anthocyanin malonyltransferase from Dendranthema morifolium and trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum (Unno et al. 2007; Garvey et al. 2008).
BAHD HCTs and monooxygenases of the CYP98A family are involved in the establishment of the caffeic acid substitution pattern of many phenylpropanoid derivatives, e.g. lignin monomers. HCTs transfer 4-coumaric acid from 4-coumaroyl-CoA to quinic and/or shikimic acid (Fig. 1; Hoffmann et al. 2003; Niggeweg et al. 2004). The 4-coumaric acid moiety is meta-hydroxylated at the ester stage leading to chlorogenic acid and caffeoylshikimic acid (Schoch et al. 2001). Chlorogenic acid is a widespread defense compound found in high concentrations in many plant species (Sondheimer 1964). Caffeoylshikimic acid is usually not accumulated to high amounts. According to recent investigations caffeic acid from caffeoylshikimate can be re-transferred to CoA for further biosynthetic transformations. HCTs accepting shikimate and/or quinate as acceptor molecules thus play different roles in plant metabolism. Down-regulation of the acyltransferase forming 4-coumaroyl-shikimate has a strong influence on lignin amount and composition (Hoffmann et al. 2004, 2005; Shadle et al. 2007; Wagner et al. 2007) whereas suppression of 4-coumaroylquinate formation leads to a reduction of the chlorogenic acid content in tomato plants and consequently to a reduced pathogen and UV resistance (Niggeweg et al. 2004; Clé et al. 2008).
Hydroxycinnamoyl-CoA:hydroxyphenyllactic acid HCT (rosmarinic acid synthase, RAS, E.C. 2.3.1.140; Fig. 1) is another member of the BAHD acyltransferase superfamily. RAS is involved in the formation of the putative defense compound rosmarinic acid (RA). RA is found not only in species of the plant families Lamiaceae (subfamily Nepetoideae) and Boraginaceae but also in many other groups of the land plants (Petersen and Simmonds 2003; Petersen et al. 2009). The first cDNA for RAS has been isolated from Coleus blumei (Berger et al. 2006). The cDNA (EMBL acc. no. AM283092) has an open reading frame (orf) of 1,290 bp encoding a protein of 430 amino acids with a calculated molecular mass of 47.9 kDa and a theoretical isoelectric point of pH 5.89. The genomic sequence revealed a single phase 0 intron of 914 bp after 405 bp. According to St. Pierre and De Luca (2000) this is a typical “Q-intron” (inserted after the conserved amino acid Q = glutamine) 17 amino acids forward of the conserved motif HxxxD(G). First screenings for substrates of heterologously expressed RAS revealed that 4-coumaroyl- and caffeoyl-CoA are accepted as donor substrates while 4-hydroxyphenyllactate (pHPL) and 3,4-dihydroxyphenyllactate (DHPL) but not shikimate and quinate are used as acceptor substrates (Berger et al. 2006).
We here present a wide substrate screening for CbRAS together with the cDNA-cloning, heterologous expression and substrate screening of a hydroxycinnamoyl-CoA:shikimate HCT (HST; E.C. 2.3.1.133) from the same plant species, C. blumei. Interestingly, both enzymes accepted a much broader range of substrates than expected and catalyzed ester as well as amide formation. This relaxed substrate acceptance of both HCTs offers the possibility to use these enzymes as catalysts to synthesize a much broader range of hydroxycinnamoyl esters and amides than present in plants with unknown biological activities.
Materials and methods
Substrates
(Hydroxy)cinnamoyl CoA-esters were synthesized and quantified essentially according to Stöckigt and Zenk (1975) and purified by thin-layer chromatography on cellulose plates (VWR, Darmstadt, Germany) with n-butanol/acetic acid/water 5:2:3 as solvent mixture. Acetyl-CoA was obtained from AppliChem (Darmstadt, Germany), malonyl-CoA, benzoyl-CoA and anthraniloyl-CoA from TransMIT (Giessen, Germany). DHPL was prepared and purified as described previously (Petersen and Alfermann 1988). Other substrates were obtained commercially: dl-phenyllactate, d-DOPA, l-DOPA, serotonin (Alfa Aesar, Karlsruhe, Germany), shikimate, quinate (Carl Roth, Karlsruhe, Germany), anthranilate, 3-hydroxyanthranilate (TransMIT, Giessen, Germany), tyramine, 3-hydroxybenzoate, 2,3-dihydroxybenzoate, 3-aminobenzoate (Acros, Geel, Belgium), tryptamine (Chempur, Karlsruhe, Germany), dopamine, l-phenylalanine, dl-tyrosine (Merck, Darmstadt, Germany), dl-pHPL, 2-phenylethylamine, 2-(4-hydroxyphenyl)ethanol, 3-(4-hydroxyphenyl)propanol, d-phenylalanine, phenol (Sigma–Aldrich, Deisenhofen, Germany).
