Abstract
The Arabidopsis genome encodes six members of microRNA395 (miR395) family previously determined to regulate the expression of ATP sulfurylase (APS) and the sulfate transporter SULTR2;1. However, the mRNA targets for the individual miR395 family members and the biological consequences produced by target gene regulation of each miR395 remain to be identified. In this study, a transgenic approach was employed to determine the mRNA targets for each miR395 family member as well as the role each member plays in plant growth under abiotic stress conditions. Overexpression of miR395c or miR395e retarded and accelerated, respectively, the seed germination of Arabidopsis under high salt or dehydration stress conditions. Despite a single nucleotide difference between miR395c and miR395e, the cleavage of mRNA targets, APS1, APS3, APS4 and SULTR2;1, was not same in miR395c- and miR395e-overexpressing plants. These results demonstrate that a given miRNA family containing a single nucleotide difference can guide the cleavage of various mRNA targets, thereby acting as a positive or negative regulator of seed germination under stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are small endogenous RNAs that play important regulatory roles in the growth, development and stress responses of plants by targeting mRNAs for cleavage or translational repression. Sequencing of the genome of several plants has resulted in the identification of many miRNAs, whose existence, biogenesis and functions have been extensively reviewed (Bartel 2004, 2009; Voinnet 2009). Despite numerous reports describing the biological roles of miRNA in the growth, development, morphogenesis and stress responses of plants (Sunkar and Zhu 2004; Chiou et al. 2006; Sunkar et al. 2006, 2007; Hirsch et al. 2006; Zhou et al. 2007; Lu et al. 2008; Schommer et al. 2008; Arenas-Huertero et al. 2009; Voinnet 2009; Zhao et al. 2009; Sunkar 2010), our understanding of the biological functions of particular miRNAs in plants is often hampered by the complexity brought by multiple targets and family members of miRNAs.
Even with hundreds of miRNAs identified in Arabidopsis thaliana, the biological roles of miRNAs during stress response have been determined only in limited cases. It was reported that miR399 regulates phosphate homeostasis in Arabidopsis and is involved in phosphate-starvation response (Fujii et al. 2005; Chiou et al. 2006; Lin et al. 2008; Pant et al. 2008). It was also reported that the regulation of Cu/Zn superoxide dismutase genes in Arabidopsis, an important event for oxidative stress tolerance, is mediated by miR398 (Sunkar et al. 2006; Jagadeeswaran et al. 2009). We have also described the stress-responsive expression patterns and functional roles of miR417 and miR402 in the seed germination and seedling growth of Arabidopsis under various abiotic stresses (Jung and Kang 2007; Kim et al. 2010).
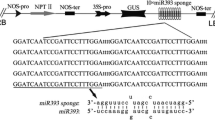
The Arabidopsis genome encodes the miR395 gene family, which exists in many dicots and monocots (Guddeti et al. 2005; Zhang et al. 2006), from six genomic loci designated a–f (Jones-Rhoades and Bartel 2004; Xie et al. 2005). The six miR395 loci are located in two clusters on chromosome 1; miR395 a, b, and c form one cluster and miR395 d, e, and f form another cluster (Kawashima et al. 2009). The nucleotide sequences of the b, c and f loci of miR395 are identical to each other, as are the a, d and e loci. Expression of the miR395c and miR395e loci was verified by 5′ RACE (Xie et al. 2005). The nucleotide sequences of miR395c and miR395e differ from each other only at position 17, where miR395c harbors a G residue and miR395e harbors an A residue (Table 1). It has been demonstrated that expression of miR395 is induced differently by low sulfate, and that its predicted targets include ATP sulfurylases (APS1, APS3 and APS4) and the sulfate transporter SULTR2;1 (Jones-Rhoades and Bartel 2004; Allen et al. 2005; Kawashima et al. 2009). This implies that miR395 potentially plays an important role in coordinating changes of sulfate assimilation and translocation (Chiou 2007; Sunkar et al. 2007). Kawashima et al. (2009) recently showed that the six Arabidopsis miR395 loci are induced differently, and that the expression of both miR395 and SULTR2;1 is increased in sulfur-starved roots. Cleavage of target mRNAs by miR395 has also been confirmed by 5′ RACE (Kawashima et al. 2009). However, the mRNA targets for each miR395 family member as well as the biological effects produced by the gene regulation of each target remain to be identified. In particular, how important a single nucleotide difference between miR395c and miR395e is has never been assessed. Here, we investigated the target genes regulated by miR395c and miR395e and provided evidence that a single nucleotide difference between miR395c and miR395e determines the cleavage of different mRNA targets, thereby functioning as a positive or negative regulator of seed germination in Arabidopsis under salt or dehydration stress.
