Abstract
A dwarf mutant, dwarf 62 (d62), was isolated from rice cultivar 93-11 by mutagenesis with γ-rays. Under normal growth conditions, the mutant had multiple abnormal phenotypes, such as dwarfism, wide and dark-green leaf blades, reduced tiller numbers, late and asynchronous heading, short roots, partial male sterility, etc. Genetic analysis indicated that the abnormal phenotypes were controlled by the recessive mutation of a single nuclear gene. Using molecular markers, the D62 gene was fine mapped in 131-kb region at the short arm of chromosome 6. Positional cloning of D62 gene revealed that it was the same locus as DLT/OsGRAS-32, which encodes a member of the GRAS family. In previous studies, the DLT/OsGRAS-32 is confirmed to play positive roles in brassinosteroid (BR) signaling. Sequence analysis showed that the d62 carried a 2-bp deletion in ORF region of D62 gene which led to a loss-of-function mutation. The function of D62 gene was confirmed by complementation experiment. RT-PCR analysis and promoter activity analysis showed that the D62 gene expressed in all tested tissues including roots, stems, leaves and panicles of rice plant. The d62 mutant exhibited decreased activity of α-amylase in endosperm and reduced content of endogenous GA1. The expression levels of gibberellin (GA) biosynthetic genes including OsCPS1, OsKS1, OsKO1, OsKAO, OsGA20ox2/SD1 and OsGA2ox3 were significantly increased in d62 mutant. Briefly, these results demonstrated that the D62 (DLT/OsGRAS-32) not only participated in the regulation of BR signaling, but also influenced GA metabolism in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dwarfism is one of the most important agronomic traits in crop breeding programs because dwarf cultivars are more resistant to lodging. During the famous “green revolution”, dwarfism was adopted to improve crop architecture which significantly increased the grain yield of cereal crops in the world (Sasaki et al. 2002; Hedden 2003). In rice, more than 60 dwarf mutants have been reported so far and several of them have been characterized thoroughly (http://www.gramene.org/rice_mutant/).
There are various factors responsible for dwarfism in plants, but gibberellin (GA) and brassinosteroid (BR) are the most intensely studied factors in determining plant height (Fujioka and Yokota 2003; Yamaguchi 2008). The GAs which control diverse biological processes including seed germination, stem elongation, leaf expansion, trichome development, pollen maturation, induction of flowering, fruit development, etc., are considered the most important bioactive growth regulators in plants (Olszewski et al. 2002). In recent years, numerous GA-related genes have been isolated from various plants (Hedden and Phillips 2000; Sun and Gubler 2004; Itoh et al. 2008) and the “green revolution” genes, such as the wheat Rht and rice sd1, are the most notable examples (Peng et al. 1999; Sasaki et al. 2002).
GA-related mutants are categorized into GA-deficient mutants and GA-insensitive mutants according to their responses to exogenous GAs (Mitsunaga et al. 1994). In rice, GA-deficient or -insensitive dwarf mutants display typical dwarfism with wide leaf blades and dark-green leaves (Ueguchi-Tanaka et al. 2000, 2005; Itoh et al. 2001; Sasaki et al. 2003). In GA-deficient dwarfs, the mutations are usually ascribed to the deficiency in GA metabolic pathway, where the GA metabolism is blocked or weakened (Hedden and Phillips 2000). In recent years, most of the genes encoding enzymes of GA metabolism have been identified in Arabidopsis, rice and other model plants (Sakamoto et al. 2004; Grennan 2006), and the complex pathways of GA metabolism have been elucidated in higher plants (Hedden and Phillips 2000; Sun and Gubler 2004; Yamaguchi 2008). In GA-insensitive dwarfs, the mutants are deficient in GA signaling and exhibit altered GA responses (Gomi and Matsuoka 2003). Also, there are slender-type mutants, such as slender rice (Ikeda et al. 2001) and slender barley (Lanahan and Ho 1988), which exhibit constitutive activation of GA responses. In recent years, the power of molecular genetics has dramatically facilitated our understanding of all aspects of GA signaling, especially the principal steps associated with GA perception and signal transduction in Arabidopsis and rice (Ueguchi-Tanaka et al. 2007).