Isolation of cDNA and genomic clones for CbRAS and CbHST
The isolation of a cDNA encoding RAS from C. blumei Benth. (Lamiaceae; syn. Solenostemon scutellarioides (L.) Codd) and its expression as His-tag fusion protein has been described earlier (Berger et al. 2006). For the isolation of a HST cDNA from C. blumei the nucleotide sequences of HCTs from A. thaliana (EMBL acc. no. BT026488), Nicotiana tabacum (EMBL acc. no. AJ507825, AJ582651) and Lycopersicon esculentum (= Solanum lycopersicon; EMBL acc. no. AJ582652) were aligned with CbRAS and a forward degenerated primer designed from a sequence stretch with high similarity within the HCTs but diverse to CbRAS (primer HCT1f AG(AG)GA(CT)GA(AT)(GC)A(AT)GG(AT)(AC)G). The reverse primer was deduced from the conserved DFGWG motif (primer DFGWGr GTC(GCT)GA(CT)AT(AGC)GC(CT)CG(AT)GG(CT)CT). PCR assays (25 μl) contained 5 μl GoTaq 5× PCR buffer, 2.5 μl 25 mM MgCl2, 1 μl 10 mM dNTP mix, 0.5 μl of each primer (100 μM), 1 μl cDNA and 0.1 μl GoTaq polymerase (5 U/μl, Promega, Mannheim, Germany). cDNA was synthesized with the Revert Aid™ First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany) using RNA isolated essentially according to Chomczynski and Sacchi (1987) from young leaves of C. blumei plants from the greenhouse of the Institut für Pharmazeutische Biologie und Biotechnologie, Philipps-Universität Marburg. A 940 bp fragment was amplified with the following program: 1 cycle 94°C 120 s, 40–48°C 60 s, 70°C 90 s; 38 cycles 94°C 30 s, 40–48°C 60 s, 70°C 90 s; 1 cycle 94°C 120 s, 40–48°C 60 s, 70°C 600 s. After ligation into pGEM®-T (Promega), transformation of E. coli DH5α, plasmid isolation and sequencing, the 5′- and 3′-ends were amplified on the basis of this first fragment using the GeneRacer™ Kit (Invitrogen, Karlsruhe, Germany) and the following gene-specific and nested primers: CbHCT5 CCACCAGGCTACTTCGGCAACGTGATCTT, CbHCT5n CGACTACTTGAGGTCAGCTCTGGATTTC, CbHCT3 CATGGCTGGAGGGGGCTGGTACT, CbHCT3n CGTCGGCTGCGTGGTGCTGCATT essentially according to the manufacturer’s protocol with the following assay (50 μl) and program: 5 μl 10× High Fidelity PCR buffer, 3 μl 25 mM MgCl2, 1 μl 10 mM dNTP mix, 1 μl GeneRacer cDNA (diluted 1:5), 3 μl of each primer (10 μM), 0.5 μl High Fidelity PCR Enzyme Mix (5 U/μl, Fermentas); 1 cycle 95°C 300 s, 45–60°C 45 s, 72°C 180 s; 38 cycles 95°C 30 s, 45–60°C 45 s, 72°C 90 s; 1 cycle, 72°C 600 s. After subcloning (as above) and sequencing the full-length cDNA could be amplified with the full-length primers CbHCTf (ATCTCGAGATGAAAATCCACGTGAGAGATTC) and CbHCTr (TAGGATCCTCAAATCTGATAGAGCAGCTTC) introducing restriction sites for XhoI and BamHI. The amplified cDNA was subcloned in pGEM®-T in E. coli DH5α, the cDNA released with XhoI and BamHI and ligated into the expression vector pET-15b (Novagen, Darmstadt, Germany) which was transferred into the expression host E. coli BL21(DE3)pLysS.
Genomic sequences of CbRAS and CbHST were isolated using total genomic DNA from C. blumei prepared essentially according to Rogers and Bendich (1985). Full-length primers were used for PCR reactions to amplify the intron-containing DNA using essentially the same PCR assays and programs as described for the cDNA sequences.
Primer synthesis as well as sequencing services were provided by Eurofins MWG Operon (Ebersberg, Germany).
Expression of CbRAS and CbHST
Protein expression in E. coli BL21(DE3)pLysS transformed with pET-15b carrying the full-length cDNA orfs of CbRAS and CbHST, respectively, was performed by cultivating the bacteria in LB medium containing 100 μg/ml ampicillin at 37°C at 220 rpm for 20 h. Bacteria were harvested by centrifugation (5,000g, 4°C, 10 min) and frozen at −80°C. Bacteria from 400 ml medium were resuspended in 16 ml His-tag binding buffer (50 mM KH2PO4/K2HPO4, pH 8.0, 300 mM NaCl, 10 mM imidazole) and disrupted by ultrasonication (90 s, 100%, 0.3 cycles). After centrifugation (10,000g, 4°C, 20 min) the extraction was repeated. The combined protein extracts were added to 1 ml His-tag resin (Ni–NTA His-Bind resin Superflow; Novagen), incubated for 1 h on ice, washed with 4 ml buffer (see above) containing 20 mM imidazole and eluted with 4 ml elution buffer (as above but containing 250 mM imidazole). The eluted protein was dialyzed against 20 mM Tris/HCl, 0.02% NaN3 pH 7.5.
Protein concentrations were determined according to Bradford (1976) using bovine serum albumin (1 mg/ml) as standard.
Removal of His-tag
For some experiments, the 6× His-tag was cleaved from CbRAS with help of thrombin. 600 μl protein solution after metal chelate chromatography, dialysis and addition of 10% DMSO were incubated with 50 μl of the Thrombin CleanCleave™ Kit (Sigma–Aldrich) and incubated for 73 h at 25°C. The protein was again applied to His-tag resin and unbound protein was collected.
Enzyme assays
All enzyme assays were performed with the same preparation of CbRAS or CbHST, respectively, purified by metal chelate chromatography as above and stored at 4°C. Standard assays for the determination of product formation contained in a total volume of 125 μl 90.5 μl 0.1 M KH2PO4/K2HPO4 pH 7.0, 5 μl 12.5 mM ascorbic acid, 12.5 μl 10 mM dithiothreitol, 5 μl 100 mM acceptor substrate, 2 μl 1–3 mM (hydroxy)cinnamoyl-CoA, 10 μl protein preparation (up to 4.2 μg RAS or 1.7 μg HST). The assays were incubated at 30°C for 2 min, then stopped by addition of 10 μl 6 N HCl and cooling on ice. Assay conditions for the determination of kinetic parameters and the concentration ranges for the substrates applied are listed in Supplementary Table 1. Reaction products were extracted by mixing with 500 μl ethyl acetate, centrifugation at 16,000g for 5 min and removal of the organic phase for three times. The ethyl acetate phases were collected and evaporated to dryness.
Controls without enzyme addition as well as control reactions omitting either the donor or the acceptor substrate were run under identical conditions. A product formation was only assumed to take place when the three controls did not result in the formation of the assumed product peak and the assumed product peak increased with time in complete reaction assays.
HPLC analyses
Dry samples were redissolved in 100 μl of the respective solvent mixture and analyzed by HPLC on an Equisil ODS column (250 × 4 mm, precolumn 20 × 4 mm; Dr. Maisch HPLC, Ammerbuch, Germany). The solvent mixtures for elution were 35–60% aqueous methanol acidified with 0.01% H3PO4 at 1 ml/min. The detection was done at 220, 280, 312 or 333 nm varying with the substrates used (see detailed information in Supplementary Table 2).