Materials and methods
Plant materials and stress treatment
Arabidopsis thaliana L. ecotype Col-0 (originally from Arabidopsis Biological Resource Center, ABRC, USA) was used in this study. Seeds sown on a 2:1:1 mixture of vermiculite, peat moss, and perlite were placed at 4°C for 3 days in the dark for stratification and transferred to normal growth conditions. Plants were grown at 23°C under long-day conditions (16-h-light/8-h-dark cycle) at ~100 μmol photons m−2 s−1. Stress treatment was performed essentially as described (Kim et al. 2005). Three-week-old Arabidopsis plants were placed in petri dishes with their roots submerged into the solutions containing 150 mM NaCl or 300 mM mannitol. The stress-treated samples and non-stressed control samples were collected at the indicated time intervals, frozen immediately into liquid nitrogen, and were used for RNA extraction and subsequent analysis. The entire experiments were repeated at least three times.
Germination and seedling growth assays under stress conditions
Transgenic plants overexpressing miR395c or miR395e under control of the cauliflower mosaic virus 35S promoter were generated by cloning the coding region of 120 nucleotide-long pre-miR395c/e into a pBI121 vector. Arabidopsis were transformed with each construct via vacuum infiltration using Agrobacterium tumefaciens GV3101. The T3 or T4 homozygous lines were selected and used for the phenotypic investigation. Germination assays were performed essentially as described (Kim et al. 2007, 2008). Seeds sown on Murashige and Skoog (MS) medium supplemented with 1.5% sucrose were placed at 4°C for 3 days in the dark and then transferred to normal growth conditions. To determine the effect of salt or dehydration stress on seed germination, the MS medium was supplemented with 100–200 NaCl or 150–300 mM mannitol, respectively. To determine the effect of ABA on seed germination, the medium was supplemented with 2–10 μM ABA. To determine the effect of cold stress on germination, the MS plates were placed in an incubator maintained at 8–11°C under white light. Seeds were regarded as germinated once the radicle protruded through the seed coat. To determine the effect of various stresses on seedling growth of the plants, the seeds were fully germinated on normal MS medium for 4 days, and the seedlings were transferred to the medium subjected to different stresses or to ABA treatment.
Detection of miRNA, reverse-transcription PCR, and real-time RT-PCR
Total RNAs were extracted from the plant samples using a Plant RNeasy extraction kit (Qiagen, Valencia, CA, USA). The RNAs were size-fractionated by 15% polyacrylamide gel electrophoresis, blotted and cross-linked to a Hybond-N nylon membrane. 32P-labeled 21 nucleotide-long miR395c or miR395e was used as a probe. To examine the miR395-guided cleavage of the putative target mRNAs listed in Table 1 and Supplemental Table S1, gene-specific primers (Supplemental Table S1) were designed across the miR395 target site of each gene. The presence and absence of corresponding RNA transcripts were determined by RT-PCR analysis. Quantitative analysis of RNA transcript levels was performed by real-time RT-PCR using the Rotor-Gene 2000 real-time thermal cycling system (Corbett Research, Sydney, Australia) and QuantiTect SYBR Green RT-PCR kit (Kim et al. 2007) with the gene-specific primers (Supplemental Table S1) which detect the precursor miRNAs. The expression levels of each gene under stress conditions were determined by comparing their expression levels in non-stressed control plants.