The identification and characterization of a dwarf mutant, designated as dwarf 62 (d62), are reported in the present study. The D62 gene was isolated via positional cloning approach and was shown to be the same locus as DLT/OsGRAS-32. In previous studies, the DLT/OsGRAS-32 has been confirmed to play positive roles in BR signaling (Tong et al. 2009). However, the d62 mutant exhibited decreased activity of α-amylase in endosperm and reduced content of endogenous GA1. The expression levels of GA biosynthetic genes including OsCPS1, OsKS1, OsKO1, OsKAO, OsGA20ox2/SD1 and OsGA2ox3 were up-regulated in the d62 mutant. These results demonstrated that the D62 (DLT/OsGRAS-32) also influenced GA metabolism in rice.
Materials and methods
Plant materials
The d62 mutant was isolated from the M2 population of rice cultivar 93-11 (Oryza sativa L. ssp. indica; supplied by the Beijing Genomics Institute, Beijing, China) by mutagenesis with γ-rays. The mapping population consisted of field grown F2 individuals derived from a cross between d62 mutant and normal rice cultivar 02428 (Oryza sativa L. ssp. japonica; supplied by the Jiangsu Academy of Agricultural Sciences, Jiangsu Province, China). A total of 546 mutant individuals were identified from the F2 mapping population. In addition, eight F1 hybrids from the cross between d62 mutant and 93-11 were obtained. Cultivar 93-11 was used as the wild-type line in the present study. All plants were grown in the paddy field of Zhejiang University.
GA-induction in shoot elongation
Elongation of shoots was measured according to the method of Ueguchi-Tanaka et al. (2000) with minor modifications. Rice seeds were divested of glumes and surface sterilized, washed three times with sterile distilled water, then imbibed at 30°C for 24 h. The seeds were placed on 1/2 MS medium containing various concentrations of GA3 and grown at 30°C under a 16-h light/8-h dark period. After an interval of 7 days, the length of second leaf sheath and the seedling height were measured.
Assay of α-amylase activity
The α-amylase assay was performed as described by Lanahan and Ho (1988). Seeds were cut transversely, and the half-seeds containing embryos were grown to identify their phenotype. The embryoless half-seeds were sterilized with 2% NaClO for 15 min and washed with sterile distilled water five times. These half-seeds were then placed on 2% agar plates containing 0.2% soluble potato starch, 10 mM sodium acetate and 2 mM CaCl2 at pH 5.3. GA plates were made by adding l μM GA3 to the cooled agar after autoclaving. Agar plates were incubated in the dark for 3 days at 30°C and then developed by flooding the plates with a solution of I2–KI (0.72 g/L I2 + 7.2 g/L KI) in 0.2 N HCl. Half-seeds which synthesized and secreted α-amylase had transparent halos around the seed resulting from the digestion of the starch by α-amylase.
GAs purification and ELISA analysis
The procedure used to purify GAs from rice plants was as described previously (Yang et al. 2001; Zhu et al. 2005). Leaf tissues from each plant were homogenized in liquid nitrogen and then extracted in 4 mL of 80% (v/v) ice-cold aqueous methanol containing butylated hydroxytoluene (1 mM) and polyvinylpyrrolidone (60 mg/g fresh weight). The samples were incubated overnight at 4°C and centrifuged at 10,000g for 10 min. The resulting supernatants were collected individually and filtered through C18 Sep-Pak cartridges (Waters, Milford, MA, USA). Efflux of each sample was collected, dried by evaporation with N2 and measured for GA1 content with enzyme-linked immunosorbent assay (ELISA) kit according to the protocols recommended by the manufacturer.