The following standards were used for identification and quantification of the corresponding reaction products: 4-coumaroyl-pHPL, caffeoyl-pHPL, 4-coumaroyl-DHPL, RA (from our laboratory collection: Petersen and Metzger 1993); 4-coumaroylquinate, 4-coumaroylshikimate and caffeoylshikimate (Dr. P. Ullmann, Strasbourg, France); chlorogenic acid (Carl Roth, Karlsruhe, Germany). These compounds were also used for co-chromatography.
For analysis of reaction products (e.g. 4-coumaroyltyrosine, caffeoylphenylalanine, 4-coumaroyl-2′,3′-dihydroxybenzoate) the compounds were hydrolyzed essentially as described by Petersen and Alfermann (1988) and analyzed again by HPLC for the presence of the expected cleavage products (e.g. 4-coumaroyltyrosine → 4-coumarate + tyrosine; caffeoylphenylalanine → caffeate + phenylalanine; 4-coumaroyl-2′,3′-dihydroxybenzoate → 4-coumarate + 2,3-dihydroxybenzoate). Furthermore spot tests with molybdate reagent and ninhydrin reagent (Clifford et al. 1989) were carried out with enzyme reaction products (caffeoyl-pHPL, caffeoyltyrosine, caffeoyl-3′-aminobenzoic acid, caffeoyl-3′-hydroxybenzoic acid) purified by HPLC.
Results
Isolation of a cDNA encoding a hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase (HST) from C. blumei
Coleus blumei (Lamiaceae) is known for its accumulation of RA and RAS has recently been cloned from this species (Berger et al. 2006). We could not detect chlorogenic acid in this plant species. In spite of this, a hydroxycinnamoyl-CoA:quinate HCT (HQT; Fig. 1) gene might still be present. As a member of the monolignol biosynthetic pathway the presence of a HST is highly probable (Fig. 1). By sequence alignment of hydroxycinnamoyl-CoA:shikimate/quinate HCTs (HCSQT) with RAS from C. blumei a degenerate forward primer was designed that was conserved in HCSQTs but differential to RAS. This primer together with a reverse primer corresponding to the DFGWG motif was used to amplify a 940 bp DNA fragment. The translated amino acid sequence had identities of around 80% to known plant HCSQTs in UniProt Knowledgebase. The 3′- and 5′-ends were amplified with the help of RACE-PCR and enabled the deduction of a full-length sequence for a HCT. The full-length orf was amplified and ligated into the expression vector pET-15b which was transferred into E. coli BL21(DE3)pLysS for protein expression. The open reading frame comprised 1,281 bp encoding a protein of 427 amino acids (EMBL acc. no. FN647681). The calculated molecular mass of the encoded protein was at 47.1 kDa and the isoelectric point at pH 5.78.
The genomic sequence for CbHST was isolated using the same full-length primers with total genomic DNA from C. blumei. The sequence reveals a phase 0 intron of 154 bp inserted after 408 bp.
Determination of the catalytic activity of the novel putative HCT
Crude protein extracts from E. coli BL21(DE3)pLysS harbouring the pET-15b vector with the orf of the putative HCT were incubated with the donor substrates 4-coumaroyl- and caffeoyl-CoA and the potential acceptor substrates shikimate, quinate, pHPL and DHPL. Reaction products were only detected in assays containing 4-coumaroyl- or caffeoyl-CoA and shikimate. The identity of the reaction products was verified by co-chromatography with authentic 4-coumaroylshikimate and caffeoylshikimate. Crude bacterial protein extracts from control cells did not show the HCT activities under investigation. The newly isolated cDNA thus encodes a HST. It is noteworthy that neither quinate, the substrate for chlorogenic acid biosynthesis, nor the substrates of RAS (pHPL, DHPL) from the same plant species could serve as substrates. The occurrence of a HST besides RAS makes it probable that the esters formed by RAS are not involved in the formation of monolignols, a role that has been assigned to 4-coumaroylshikimate, the ester synthesized by HST (Hoffmann et al. 2004, 2005; Shadle et al. 2007; Wagner et al. 2007).
General enzyme characteristics of heterologously expressed CbRAS and CbHST
Recombinant CbRAS and CbHST proteins produced in E. coli strains expressing each gene were isolated and purified to near homogeneity by metal chelate chromatography with the help of the N-terminally attached 6× His-tag (Supplementary Fig. 1). As usual with this purification method, minor bands of histidine-rich E. coli proteins were co-purified (Bolanos-Garcia and Davies 2006). His-tag-purified protein preparations were used in standard enzyme assays for the determination of general enzyme characteristics. The temperature optima of RAS and HST were at 40–45°C. The pH optimum for RAS was at pH 8.4 for the protein still carrying the N-terminal His-tag and slightly lower at pH 7.8 after its removal. The His-tagged HST displayed a pH optimum of 7.2.
Acyl-CoA substrates for RAS and HST from C. blumei
First experiments had shown that RAS accepts 4-coumaroyl- and caffeoyl-CoA as donor and pHPL and DHPL as acceptor substrates for ester formation (Berger et al. 2006) while HST used shikimate as acceptor substrate along with the same hydroxycinnamoyl-CoAs as donors. To further elucidate the substrate specificities of both enzymes a large variety of potential donor and acceptor substrates were tested.
Several CoA-activated acids were applied (Tables 1, 2), but only activated phenylpropenoic acids (cinnamoyl-, 4-coumaroyl-, caffeoyl-, feruloyl-, sinapoyl-CoA) could serve as substrates for both, RAS and HST (except sinapoyl-CoA for RAS), while no reaction products were observed with acetyl-, malonyl-, benzoyl- and anthraniloyl-CoA. The K m values determined for the hydroxycinnamoyl donor substrates (Table 3) varied with the acceptor substrates used.
RAS displayed the highest affinities with pHPL as acceptor for caffeoyl-CoA (app. K m 2.1 μM) and 4-coumaroyl-CoA (app. K m 9.2 μM), while cinnamoyl-CoA (app. K m 25.5 μM) and feruloyl-CoA (app. K m 36.4 μM) showed considerably lower affinities and sinapoyl-CoA did not lead to product formation. After removal of the N-terminal His-tag the K m value for caffeoyl-CoA was at 3.4 μM and for 4-coumaroyl-CoA at 4.7 μM. With DHPL as acceptor substrate the affinities towards 4-coumaroyl-CoA (app. K m 1.8/1.3 μM with/without His-tag) were higher compared to caffeoyl-CoA (app. K m 3.9/4.0 μM with/without His-tag). The data determined for CbRAS with and without His-tag did not differ significantly indicating that the N-terminal His-tag does not influence the enzyme performance strongly. With both protein preparations the highest V max/K m values were observed for the combination of caffeoyl-CoA with pHPL. Nevertheless 4-coumaroyl-CoA was the second best substrate and the formation of 4-coumaroyl-pHPL in the cytoplasm can be anticipated considering the abundance of 4-coumaroyl- and caffeoyl-CoA in this environment.