Results
miR395c and miR395e have negative and positive effects, respectively, on the seed germination of Arabidopsis under salt or dehydration stress conditions
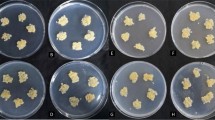
To determine whether miR395c or miR395e influences plant growth under stress conditions, we generated transgenic Arabidopsis plants overexpressing miR395c or miR395e under control of the 35S promoter (35S::miR395c and 35S::miR395e plants), and examined the seed germination and seedling growth of miR395-overexpressing Arabidopsis under high salt or dehydration stress conditions. The transcript levels of the 120-mer precursor-miRNA and the 21-mer mature miRNA were highly enriched in transgenic plants (Fig. 1), indicating that the 35S::miR395c and 35S::miR395e plants overexpress miR395c and miR395e, respectively. When the plants were grown under normal conditions, no noticeable differences in seed germination and seedling growth were observed between wild-type and transgenic plants (Supplemental Fig. S1). However, noticeable differences in seed germination were observed between the genotypes when the seeds of the wild-type and transgenic plants were germinated on MS media supplemented with various concentrations of NaCl or mannitol. As shown in Fig. 2, seed germination of 35S::miR395c plants was significantly retarded compared with wild-type plants under salt or dehydration stress. In contrast, seed germination of 35S::miR395e plants was noticeably accelerated compared with wild-type plants under salt or dehydration stress (Fig. 2). No noticeable differences in seed germination and seedling growth were observed between wild-type and transgenic plants under cold stress (data not shown). These results demonstrate that miR395c and miR395e have negative and positive effects, respectively, on the seed germination of Arabidopsis under high salt or dehydration stress.
Confirmation of transgenic Arabidopsis plants. Overexpression of 120 nucleotide-long precursor-miRNA and 21-mer mature miRNAs was verified in miR395c/e-overexpression lines by RT-PCR and Northern analysis. Ribosomal RNA (rRNA) and actin were used as a reference to ensure that equal amounts of RNA were present in the samples
Effect of salt or dehydration stress on the seed germination of wild-type and miR395c/e-overexpressing plants. Germination rates of the wild-type (Col-0), 35S::miR395c plants (C1, C4, and C7), and 35S::miR395e plants (E1, E11, and E13) were measured at the indicated days in medium supplemented with 150 mM NaCl or 300 mM mannitol. Mean values and stand errors were obtained from five independent experiments (n = 50–55)
To get some clue about how miR395c and miR395e affect oppositely the seed germination of Arabidopsis under salt or dehydration stress, the transcript levels of target mRNAs in the overexpression lines were determined during seed germination under stress conditions (Fig. 3). During seed germination under salt stress, it was evident that the transcript levels of APS1, APS4 and SULTR2;1 were decreased in 35S::miR395c plants compared with wild-type plants, while only SULTR2;1 level was decreased in 35S::miR395e plants compared with wild-type plants. The transcript level of APS4 was marginally decreased and increased in 35S::miR395c and 35S::miR395e plants, respectively, compared with wild-type plants. During seed germination under dehydration stress, the transcript levels of APS1 and APS4 were markedly decreased in 35S::miR395c plants compared with wild-type plants, whereas slightly increased in 35S::miR395e plants compared with wild-type plants. The transcript levels of APS3 were markedly decreased only in 35S::miR395e plants but not in 35S::miR395c plants, and the transcript level of SULTR2;1 was increased in both 35S::miR395c and 35S::miR395e plants.
Expression levels of target mRNAs during seed germination under stress conditions. Transcript levels of target mRNAs in the wild-type (Col-0), 35S::miR395c plants (C1, C4, and C7), and 35S::miR395e plants (E1, E11, and E13) were measured by real-time RT-PCR on the third day in medium supplemented with 150 mM NaCl or 300 mM mannitol, and the plots represent the relative expression (fold) of each gene in the transgenic plants compared with the expression in wild-type plants. Mean values and standard errors were obtained from three independent experiments
Response of miR395c- or miR395e-overexpressing plants to ABA
As ABA is a plant hormone involved in the stress response of plants, we examined the response of 35S::miR395c and miR395e plants to ABA treatment. The seed germination and post-germination growth of 35S::miR395c plants were significantly decreased compared with wild-type plants when performed on MS medium supplemented with 2.5 μM ABA (Fig. 4). In contrast, no differences in seed germination and post-germination growth were found between 35S::miR395e plants and wild-type upon ABA treatment (Fig. 4). These results suggest that miR395c and miR395e affect seed germination and growth of Arabidopsis under high salt or dehydration stress conditions in an ABA-dependent and ABA-independent manner, respectively.