Mapping and cloning of the D62 gene
Rice genomic DNA was extracted from fresh leaves of rice plants according to the method of Murray and Thompson (1980) with minor modifications. The D62 gene was mapped primarily with SSR markers (http://www.gramene.org/), by using 136 mutant individuals of F2 mapping population. To fine map the D62 gene, an InDel marker M14 (forward: 5′-CTCAGGAGCAAGAAGAGGAATA-3′; reverse: 5′-CACTAATGTTGTAGCAAACTGAT-3′) was newly developed based on the sequence difference between Nipponbare (spp. japonica) and 93-11 (spp. indica). A SSR marker MM0124 (forward: 5′-ATCAAGGAGGAGAAGGAGCC-3′; reverse: 5′-CGCACCTAGAGGAGATGAGG-3′) from Zhang et al. (2007) was also used in fine mapping. Thus, the D62 gene was further narrowed to a region between the markers MM0124 and RM19320 by using 546 mutant individuals of the F2 mapping population. This region of 131-kb in length was present in three BAC contigs, AP001168, AP002838 and AP000391. To identify the mutation site of the d62 mutant, the candidate D62 gene was amplified using genomic DNA extracted from d62 mutant and wild-type pants by using five pairs of primers, P1, P2, P3, P4 and P5 (Table 1). The amplified DNA fragments were cloned into pMD19-T vector (TaKaRa, Dalian, China) and sequenced.
RNA isolation and RT-PCR analysis
Total RNA was extracted from various rice tissues of wild type and d62 mutant by using RNAisoTM Plus (TaKaRa). The extracted RNA was treated with RNase-free DNaseI (Fermentas, Beijing, China) to eliminate genomic DNA contamination according to the protocols recommended by the manufacturer. The first strand of cDNA was synthesized from 4 μg of total RNA using oligo(dT)18 (TaKaRa) as primers. The first-strand cDNA products equivalent to 320 ng of total RNA were used as templates in a 20 μl PCR reaction system. To amplify the ORF region of D62 gene, the primers of DORF (forward: 5′-GAGGAGGTCTCTTCTTGGCACG-3′; reverse: 5′-CACCATTCATCGTCAGCTTAGCT-3′) were used in RT-PCR. The amplified cDNA fragments were sequenced as described above.
To investigate the effects of GA3 on the expression of D62 and GA biosynthetic genes, the seedlings at 5-leaf stage were incubated in 100 μM GA3 solution or control solution (distilled water) for 4 days and sprayed with the solution per day. After 4 days of incubation, total RNA was isolated from leaves as described above. For expression analysis of D62, RT-PCR was performed using the primers RT-D62 (Table 1). For expression analysis of the GA biosynthetic genes including OsCPS1, OsKS1, OsKO1, OsKAO, OsGA20ox2/SD1, OsGA3ox2/D18 and OsGA2ox3, the primers used in RT-PCR analysis are listed in Table 1. The rice Actin1 gene was used as an internal control in RT-PCR analysis.
Real-time qPCR analysis
Real-time qPCR was performed with SYBR Premix Ex Taq™ HS (Takara) on the Applied Biosystems 7500 Real-Time PCR System. The relative expression levels of each transcript were obtained by normalization to rice Actin1 gene. PCR was carried out with the two-step protocol as follows: activation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s and annealing/extension at 62°C for 34 s. For real-time qPCR analysis of the GA biosynthetic genes OsCPS1, OsKS1, OsKAO, OsGA20ox2/SD1 and OsGA3ox2/D18, the primers for each gene are listed in Table 1. The rice Actin1 gene was used as an internal control in real-time qPCR analysis.
Complementation experiment
A 6.1-kb DNA fragment containing the entire D62 coding region, 3,093 bp of the sequence upstream of the start codon and 1,153-bp downstream of the stop codon, was amplified from wild-type genomic DNA by HiFi DNA polymerase with the primers CT6.1 (forward: 5′-CTGGTGGTTTGGGGATTGGAGTTG-3′; reverse: 5′-CTGGAGATTTGGAGTGCGTGATGG-3′). The amplified fragment was cloned into the pMD19-T vector and sequenced, then digested with EcoRI and HindIII, and inserted into the binary vector pCAMBIA1301 to generate a transformation plasmid pCT6.1. The binary plasmid was introduced into Agrobacterium tumefaciens EHA105, and then transformed into d62 callus as described by Hiei et al. (1994). The empty vector pCAMBIA1301 was introduced into d62 callus as a control.