The preferred substrate for HST with shikimate as acceptor substrate was 4-coumaroyl-CoA (app. K m 4.3 μM) (Table 4). High affinities were observed to caffeoyl-CoA (app. K m 8.8 μM) and feruloyl-CoA (app. K m 5.1 μM) as well, but cinnamoyl- (app. K m 26.5 μM) and sinapoyl-CoA (K m 20.0 μM) were poorly accepted. The calculated V max/K m values revealed a slight preference for 4-coumaroyl-CoA in combination with shikimate as acceptor substrate.
Acceptor substrate specificity of CbRAS
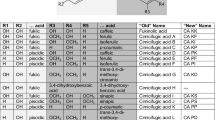
Investigations of the acceptor substrate variety for RAS (see Fig. 2 for structures) brought some unexpected results since not only esters—as anticipated from the known in vivo activities—were formed by RAS but putatively also amides. As expected, RAS formed esters with several d-(hydroxy)phenyllactates: phenyllactic acid (PL), pHPL and DHPL (Table 1). The affinities decreased from pHPL (app. K m 25 μM) over DHPL (app. K m 35 μM) to PL (app. K m 490 μM) when 4-coumaroyl-CoA was used as hydroxycinnamoyl donor (Table 3). Higher values were determined when caffeoyl-CoA was used as second substrate (app. K m for pHPL 163 μM, DHPL 193 μM, PL 1,035 μM). After that, we set out to test several substrates that were structurally related to (hydroxy)phenyllactates. Unexpectedly, reaction products of RAS were observed with dl-tyrosine, dl-phenylalanine and dl-3,4-dihydroxyphenylalanine (DOPA) (Table 1). Since the respective l-stereoisomers did not result in reaction products, it can be concluded that the D-isoforms served as substrates. Previous experiments had already shown that l-DHPL, l-pHPL and l-PL were not accepted by RAS, thus demonstrating a strict stereoselectivity of the enzyme. The reaction products with the amino acids were detected with cinnamoyl-, 4-coumaroyl- and caffeoyl-CoA and they were verified by enzymatic hydrolysis of the products and detection of the respective cinnamic acids and the acceptor substrates used in the assays by HPLC. Furthermore, reaction products formed from caffeoyl-CoA and pHPL or tyrosine were purified by HPLC and subjected to spot tests as described by Clifford et al. (1989). Using molybdate reagent, the putative caffeoyltyrosine as well as caffeoyl-pHPL gave yellow spots showing the aromatic ortho-dihydroxy substitution. Only the putative caffeoyltyrosine showed a slight purple color with ninhydrin reagent due to the presence of an –NH (or –NH2) group. The positive reactions with both ninhydrin and molybdate reagent suggests the formation of an amide between caffeic acid and tyrosine. Similarly, since phenylalanine has no free hydroxyl group for ester formation, an amide formation must be assumed. This is also anticipated with DOPA as substrate. The apparent K m values for tyrosine are considerably higher than the respective values for (hydroxy)phenyllactates, namely 1.0 mM with 4-coumaroyl-CoA and 17.8 mM with caffeoyl-CoA (Table 3).
Time courses of RAS with different substrate combinations reflected best the relative suitability of substrate combinations (see examples in Supplementary Fig. 2a). These examples demonstrate that the formation of 4-coumaroyl-pHPL is faster than the formation of RA under comparable reaction conditions. Substrate combinations like feruloyl-CoA and pHPL are less favourable. The putative amide formation is a rather slow reaction even with increased enzyme concentrations.
No reaction products were detected by HPLC when 4-coumaroyl-CoA and the following putative acceptor substrates were incubated with RAS (Table 1): shikimate, quinate, anthranilate, 3-hydroxyanthranilate, tyramine, dopamine, tryptamine, serotonin, 2-phenylethylamine, 2-(4-hydroxyphenyl)ethanol and 3-(4-hydroxyphenyl)propanol (see Fig. 2 for structures).
Acceptor substrate specificity of CbHST
As described above, CbHST accepted shikimic acid as acceptor substrate while quinic acid was not converted to an ester even with very high enzyme concentrations and prolonged incubation times. PL derivatives, the substrates for CbRAS, were not converted, neither were amines (2-phenethylamine, tyramine, dopamine, tryptamine, serotonin), DOPA or phenol (Table 2; see Fig. 2 for structures). Shikimate caused severe substrate inhibition at concentrations exceeding 40 mM. Thus, the determination of V max values was overlaid by the increasing inhibition already at lower substrate concentrations. The K m values were strongly dependent on the (hydroxy)cinnamoyl-CoA applied as donor substrate (Table 4). The lowest apparent K m value for shikimate was observed with 4-coumaroyl-CoA at 332 μM, followed by cinnamoyl-CoA (2.2 mM) and caffeoyl-CoA (6.5 mM). The affinity towards shikimate was very low when feruloyl- and sinapoyl-CoA were used as donor substrates (app. K m 22 mM/26 mM).
Since one of the earliest identified members of the BAHD acyltransferase family is anthranilate N-hydroxycinnamoyl/benzoyltransferase (HCBT) from Dianthus caryophyllus (Yang et al. 1997), we also tested anthranilate as possible acceptor substrate but did not detect any reaction product. 3-Hydroxyanthranilate, however, resulted in the formation of a reaction product, putatively an ester, with all five cinnamic acid CoA-esters available (Table 2). In order to get further support for the ester formation with 3-hydroxyanthranilate, 3-hydroxybenzoate was used as substrate and also resulted in product formation with cinnamoyl-, 4-coumaroyl-, caffeoyl- and feruloyl-CoA. Similarly, 2,3-dihydroxybenzoate was accepted. Interestingly, a benzoic acid derivative without a free OH-group, namely 3-aminobenzoate could serve as substrate as well. Here the formation of an amide must be anticipated. Compared to shikimate as acceptor substrate, 3-hydroxyanthranilate and 2,3-dihydroxybenzoate were accepted with a similar affinity or even better (Table 4). The affinities for 3-hydroxybenzoate and 3-aminobenzoate were considerably lower. 4-Coumaroyl-2′,3′-dihydroxybenzoate was further analysed by HPLC purification followed by enzymatic hydrolysis resulting in the presence of the expected cleavage products (4-coumarate, 2,3-dihydroxybenzoate). Spot tests according to Clifford et al. (1989) with molybdate reagent and ninhydrin reagent were performed with the putative ester caffeoyl-3′-hydroxybenzoate and the putative amide caffeoyl-3′-aminobenzoate purified by HPLC. Both products reacted positively with molybdate reagent, but only caffeoyl-3′-aminobenzoate showed a purple spot with ninhydrin reagent. The substrate specificities reported above would suggest that hydroxycinnamoyl transfer can only occur in meta-position to the carboxyl group.