Effect of ABA treatment on the seed germination and post-germination growth of wild-type and miR395-overexpressing plants. Germination rates of the wild-type (Col-0), 35S::miR395c plants (C1, C4, and C7), and 35S::miR395e plants (E1, E11, and E13) were measured at the indicated days in medium supplemented with 2.5 μM ABA. The pictures were taken at day 10 and day 14, respectively, for 35S::miR395c and 35S::miR395e plants. Mean values and stand errors were obtained from five independent experiments (n = 50–55)
Cleavage of target mRNAs by miR395c and miR395e
To further understand whether the opposite effect of miR395c and miR395e on seed germination of Arabidopsis under salt or dehydration stress is due to a different cleavage of target mRNAs by miR395c and miR395e, the transcript levels of each putative target mRNAs were analyzed in the overexpression lines. The mRNA targets for the miR395 family have been identified as APS1, APS3, APS4 and SULTR2;1, the proteins of which function in sulfate assimilation and transport (Takahashi et al. 2000). It was also reported that sulfate starvation increases the expression of endogenous miR395 while simultaneously decreasing APS1 transcript levels, and that APS1, APS3, APS4 and SULTR2;1 are cleaved by miR395 (Jones-Rhoades and Bartel 2004; Allen et al. 2005; Kawashima et al. 2009). Interestingly, Kawashima et al. (2009) have recently demonstrated that the cleavage of APS3 was detected only in leaves but not in roots. To determine whether miR395c and miR395e mediate the cleavage of the above-mentioned mRNA targets, we analyzed the expression level of each putative target mRNA by RT-PCR in the transgenic plants. Since Kawashima et al. (2009) have demonstrated the different cleavage of APS3 in leaves and roots, we extensively analyzed the cleavage of mRNA targets in leaves and roots of 35S::miR395c and 35S::miR395e plants, and Fig. 5 shows the target mRNA cleavage in leaves of the plants. It was evident that the transcript levels of APS1, APS4, and SULTR2;1 were downregulated in both leaves (Fig. 5) and roots (data not shown) by the overexpression of miR395c, and that the transcript level of APS3 was partially downregulated only in leaves but not in roots by the overexpression of miR395c (Fig. 5). It was also found that miR395e guide the cleavage of SULTR2;1 in both leaves (Fig. 5) and roots (data not shown), and that the transcript level of APS3 was partially downregulated only in leaves but not in roots by the overexpression of miR395e (Fig. 5). Our extensive analysis of the cleavage of APS1 and APS4 in leaves and roots demonstrated that the transcript levels of APS1 and APS4 are not reduced in 35S::miR395e plants (Fig. 5 and data not shown). Additional putative mRNA targets for miR395c and miR395e were searched for using the web-based computer program (http://bioinfo3.noble.org/miRNA/miRU.htm) and analyzed for their expression levels in the transgenic plants. Allowing two or three mismatches along with G·U wobble base pairs between miR395 and its target mRNAs resulted in the program predicting several additional mRNA targets. However, no changes in the expression levels of these predicted mRNA targets were observed by the overexpression of miR395c or miR395e (Supplemental Fig. S2).
Cleavage of target mRNAs in transgenic plants. Expression levels of the putative target mRNAs for miR395c and miR395e were analyzed by RT-PCR using gene-specific primers designed across the miR395c/e target site of each gene. Actin was used as a reference to ensure that equal amounts of RNA were present in the samples. L leaves, R roots
Discussion
miR395 family members have been predicted to target several mRNAs for proteins involved in sulfate assimilation and translocation, and are therefore implicated as regulators in the responses of plants to changing environmental factors. However, the cellular targets of each miR395 member as well as the functional consequences of their regulation remain largely unknown. Our results show that miR395c and miR395e have different effects on the seed germination of Arabidopsis under high salt or dehydration stress conditions by targeting different mRNAs. The precise correlation between target mRNA regulation by the overexpression of miR395c or miR395e and the stress-responsive seed germination of Arabidopsis under salt or dehydration stress is currently unclear. APS is the first enzyme of the sulfate assimilation pathway, and is present in the chloroplasts and cytosol of plants. SULTR2;1 is a low-affinity sulfate transporter located in the vascular tissues of leaves and roots and is involved in the internal translocation of sulfate from roots to shoot (Takahashi et al. 1997, 2000). It has been determined that the allocation and assimilation of sulfate are important for the adaptation of plants to adverse environmental conditions (Mittova et al. 2003; Koprivova et al. 2008). Although it is not clearly understood how the regulation of target mRNAs by miR395c or miR395e affects differently seed germination under stress conditions, it is likely that the down-regulation of APS1, APS4, or SULTR2;1 by miR395c in the transgenic plants (Fig. 3) results in decreased levels of sulfate assimilation and transport, leading to retarded seed germination under salt or dehydration stress. In comparison, the up-regulation of APS1, APS4, or SULTR2;1 and down-regulation of APS3 by miR395e in the transgenic plants (Fig. 3) result in increased levels of sulfate assimilation and transport, leading to accelerated seed germination under salt or dehydration stress. We do not know at present to what extent sulfur metabolism is affected by miR395c or miR395e. Determination of the amounts and distribution of sulfur in the transgenic plants would be important to fully understand the regulation of sulfur metabolism by miR395c or miR395e and subsequent impact on seed germination under stress conditions.