Promoter activity analysis
For the construction of the fusion between the D62 promoter and GUS coding sequence (D62 promoter::GUS), the 1.6-kb upstream of D62 gene was amplified from wild-type genomic DNA and inserted into the 5′-end of the GUS gene (gusA) in pCAMBIA1301 at the EcoRI and NcoI sites to create a transformation plasmid of GUS driven by D62 promoter. The primers used in PCR amplification were DP (forward: 5′-ATGAATTCCTACTCATCACCGTCGCATTTCTT-3′; reverse: 5′-AATCCATGGGACGTGCCAAGAAGAGACCTCCT-3′; EcoRI and NcoI sites are italicized, respectively). The plasmid was introduced into Agrobacterium tumefaciens EHA105, and then transformed into Nipponbare (Oryza sativa L. ssp. japonica; supplied by the College of Life Sciences, Zhejiang University) callus as described by Hiei et al. (1994). Transgenic plants harvested at different developmental stages were incubated with X-gluc buffer overnight at 37°C to ensure that they develop blue color (Jefferson et al. 1987). Green tissues were incubated with 70–100% ethanol at room temperature to remove chlorophyll before their observation under a dissecting microscope.
Phylogenetic analysis
Protein BLAST search was performed with the BLASTP program (http://blast.ncbi.nlm.nih.gov/). The deduced amino acid sequence was aligned using the Clustal X program (Thompson et al. 1997) with the default parameters. A phylogenetic tree was generated using MEGA4 (Tamura et al. 2007) with the neighbor-joining method. GenBank/EMBL/DDBJ and Swiss-Prot accession numbers for the genes mentioned in present study are as follows: D62 (BAC24836 or BAA90816), At RGA (CAA72177), At GAI (CAA75492), At RGL1 (AAL05911), At RGL2 (NP_186995), At RGL3 (NP_197251), At SCR (AAB06318), At SCL28 (AAG51600), Br RGA1 (AAX33297), Br RGA2 (AAX33298), Cs SCR (CAI30892), Hv SLN1 (AAL66734), La SCR1 (ACQ84011), La SCR2 (ACR48080), Ls DELLA1 (BAG71200), Ls DELLA2 (BAG71201), Os SCR (BAD22576), Os SLR1 (BAE96289), Os SLRL1 (AAR31213), Os SLRL2 (AAT69589), Pn SCR (BAE48702), Pp DELLAL1 (ABX10764), Pp DELLAL2 (ABX10765), Ps SCR (ABH85406), Pt GRAS18 (EEE84364), Pt GRAS19 (EEE78544), Rc GAI1 (EEF49399), Rc RGL1 (EEF51101), RHT-D1A (CAB51555), Sm DELLA1 (ABX10758), Vv GAI1 (AAM19210), Zm DWARF8 (CAB51557), Zm DWARF9 (ABI84226) and Zm SCR (AAG13663).
Results
Characterization of the d62 mutant
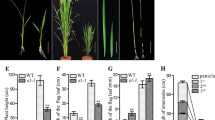
Before the 3-leaf stage, no obvious morphological differences could be observed between the d62 mutant and wild type. Beginning from the 4-leaf stage, the d62 mutant showed some characteristic phenotypes, such as dwarfism, abnormal leaf blade morphology, dark-green leaves, etc. (Fig. 1). Figure 1a–c displayed the gross morphology of d62 mutant at seedling stage (Fig. 1a), tillering stage (Fig. 1b) and grain-filling stage (Fig. 1c). After the heading stage, the mutant line reached ~66% of the height of wild type (Fig. 1c, Table 2). All internodes of d62 mutant were shorter than that of the wild type, but the first (uppermost) and second internodes of the mutant were less affected (Table 2). Compared with wild type, the d62 mutant showed reduced tiller numbers, late and asynchronous heading and partial male sterility. The leaf blades of d62 mutant were shorter and wider (Fig. 1d), and edges of leaf tips were more rounded than that of the wild type (Fig. 1e). In d62 mutant, crinkly leaf phenotype was observed after heading stage (Fig. 1f). The d62 mutant also showed smaller panicles and shorter rachis-branches (Fig. 1g). The grains of d62 became shorter and wider than those of the wild type (Fig. 1h), and increased with larger chalkiness was observed in its brown rice (Fig. 1i). In addition, the d62 mutant had short roots (Fig. 1j). These observations indicated that the d62 mutant had multiple morphological defects.