Molecular and phylogenetic analysis of CbRAS and CbHST
On the basis of the nucleotide sequences the identity of CbRAS and CbHST to each other was 66.1%, on the amino acid level identities/similarities of 56%/73% were found. Data bank searches for CbHST revealed highest identities of around 82% on amino acid level to HCTs and HSTs from Coffea canephora (UniProt acc. no. A4ZKE4, A4ZKM2), Coffea arabica (Q05HB0, A4ZKI0) and N. tabacum (Q8GSM7). Alignment of the amino acid sequences of CbRAS and CbHST revealed similarities throughout the sequence. Only rather short stretches showed pronounced dissimilarities (Fig. 3). The conserved sequence motifs HxxxDG and DFGWG are surrounded by differing amino acid environments which show typical features for RAS and HST/HQT. Whether this is important for the diverse substrate specificity must be clarified in future investigations.
Aligned amino acid sequences of CbRAS and CbHST. Conserved sequence motifs of BAHD acyltransferases are shaded in grey, the position of the conserved amino acid Q after which a conserved intron can be inserted is marked in bold letters. Identical amino acids are marked with asterisks, similar amino acids with colons and dots
Discussion
Cloning of a HST from C. blumei
HSTs are now widely acknowledged as enzymes involved in the biosyntheses of diverse cinnamic acids and derivatives with more elaborated substitution patterns in the aromatic ring, namely caffeic, ferulic and sinapic acids (Hoffmann et al. 2003; Niggeweg et al. 2004). After reduction to alcohols these may serve as monolignols in the biosynthesis of lignin or lignans. In a biosynthetic scheme postulated recently, the 4-coumarate moiety of 4-coumaroyl-CoA is transferred to shikimic acid, then a 3-hydroxyl group is introduced by a CYP98A monooxygenase on the ester stage resulting in the formation of caffeoylshikimate (Fig. 1). The caffeic acid moiety then can be re-transferred to CoA and serve for further biosynthetic reactions (Schoch et al. 2001). To date it is not clarified whether the formation of monolignols by this pathway is ubiquitous in all lignin-containing land plants. The presence of lignin in tracheophytes as well as in red algae (Waters 2003; Martone et al. 2009) has been suggested to occur as a result of convergent evolution, although the enzymes involved in red algae are not yet characterized in detail. Bryophytes including the hornworts (some of which accumulate high amounts of RA) are free of lignin (Popper and Tuohy 2010).
In a similar pathway, quinate is used as acceptor molecule by a HQT and 4-coumaroylquinate is formed, which is hydroxylated by a member of the CYP98A family to chlorogenic acid (Fig. 1), a widely occurring defense compound and UV-protectant in plants (Niggeweg et al. 2004). The relative preferences for shikimate and/or quinate vary between enzymes from different plant species. The HCT cloned from tobacco by Hoffmann et al. (2003) preferred shikimate over quinate, while the HCT from tomato accepted quinate better than shikimate (Niggeweg et al. 2004). Comino et al. (2009) reported a HCT from artichoke that only accepted quinate as substrate. Strict specificity for either quinate or shikimate had previously already been reported for purified proteins from, e.g. Stevia rebaudiana and Nicotiana alata (quinate) or Cichorium endivia (shikimate) (Ulbrich and Zenk 1979, 1980).
Our search for a CbHST sequence from C. blumei was based on the similarity of HST and HCSQT sequences from other plant species and differences to RAS. The resulting CbHST amino acid sequence showed considerably higher identities to HCSQT sequences from other plant species (>80%) than to RAS from the same species (56%) (see above). Typical BAHD acyltransferase sequence motifs (HxxxDG, DFGWG) are conserved (Fig. 3). Another conserved feature is the position of the so-called “Q-intron” (St. Pierre and De Luca 2000). In the genomic CbHST sequence the intron starts after 408 bp in comparison to 405 bp in CbRAS. However, the intron lengths are largely different: the CbRAS intron is 914 bp long, the CbHST intron only 154 bp. Sequence comparison and alignment of the two intron sequences shows that the short CbHST intron is 46% identical to a sequence stretch between bp 252 and 466 of the CbRAS intron. According to St. Pierre and De Luca (2000) about 75% of all BAHD acyltransferase genes have a conserved “Q-intron”. Yu et al. (2009) analyzed the BAHD acyltransferase sequences in the genomes of A. thaliana and P. trichocarpa and stated that some clades within the superfamily have no introns. However, clade I (according to Yu et al. (2009) and corresponding to D’Auria’s (2006) clade V) where HCTs are located, typically carry at least one intron which shows highly differing lengths.