Small RNAs were found to be positive or negative regulators in plant hormone signaling, which plays a prominent role in the growth, development and stress responses of plants (Achard et al. 2004; Fujii et al. 2005; Mallory et al. 2005; Zhang et al. 2008). We show that miR395c and miR395e affect seed germination and growth of Arabidopsis under high salt or dehydration stress conditions in an ABA-dependent and ABA-independent manner, respectively (Fig. 4). Although it is currently unclear how miR395c and miR395e affect differently the ABA response, it is interesting to note that miR395c expression is upregulated by ABA treatment but the expression of miR395e is not significantly affected by ABA treatment in Arabidopsis (Supplemental Fig. S3). A recent report demonstrating that the expression of miR395d in rice, the nucleotide sequences of which is identical to miR395e in Arabidopsis, was not affected by ABA treatment (Liu et al. 2009) has implications on our current findings. The importance of miRNAs in ABA and stress responses has been demonstrated from the findings that the disturbance of miRNA pathways alters ABA response and multiple stress responses (Zhang et al. 2008). The plant hormones cytokinin and auxin have been determined to play an important role in sulfate deficiency response (Ohkama et al. 2002; Dan et al. 2007). Although the role of ABA in sulfate deficiency response has not been characterized in detail, Dan et al. (2007) suggested that ABA has a negative role in part of sulfate deficiency response. Based on these considerations, we propose that ABA-regulated expression of miR395c modulates sulfate metabolism and thereby affects seed germination and growth of Arabidopsis under high salt or dehydration stress conditions. However, it is noteworthy that the transcript level of adenosine 5′-phosphosulfate reductase, another key enzyme in sulfate metabolism, is induced under high salinity stress through an ABA-independent pathway (Koprivova et al. 2008). These results suggest that an ABA-independent pathway induces sulfate reduction, counteracting with the ABA-dependent miR395c pathway that regulates APS under stress conditions.
It is interesting to note that the transcript levels of target mRNAs are different in miR395c- and miR395e-overexpressing plants. miR395c and miR395e are two representative miR395 family members whose nucleotide sequences differ from each other by only one nucleotide. Specifically, miR395c harbors a G residue at position 17 while miR395e harbors an A residue. The present findings suggest that a single nucleotide difference between miR395c and miR395e may mediate the cleavage of different mRNA targets; miR395c targets APS1, APS3, APS4, and SULTR2;1 for cleavage, whereas miR395e cleaves only APS3 and SULTR2;1. Our present data further support the notion that miR395 mediates the cleavage of APS3 only in leaves but not in roots, as demonstrated by Kawashima et al. (2009). We do not know at present why the G·U base pair between miR395c and the targets, APS1 and APS4, results in the cleavage of these targets, whereas the Watson–Crick A·U base pair between miR395e and these targets does not result in target cleavage. As miR395 and some of its mRNA targets are expressed in different cells, it is likely that the cleavage of target mRNAs by miR395 is also regulated in a spatially or temporally dependent manner. More precise analysis of target gene cleavage by miR395 in different tissues or cells is required for better understanding of target mRNA regulation by miR395 under stress. In addition, analysis of the levels of mature miR395c/e and any posttranscriptional regulation of miR395 under stress conditions are required for a much complete understanding of miR395-guided target cleavage and stress-responsive roles of miR395 family members.
In conclusion, a single nucleotide difference between miR395c and miR395e guides the cleavage of different mRNA targets, APS1, APS3, APS4 and SULTR2;1, whose coordinate functions are in sulfate translocation and assimilation pathways. It is clear that the miR395-regulated allocation of sulfate to roots and shoots, as well as the assimilation of sulfate, affects the seed germination of plants under high salt or dehydration stress. Our current findings demonstrating the miR395-mediated regulation of APS1, APS3, APS4 and SULTR2;1, and its effect on seed germination under stress conditions provide new approaches for the functional examination of miR395 in the response of plants to adverse environmental conditions.