Morphological characterization of the wild type (left image of each part) and d62 mutant (right). a–c Gross morphology of the d62 mutant at seedling stage (a); tillering stage (b); grain-filling stage (c); d leaf blades; e leaf tips; f partial enlarged morphology of leaf blades; g panicles; h grains; i the brown rice observed under a X-ray viewer. Bars 2 cm (e, f), 5 cm (g), 10 cm (a, d, h, i, j), 15 cm (b), 20 cm (c)
Positional cloning of the D62 gene
To determine whether d62 was controlled by a single gene or multiple genes, in 576 F2 individuals from the cross between d62 mutant and japonica cultivar 02428, 440 wild-type individuals and 136 mutants were identified, respectively. The segregation of normal plants and mutants was fitted to a 3:1 segregation ratio (χ2 = 0.59 < χ 20.05 = 3.84). Also, all F1 hybrids derived from the crosses between d62 mutant and 93-11, or d62 mutant and 02428, showed a normal phenotype. So these results indicated that the d62 mutation was controlled by a single recessive nuclear gene.
To elucidate the molecular function of D62, the gene was isolated by positional cloning. The D62 gene was primarily mapped at the short arm of chromosome 6 by SSR markers RM19289 and RM19320 (Fig. 2a), and was subsequently fine mapped at an interval of 131 kb bracketed by SSR markers MM0124 and RM19320 (Fig. 2b). Using the rice Gramene database (http://www.gramene.org/) and Rice Annotation Project Database (RAP-DB) (http://rapdb.dna.affrc.go.jp/viewer/gbrowse/build4/), 20 putative genes were annotated in this region (Fig. 2c, Suppl. Table 1). Among them, the gene (IRGSP locus: Os06g0127800; TIGR locus: Os06g03710) which encodes a putative GA response modulator of GRAS transcription factor was specially focused in present experiment. The results of BLASTP search indicated that Os06g0127800 was homologous to DELLA and DELLA-like proteins of the GRAS family. In rice, the overexpression of DELLA-like protein SLRL1 induced a dwarf phenotype with increased levels of OsGA20ox2/SD1 gene expression (Itoh et al. 2005). These results elicited the possibility that Os06g0127800 was the D62 gene. In fact, recent findings indicate that Os06g0127800 is DLT/OsGRAS-32, which plays positive roles in BR signaling (Tian et al. 2004; Tong et al. 2009). The dlt mutant produces dwarf and low-tillering phenotypes and exhibits insensitivity or less response to BR application (Tong et al. 2009).
Isolation of D62 gene by positional cloning. a The D62 gene was primarily mapped on chromosome 6 based on the genotyping of 136 F2 mutant individuals. Marker names are indicated above the vertical lines and the numbers of recombinants are shown below the corresponding markers. The genetic distances (cent Morgan [cM]) between adjacent markers are indicated. b The D62 gene was fine mapped to an interval of 131-kb by genotyping of 546 F2 mutant individuals. Numbers of recombinants are shown below the corresponding markers. BAC contigs covering the D62 gene are shown below the linkage map. The physical distance of 131 kb was calculated from the map-based sequence published by IRGSP (2005). c In the 131 kb region, 20 putative genes were annotated in RAP-DB database. The candidate gene 8 (IRGSP locus: Os06g0127800; TIGR locus: Os06g03710) that encodes a putative gibberellin response modulator was suspected to be the D62 gene. d The structure of D62 gene and the mutation site. The start codon (ATG) and stop codon (TAA) are indicated. Black and gray boxes indicate the ORF region and untranslated region (UTR) of D62 gene, respectively. The d62 had a 2-bp deletion at the ORF region
The locus Os06g0127800 was therefore amplified from d62 mutant and wild-type plants by PCR and then sequenced. The results of DNA sequencing revealed that the d62 mutant had a 2-bp deletion of GC at the ORF region of locus Os06g0127800 (Fig. 2d). To confirm the mutation site, the ORF region of locus Os06g0127800 was amplified by RT-PCR. Sequencing of the cDNA fragment further confirmed the 2-bp deletion in the d62 mutant. These results indicated that the locus Os06g0127800 was the D62 gene.