Determination of kinetic parameters for CbRAS and CbHST
The determination of kinetic data for CbRAS and CbHST proved to be difficult since quantification of the reaction products and thus the determination of reliable V max values could not be done due to missing reference compounds for most of the unusual products formed. Some of the reported reaction products have never been described before and thus are novel compounds which might be interesting for further pharmacological studies. Moreover, with many substrate combinations a strong substrate inhibition was observed, making the determination of reliable V max values impossible. As can be seen in Tables 3 and 4, the K m values for a specific substrate were dependent on the second substrate used in the assay. Similar observations have been found (but not discussed) for other HCTs. Hoffmann et al. (2003) showed this feature in HCT from tobacco with K m values of 150 and 600 μM for 4-coumaroyl-CoA with quinate and shikimate, respectively. The HCSQT from Cynara cardunculus displayed a K m value for 4-coumaroyl-CoA of 53 μM with quinate and of 702 μM with shikimate as acceptor (Comino et al. 2007). The overall suitability of substrate combinations (donor + acceptor) can be judged from time courses as depicted in Supplementary Fig. 2. According to the graphs for CbRAS the substrate combination 4-coumaroyl-CoA and pHPL is better than caffeoyl-CoA and DHPL. Both are considerably preferred over feruloyl-CoA plus pHPL. While the putative formation of amides, such as caffeoyltyrosine, occurs, it is a very slow process and takes hours for measurable product formation. Similar observations have been made for CbHST. With shikimate as acceptor substrate 4-coumaroyl-CoA was slightly preferred over caffeoyl-CoA. The formation of feruloylshikimate is much slower although a 25-fold protein concentration was applied in the assays. Ester formation with 4-coumaroyl-CoA and 3-hydroxybenzoic acid was even slower. Again the supposed amide formation with 3-aminobenzoate took place only very slowly (Supplementary Fig. 2).
Substrate specificities of CbRAS
RAS was described as a specific enzyme of RA biosynthesis and it was first characterized and cloned from C. blumei (syn. S. scutellarioides) from the family Lamiaceae (Petersen and Alfermann 1988; Petersen 1991; Berger et al. 2006). The native enzyme from suspension cultures of C. blumei accepted cinnamoyl-, 4-coumaroyl- and caffeoyl-CoA as hydroxycinnamoyl donors with good activities, while the usage of sinapoyl- and feruloyl-CoA could not be shown unequivocally. Acceptor substrates were pHPL and DHPL as well as 3-methoxy-4-hydroxyphenyllactate. Phenyllactate and 3,4-dimethoxyphenyllactate did not serve as substrates. Substrates like shikimic or quinic acid have not been applied to this native enzyme and amide formation has not been tested (Petersen 1991). Testing the substrate specificity of the heterologously synthesized RAS revealed a much broader range of acceptor substrates than expected (Table 1). Except sinapoyl-CoA all (hydroxy)cinnamoyl-CoA thioesters could serve as (hydroxy)cinnamoyl donor with preference for 4-coumaroyl- and caffeoyl-CoA. This fits with the substitution patterns of the RA-related phenolic esters in C. blumei, namely 4-coumaroyl-3′,4′-dihydroxyphenyllactate (which we have found in rather low concentrations in cell extracts from suspension-cultured C. blumei; own unpublished result) and RA. A RA derivative with a 3-methoxy-4-hydroxy substitution pattern at the phenylalanine-derived aromatic ring has been reported as metabolite of orally administered RA in rats, whereas 3-methoxylation at the tyrosine-derived aromatic ring was shown in clinopodic acid A from Clinopodium chinense var. parviflorum (Lamiaceae) (Baba et al. 2004; Murata et al. 2009). We could not detect reaction products with other CoA-activated acids (acetyl-CoA, malonyl-CoA, benzoyl-CoA and anthraniloyl-CoA). This suggests the necessity of a C6C3-skeleton while the substitution pattern at the aromatic ring was not determinative.
The expected acceptor substrates pHPL and DHPL were converted by the heterologously synthesized RAS with high affinities and activities. Phenyllactate, which did not result in a detectable reaction product with the native RAS, was also accepted as substrate but with a more than tenfold lower affinity. Interestingly, we also found reaction products when hydroxycinnamoyl-CoA was incubated with amino acids such as racemic phenylalanine, tyrosine and 3,4-dihydroxyphenylalanine (DOPA). The same incubations with the respective l-enantiomers did not result in product formation, showing the specificity for d-enantiomers. The presence of the non-proteinogenic d-amino acids inside the plant cell, however, should be rather limited. Thus, the in vivo relevance of these reactions might be low. Since there is no OH-group present in the side chain and no OH-group at all in phenylalanine, the reaction with the amino acids must have been the formation of amides. Since amides are much slower metabolized than esters, these hydroxycinnamoylamides might be pharmacologically interesting. Due to the high similarity to RA they might have the same biological activities but might be metabolized more slowly. N-Caffeoyl-l-phenylalanine has been isolated from the fern Athyrium filix-femina (Adam 1995). Coffee beans, cocoa or clover contain a variety of hydroxycinnamic acid conjugates with l-phenylalanine, l-tyrosine, l-DOPA (e.g. clovamide) or l-tryptophan (Sanbongi et al. 1998; Tebayashi et al. 2000; Clifford and Knight 2004 and literature cited therein; Stark and Hofmann 2005), but conjugates with d-amino acids have to our knowledge not been reported before. N-Hydroxycinnamoyl transfer by BAHD acyltransferases has previously been reported in the biosyntheses of avenanthramides, hydroxycinnamoylagmatine or hydroxycinnamoylpolyamines (Burhenne et al. 2003; Yang et al. 2004; Fellenberg et al. 2009; Grienenberger et al. 2009; Luo et al. 2009). Recently, Landmann reported that an alcohol acyltransferase from Lavandula angustifolia was able to form esters as well as amides (Landmann 2007). Polyamine conjugation with (hydroxy)cinnamic acids has formerly been attributed to THT-like enzymes as has been the formation of hydroxycinnamoyl amides with tyramine or serotonin (Facchini et al. 2002; Kang et al. 2006). Tyramine, dopamine, tryptamine, serotonin and phenethylamine (Table 1; Fig. 2) have not been accepted by RAS for amide formation. This suggests that the enzyme requires a carboxyl group and an OH- or NH2-group in α-position for (hydroxy)cinnamoyl transfer. A hydroxyl group alone in the side chain is not sufficient since 2-(4-hydroxyphenyl)ethanol and 3-(4-hydroxyphenyl)propanol did not serve as hydroxycinnamoyl acceptors.
Substrate specificities of CbHST
Although many cloned and heterologously expressed HCTs were able to accept quinate as well as shikimate (see above), the newly isolated HST from C. blumei displayed a strict substrate specificity for shikimate (Table 2). PL derivatives or quinate were not accepted, even at prolonged incubation times with excess protein. To elucidate the substrate variability of CbHST we tried all possible (hydroxy)cinnamoyl-CoA-esters as substrate and could show product formation with shikimate in all cases. Cinnamoyl-CoA and sinapoyl-CoA were accepted with the lowest affinities (Tables 2, 4). This restriction may be based in the architecture of the active centre of HST. As observed for CbRAS, product formation could not be shown for CoA-esters of acids not belonging to the phenylpropenoic acids. This again shows the importance of a C6C3-skeleton and suggests that not just the CoA-activated carboxyl part of the molecule is necessary for binding to the protein.