Abbreviations
- APS:
-
ATP sulfurylase
- miRNA:
-
MicroRNA
- SULTR2;1:
-
Sulfate transporter
References
Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131:3357–3365
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Arenas-Huertero C, Pérez B, Rabanal F, Blanco-Melo D, De la Rosa C, Estrada-Navarrete G, Sanchez F, Covarrubias AA, Reyes JL (2009) Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol 70:385–401
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Chiou T-J (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30:323–332
Chiou T-J, Aung K, Lin S-I, Wu C-C, Chiang S-F, Su C-L (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18:412–421
Dan H, Yang G, Zheng Z-L (2007) A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Mol Biol 63:221–235
Fujii H, Chiou T-J, Lin S-I, Aung K, Zhu J-K (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043
Guddeti S, Zhang DC, Li AL, Leseberg CH, Kang H, Li XG, Zhai WX, Johns MA, Mao L (2005) Molecular evolution of the rice miR395 gene family. Cell Res 15:631–638
Hirsch J, Lefort V, Vankersschaver M, Boualem A, Lucas A, Thermes C, d’Aubenton-Carafa Y, Crespi M (2006) Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol 140:1192–1204
Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229:1009–1014
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cells 14:787–799
Jung HJ, Kang H (2007) Expression and functional analyses of microRNA417 in Arabidopsis thaliana under stress conditions. Plant Physiol Biochem 45:805–811
Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T (2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57:313–321
Kim YO, Kim JS, Kang H (2005) Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J 42:890–900
Kim JY, Park SJ, Jang B, Jung C-H, Ahn SJ, Goh C-H, Cho K, Han O, Kang H (2007) Functional characterization of a glycine-rich RNA-binding protein2 in Arabidopsis thaliana under abiotic stress conditions. Plant J 50:439–451
Kim JS, Jung HJ, Lee HJ, Kim KA, Goh C-H, Woo Y, Oh SH, Han YS, Kang H (2008) Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55:455–466
Kim JY, Kwak KJ, Jung HJ, Lee HJ, Kang H (2010) MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE protein3 mRNA. Plant Cell Physiol 51:1079–1083
Koprivova A, North KA, Kopriva S (2008) Complex signaling network in regulation of adenosine 5′-phosphosulfate reductase by salt stress in Arabidopsis roots. Plant Physiol 146:1408–1420
Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147:732–746
Liu Q, Zhang Y-C, Wang C-Y, Luo Y-C, Huang Q-J, Chen S-Y, Zhou H, Qu L-H, Chen Y-Q (2009) Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 583:723–728
Lu S, Sun YH, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55:131–151
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Mittova V, Theodoulou FL, Kiddle G, Gomez L, Volokita M, Tal M, Foyer CH, Guy M (2003) Coordinate induction of glutathione biosynthesis and glutathione-metabolizing enzymes is correlated with salt tolerance in tomato. FEBS Lett 554:417–421
Ohkama N, Takei K, Sakakibara H, Hayashi H, Yoneyama T, Fujiwara T (2002) Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol 43:1493–1501
Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53:731–738
Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6:e230
Sunkar R (2010) MicroRNAs with macro effects on plant stress responses. Semin Cell Dev Biol. doi:10.1016/j.semcdb.2010.04.001
Sunkar R, Zhu J-K (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Sunkar R, Kapoor A, Zhu J-K (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Sunkar R, Chinnusamy V, Zhu J, Zhu J-K (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12:301–309
Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JA, Engler G, Van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94:11102–11107
Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23:171–182
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis miRNA genes. Plant Physiol 138:2145–2154
Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Zhang JF, Yuan LJ, Shao Y, Du W, Yan DW, Lu YT (2008) The disturbance of small-RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ 31:562–574
Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10:29
Zhou G-K, Kubo M, Zhong R, Demura T, Ye Z-H (2007) Overexpression of miR165 affects apical meristem formation, organ polarity establishment, and vascular development in Arabidopsis. Plant Cell Physiol 48:391–404
Acknowledgments
This work was supported in part by a grant from the National Research Foundation (NRF) of Korea to the Agricultural Plant Stress Research Center (APSRC, R11-2001-092-04002-0) of Chonnam National University and by World Class University program through the NRF of Korea funded by the Ministry of Education, Science and Technology (R32-20047-0).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, J.Y., Lee, H.J., Jung, H.J. et al. Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta 232, 1447–1454 (2010). https://doi.org/10.1007/s00425-010-1267-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1267-x