The full-length cDNA of D62 (Genbank Accession: AK106449) was 3,084-bp in length containing an ORF of 1,854-bp; the 5′-UTR was 770-bp long, and the 3′-UTR was 460-bp long; the ORF was predicted to encode a polypeptide of 617-amino acids with calculated molecular mass of 65.85 kD and a pI of 6.02 (Suppl. Fig. 1). There was no intron in D62 gene, as is the case of most members of the GRAS family in rice and Arabidopsis (Tian et al. 2004). In the d62 mutant, a 2-bp deletion at the position +822–823 (start codon is position +1) of the ORF caused frame shift mutation (Suppl. Fig. 1).
Confirmation of the 2-bp deleted ORF as the d62 was achieved by complementation experiment. A genomic DNA fragment of 6.1-kb in length, including the entire sequence of the D62 gene, was introduced into d62 mutant with Agrobacterium tumefaciens-mediated transformation method (Hiei et al. 1994). Six independent transgenic lines were obtained by identifying the expression of GUS reporter gene (Suppl. Fig. 2). The mutative phenotypes of d62 were rescued in these transgenic plants (Fig. 3), which indicated that the locus Os06g0127800 was indeed the D62 gene and the d62 was a loss-of-function mutation in this locus. Based on the fact that Os06g0127800 was DLT/OsGRAS-32, these results also indicated that the D62 gene was the same locus as DLT/OsGRAS-32 and the d62 was allelic to the dlt mutant reported by Tong et al. (2009).
Phenotypic complementation by the introduction of D62 gene. Regenerated d62 mutant plants containing the empty vector pCAMBIA1301 (left) and pCT6.1 vector encompassing the entire D62 gene (right) are shown. Bar 20 cm. The image superimposed at the top middle is a close-up view of the leaf blades. Bar 10 cm
Phylogenetic analysis of D62 and its homologous proteins
The results of BLAST search indicated that the D62 protein shared homology (33–37% identity and 49–53% similarity) with DELLA or DELLA-like proteins, the repressors of GA signaling pathway. The D62 protein also showed homology (34–37% identity and 51–53% similarity) to SCR (SCARECROW) proteins, another GRAS subfamily involved in asymmetric cell divisions in plants (Di Laurenzio et al. 1996; Lim et al. 2000; Kamiya et al. 2003). Sequence analysis found that the N-terminal region of D62 protein contained no DELLA or TVHYNP motifs which were conserved in DELLA proteins, but the D62 protein shared GRAS domains with DELLA proteins, such as LHRI, VHIID, LHR µ, PFYRE and SAW motifs, with high similarity (Fig. 4a). The sequence alignment of D62 protein with DELLA-like and SCR proteins also showed that the N-terminal of D62 protein was highly variable, but the C-terminal of D62 protein was highly similar to those of DELLA-like or SCR proteins (Suppl. Figs. 3, 4). However, phylogenetic analysis demonstrated that the D62 protein could be categorized into a novel group distinct from DELLA, DELLA-like or SCR groups (Fig. 4b).
Sequence and phylogenic analysis of D62 protein. a Alignment of D62 and DELLA proteins. Full-length amino acid sequences of D62 and DELLA proteins from rice, Arabidopsis and wheat were aligned using the Clustal X program. Black and gray boxes indicate identical and similar amino acids, respectively. The lines above the alignment indicate the locations of the conserved regions in GRAS proteins as defined by Pysh et al. (1999). b Phylogenetic tree of GRAS proteins. The phylogenetic analysis was performed with the Clustal X program and phylogenetic tree was constructed using the MEGA4 program with the neighbor-joining method by 1,000 bootstrap replicates. Scale 0.1 nucleotide substitutions per site
Expression patterns of the D62 gene
To study the functions of D62, expression patterns of D62 gene were analyzed by using semi-quantitative RT-PCR. Total RNA was extracted from leaves, stems and roots at 5-leaf stage and young panicles of wild-type plants. The results showed that the D62 gene expressed in leaves, stems, roots and panicles (Fig. 5a). The expression levels in roots were lower than those in other tissues. For reconfirmation of the expression patterns, a vector of GUS driven by D62 promoter was constructed and introduced into rice plants. The GUS staining showed that Gus gene expressed in roots, stems, leaves and panicles of the transgenic lines (Fig. 5b–e). These results indicated that the D62 gene expressed in all tested tissues including roots, stems, leaves and panicles of rice plant.