Other commercially available possible acceptor substrates were tested as well. Although an anthranilate hydroxycinnamoyl/benzoyltransferase was shown by Yang et al. (1997), HST from C. blumei was not able to acylate anthranilate. 3-Hydroxyanthranilate, however, was transformed readily with all tested (hydroxy)cinnamoyl-CoAs as were 3-hydroxybenzoate and 2,3-dihydroxybenzoate (except with sinapoyl-CoA; Table 2). This suggests that the OH-group in meta-position to the carboxyl group is the acylated site. Phenol containing only an OH-group did not serve as acceptor. HST from tobacco has been tested with anthranilate and benzoate, but did not show activity (Hoffmann et al. 2003). The authors, however, did not test related substrates such as 3-hydroxybenzoate and 2,3-dihydroxybenzoate which were accepted by CbHST. To test the capacity of CbHST to form amides, 3-aminobenzoate was applied as substrate and product formation could easily be shown, although with considerably lower activity compared to 3-hydroxybenzoate (Supplementary Fig. 2). Here again the acylation takes place in meta-position of the carboxyl group. The putative amide formation is not favored compared to esterification. In summary, HST requires a carboxyl group in the acceptor substrate and then catalyzes the acyl transfer to the OH- or NH2-group in meta-position.
Conclusion
Two HCTs of the BAHD superfamily have been cloned and biochemically characterized from C. blumei, one catalyzing the biosynthesis of RA, the other the formation of 4-coumaroylshikimate. The HCTs displayed a donor substrate specificity limited to (hydroxy)cinnamoyl-CoAs but a much broader range of acceptor substrates. Both HCTs putatively also catalyzed the formation of amides besides ester formation. CbRAS mediates the hydroxycinnamoyl transfer to an OH- or NH2-group in α-position to a carboxyl group while CbHST catalyzes the acylation of an OH- or NH2-group in meta-position to a carboxyl group. In nature, most of the reported reactions may be of limited importance due to the low abundance of the acceptor substrates in plant cells. The rather broad substrate specificity may reflect a relaxed architecture in the active centre of the HCTs and might open up the possibility for an evolutionary development of new HCTs catalyzing the formation of novel plant secondary compounds. The reported enzymes can be used for the biotechnological production of novel compounds with unknown biological activities. The biotechnological production of the novel hydroxycinnamoyl esters and amides and successively pharmacological testing would be an interesting future task.
Abbreviations
- DHPL:
-
3,4-Dihydroxyphenyllactic acid
- HCT:
-
Hydroxycinnamoyltransferase
- HCSQT:
-
Hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyltransferase
- HQT:
-
Hydroxycinnamoyl-CoA:quinate hydroxycinnamoyltransferase
- HST:
-
Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase
- pHPL:
-
4-Hydroxyphenyllactic acid
- RAS:
-
Rosmarinic acid synthase = hydroxycinnamoyl-CoA:hydroxyphenyllactate hydroxycinnamoyltransferase
References
Adam KP (1995) Caffeic acid derivatives in fronds of the lady fern (Athyrium filix-femina). Phytochemistry 40:1577–1578
Baba S, Osakabe N, Natsume M, Terao J (2004) Orally administered rosmarinic acid is present as the conjugated and/or methylated forms in plasma, and is degraded and metabolized to conjugated forms of caffeic acid, ferulic acid and m-coumaric acid. Life Sci 75:165–178
Berger A, Meinhard J, Petersen M (2006) Rosmarinic acid synthase is a new member of the superfamily of BAHD acyltransferases. Planta 224:1503–1510
Bolanos-Garcia VM, Davies OR (2006) Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim Biophys Acta 1760:1304–1313
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal Biochem 72:248–254
Burhenne K, Kristensen BK, Rasmussen SK (2003) A new class of N-hydroxycinnamoyltransferases. Purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J Biol Chem 278:13919–13927
Chomczynski P, Sacchi N (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate phenol–chloroform extraction. Anal Biochem 162:156–159
Clé C, Hill LM, Niggeweg R, Martin CR, Guisez Y, Prinsen E, Jansen MAK (2008) Modulation of chlorogenic acid biosynthesis in Solanum lycopersicon: consequences for phenolic accumulation and UV-tolerance. Phytochemistry 69:2149–2156
Clifford MN, Knight S (2004) The cinnamoyl-amino acid conjugates of green robusta coffee beans. Food Chem 87:457–463
Clifford MN, Kellard B, Ah-Sing E (1989) Caffeoyltyrosine from green robusta coffee beans. Phytochemistry 28:1989–1990
Comino C, Lanteri S, Portis E, Acquadro A, Romani A, Hehn A, Larbat R, Bourgaud F (2007) Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L. BMC Plant Biol 7:14
Comino C, Hehn A, Moglia A, Menin B, Bourgaud F, Lanteri S, Portis E (2009) The isolation and mapping of a novel hydroxycinnamoyltransferase in the globe artichoke chlorogenic acid pathway. BMC Plant Biol 9:30
D’Auria JC (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9:331–340
Facchini PJ, Hagel J, Zulak K (2002) Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot 80:577–589
Fellenberg C, Boettcher C, Vogt T (2009) Phenylpropanoid polyamine conjugate biosynthesis in Arabidopsis thaliana flower buds. Phytochemistry 70:1392–1400
Garvey GS, McCormick SP, Rayment I (2008) Structural and functional characterization of the TRI101 trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: kinetic insights to combating Fusarium head blight. J Biol Chem 283:1660–1669
Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58:246–259
Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278:95–103
Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxy-cinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyl-transferase affects phenylpropanoid biosynthesis. Plant Cell 16:1446–1465
Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2005) Acyltransferase-catalysed p-coumarate ester formation is a committed step of lignin biosynthesis. Plant Biosyst 139:50–53