Expression patterns of D62 gene. a Detection of D62 transcript by RT-PCR. Total RNA was isolated from roots, stems and leaves at 5-leaf stage, and panicles at heading stage of wild type. The rice Actin1 gene was used as a control. b–e GUS activity was detected in D62 promoter::GUS transgenic plants: lateral root (b), stem (c) and leaf blade (d) at tillering stage, and young spikelet (e)
Reduced α-amylase activity and GA1 contents in the d62 mutant
To investigate the possible relationship between D62 and the GA-related pathway, the shoot elongation and α-amylase activity induction by GA application, both of which are GA-mediated physiological processes, were examined in this experiment. To examine the GA-induction of α-amylase activity, embryoless half-seeds were placed on the starch plate with or without GA3 for 3 days, and the starch plate was stained with iodine. Production of α-amylase was observed as plaques both in wild-type and in d62 mutant on the plate containing GA3, but the activity of α-amylase was significantly decreased in the d62 mutant (Fig. 6). This indicated that the d62 mutant was related to the GA pathway.
GA-induction in shoot elongation was also analyzed in the d62 mutant. When the second leaf sheaths of d62 and wild-type plants were compared, the d62 seedlings were found to be normally responsive to exogenous GA3 treatment (Suppl. Fig. 5). The shoot elongation was triggered both in the d62 mutant and in the wild type by treatment with GA3 concentrations higher than 10−8 M. The elongation ratios were similar between d62 and wild-type plants, and the response curve in d62 mutant was also more or less parallel with that of wild-type plants. This indicated that the dwarf phenotype of d62 mutant was limited by factors independent of GA signaling.
For further characterization of d62, levels of endogenous GA1 (a major active GA in rice vegetative tissues) were measured in the d62 mutant and wild type. GAs were extracted from fresh leaf tissues at 4-leaf stage and measured for GA1 content with ELISA kit. The result showed that the levels of endogenous GA1 in d62 mutant were lower than those in wild-type plants (Fig. 7). This further confirmed that the d62 mutant was related to the GA pathway.
Increased expression of GA biosynthetic genes in the d62 mutant
To further investigate the relationship between the D62 gene and GA metabolic pathway, the expression of GA biosynthetic genes was examined by using semi-quantitative RT-PCR. First, two important genes involved in GA biosynthesis, the OsGA20ox2/SD1 and OsGA3ox2/D18, were analyzed and the expression of which was negatively regulated by the levels of GA in a feedback manner (Itoh et al. 2001; Sasaki et al. 2003). As expected, the expression levels of the two genes were decreased by GA3 treatment (Suppl. Fig. 6). Compared with the wild-type, the expression of OsGA3ox2/D18 was not affected in d62 mutant, but the expression level of OsGA20ox2/SD1 significantly increased in d62 mutant (Suppl. Fig. 6). The expression levels of other GA biosynthetic genes, including OsCPS1, OsKS1, OsKO1 and OsKAO were also examined by using semi-quantitative RT-PCR. The results showed that the expression levels of OsCPS1, OsKS1, OsKO1 and OsKAO significantly enhanced in the d62 mutant (Suppl. Fig. 6). The expression level of OsGA2ox3 which encodes GA 2-oxidase (GA2ox) was also analyzed. GA2ox proteins function in the control of GA levels by inactivating the bioactive GAs. The results from semi-quantitative RT-PCR showed that the expression level of OsGA2ox3 increased in d62 mutant (Suppl. Fig. 6). Furthermore, the expression levels of OsCPS1, OsKS1, OsKAO, OsGA20ox2/SD1 and OsGA3ox2/D18 were analyzed by real-time qPCR (Fig. 8, Suppl. Fig. 7), and the result showed that the mRNA level of OsGA3ox2/D18 in d62 mutant is slightly higher than that in wild-type plants (Suppl. Fig. 7). However, the mRNA levels of OsCPS1, OsKS1, OsKAO and OsGA20ox2/SD1 in d62 mutant were 4–8 times higher than those in wild-type plants (Fig. 8). These results indicated that D62 gene influenced the expression of GA biosynthetic genes.