Kang S, Kang K, Chung GC, Choi D, Ishihara A, Lee DS, Back K (2006) Functional analysis of the amine substrate specificity domain of pepper tyramine and serotonin N-hydroxycinnamoyltransferases. Plant Physiol 140:704–715
Landmann C (2007) Funktionelle Charakterisierung von Enzymen des Sekundärstoffwechsels in Lavendel (Lavandula angustifolia) und Erdbeere (Fragaria × ananassa). Dissertation, Technical University of Munich (Germany)
Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12:1295–1306
Luo J, Fuell C, Parr A, Hill L, Bailey P, Elliott K, Fairhurst SA, Martin C, Michael AJ (2009) A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21:318–333
Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J (2005) Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J Biol Chem 280:13576–13583
Martin-Tanguy J, Cabanne F, Perdrizet E, Martin C (1978) The distribution of hydroxycinnamic acid amides in flowering plants. Phytochemistry 17:1927–1928
Martone PT, Estevez JM, Lu F, Ruel K, Denny MW, Somerville C, Ralph J (2009) Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol 19:169–175
Milkowski C, Strack D (2004) Serine carboxypeptidase-like acyltransferases. Phytochemistry 65:517–524
Mugford ST, Qi X, Bakht S, Hill L, Wegel E, Hughes RK, Papadopoulou K, Melton R, Philo M, Sainsbury F, Lomonossoff GP, Roy AD, Goss RJM, Osbourn A (2009) A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21:2473–2484
Murata T, Sasaki K, Sato K, Yoshizaki F, Yamada H, Mutoh H, Umehara K, Miyase T, Warashina T, Aoshima H, Tabata H, Matsubara K (2009) Matrix metalloproteinase-2 inhibitors from Clinopodium chinense var. parviflorum. J Nat Prod 72:1379–1384
Niggeweg R, Michael A, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nature Biotechnol 22:746–754
Petersen M (1991) Characterization of rosmarinic acid synthase from cell cultures of Coleus blumei. Phytochemistry 30:2877–2881
Petersen M, Alfermann AW (1988) Two new enzymes of rosmarinic acid biosynthesis from cell cultures of Coleus blumei: hydroxyphenylpyruvate reductase and rosmarinic acid synthase. Z Naturforsch 43c:501–504
Petersen M, Metzger JW (1993) Identification of the reaction products of rosmarinic acid synthase from cell cultures of Coleus blumei by ion spray mass spectrometry and tandem mass spectrometry. Phytochem Anal 4:131–134
Petersen M, Simmonds MSJ (2003) Molecules of interest: rosmarinic acid. Phytochemistry 62:121–125
Petersen M, Abdullah Y, Benner J, Eberle D, Gehlen K, Hücherig S, Janiak V, Kim KH, Sander M, Weitzel C, Wolters S (2009) Evolution of rosmarinic acid biosynthesis. Phytochemistry 70:1663–1679
Popper ZA, Tuohy MG (2010) Beyond the Green: understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiol 153:373–383
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified tissues. Plant Mol Biol 5:69–76
Sanbongi C, Osakabe N, Natsume M, Takizawa T, Gomi S, Osawa T (1998) Antioxidative polyphenols isolated from Theobroma cacao. J Agric Food Chem 46:454–457
Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276:36566–36574
Shadle G, Chen F, Reddy MSS, Jackson L, Nakashima J, Dixon RA (2007) Down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68:1521–1529
Sondheimer E (1964) Chlorogenic acid and related depsides. Bot Rev 30:667–712
St. Pierre B, De Luca V (2000) Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. Rec Adv Phytochem 34:285–316
Stark T, Hofmann T (2005) Isolation, structure determination, synthesis, and sensory activity of N-phenylpropenoyl-l-amino acids from cocoa (Theobroma cacao). J Agric Food Chem 53:5419–5428
Steffens JC (2000) Acyltransferases in protease’s clothing. Plant Cell 12:1253–1256
Stehle F, Brandt W, Schmidt J, Milkowski C, Strack D (2008a) Activities of Arabidopsis sinapoylglucose:malate sinapoyltransferase shed light on functional diversification of serine carboxypeptidase-like acyltransferases. Phytochemistry 69:1826–1831
Stehle F, Stubbs MT, Strack D, Milkowski C (2008b) Heterologous expression of a serine carboxypeptidase-like acyltransferase and characterization of the kinetic mechanism. FEBS J 275:775–787
Stöckigt J, Zenk MH (1975) Chemical syntheses and properties of hydroxycinnamoyl‐coenzyme A derivates. Z Naturforsch 30c:352–358
Tebayashi S, Ishihara A, Tsuda M, Iwamura H (2000) Induction of clovamide by jasmonic acid in red clover. Phytochemistry 54:387–392
Ulbrich B, Zenk MH (1979) Partial purification and properties of hydroxycinnamoyl-CoA:quinate hydroxycinnamoyl transferase from higher plants. Phytochemistry 18:929–933
Ulbrich B, Zenk MH (1980) Partial purification and properties of p-hydroxycinnamoyl-CoA:shikimate-p-hydroxycinnamoyl transferase from higher plants. Phytochemistry 19:1625–1629
Unno H, Ichimaida F, Suzuki H, Takahashi S, Tanaka Y, Saito A, Nishino T, Kusunoki M, Nakayama T (2007) Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J Biol Chem 282:15812–15822
Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci USA 104:11856–11861
Waters ER (2003) Molecular adaptation and the origin of land plants. Mol Phylogen Evol 29:456–463
Yang Q, Reinhard K, Schiltz E, Matern U (1997) Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35:777–789
Yang Q, Trinh HX, Imai S, Ishihara A, Zhang L, Nakayashiki H, Tosa Y, Mayama S (2004) Analysis of the involvement of hydroxyanthranilate hydroxycinnamoyltransferase and caffeoyl-CoA 3-O-methyltransferase in phytoalexin biosynthesis in oat. Mol Plant Microbe Interact 17:81–89
Yu XH, Gou JY, Liu CJ (2009) BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: bioinformatics and gene expression. Plant Mol Biol 70:421–442
Acknowledgments
This work was funded by the Deutsche Forschungsgemeinschaft (grant PE 360/21-1). We want to thank Dr. S. Martens and Prof. Dr. S.-M. Li for providing us with some of the substrates.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sander, M., Petersen, M. Distinct substrate specificities and unusual substrate flexibilities of two hydroxycinnamoyltransferases, rosmarinic acid synthase and hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl-transferase, from Coleus blumei Benth.. Planta 233, 1157–1171 (2011). https://doi.org/10.1007/s00425-011-1367-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1367-2