Discussion
This investigation was carried out for the characterization of the rice mutant dwarf62 (d62) and the isolation of D62 gene. Under normal growth conditions, the d62 mutant produced many abnormal phenotypes throughout development, such as dwarfism, wide and short leaf blades, dark green and crinkly leaves, round leaf tips, reduced tiller numbers, short roots, late and asynchronous heading, partial male sterility, smaller panicles, shorter rachis-branches, etc. Genetic analysis indicated that the d62 mutation was controlled by a single recessive nuclear gene. The D62 gene was primarily mapped to the short arm of chromosome 6 by SSR markers RM19289 and RM19320, and was subsequently bracketed in the 131-kb region by newly developed molecular markers. Positional cloning of the D62 gene had shown that it encodes a GRAS protein homologous to DELLA, DELLA-like and SCR proteins of the GRAS (GAI-RGA-SCR) family. The D62 gene is the same locus as DLT/OsGRAS-32, which has been shown to act down-stream of the BR signaling pathway by Tong et al. (2009). However, several findings led toward the conclusion that the D62 gene was related to the pathway of GA metabolism in rice. First, the d62 mutant showed reduced activity of α-amylase in endosperm induction by GA application. Second, the d62 mutant had decreased levels of endogenous bioactive GA. Third, the expression levels of GA biosynthetic genes, including OsCPS1, OsKS1, OsKO1, OsKAO, OsGA20ox2/SD1 and OsGA2ox3 were up-regulated in the d62 mutant.
Positional cloning of the target gene revealed that D62 encodes a member of the GRAS family. Plant-specific GRAS proteins are suggested to play important roles in all aspects of plant growth and development (Pysh et al. 1999; Bolle 2004; Tian et al. 2004). As one of the most important subfamilies of the GRAS family, DELLA proteins are considered as the repressors in GA-related biological processes. In Arabidopsis, there are five distinct DELLA proteins (RGA, GAI, RGL1, RGL2 and RGL3), which have overlapping functions as repressors of GA signaling during GA-regulated plant growth processes, such as stem elongation, flower development and seed germination (Peng et al. 1997; Dill and Sun 2001; King et al. 2001; Lee et al. 2002; Cheng et al. 2004). Loss-of-function in any of these five genes shows a reduction in GA responsiveness (Silverstone et al. 1998; Wen and Chang 2002). In rice, SLR1 is considered as the sole DELLA protein suppressing GA signals and controls almost all GA-regulated events because SLR1 loss-of-function mutant shows the typical GA constitutive responses of slender phenotype (Ikeda et al. 2001). In addition, Itoh et al. (2005) identified two DELLA-like proteins (SLRL1 and -L2), which contain regions highly similar to the C-terminal GRAS domains of SLR1, but lacking the N-terminal DELLA domain that is unique to DELLA proteins. The overexpression of SLRL1 in wild-type rice plants produces a dwarf phenotype with increased expression levels of OsGA20ox2 and diminished the GA-induced shoot elongation (Itoh et al. 2005). It is suggested that SLRL1 functions as a repressor of GA signaling and prevents an excessive response to GA (Itoh et al. 2005). Another representative of GRAS family that has been well-characterized is SCR proteins, the members of which function as specifying asymmetric cell divisions throughout development and are especially crucial for root and shoot cell radial patterning (Di Laurenzio et al. 1996; Dolan 1997; Lim et al. 2000; Kamiya et al. 2003). Molecular analyses of D62 gene demonstrated that it encodes a GRAS domain protein homologous to DELLA, DELLA-like or SCR proteins. The D62 (OsGRAS-32) shared C-terminal GRAS domains with DELLA, DELLA-like and SCR proteins, with high similarity, but in fact the N-terminal of D62 protein was highly variable. However, phylogenetic analysis showed that D62 could be categorized into a novel group distinct from DELLA, DELLA-like or SCR groups. These results suggested that the D62 might play different roles in the control of development of rice plant.
Tian et al. (2004) performed a genome-wide analysis of the GRAS gene family in rice and Arabidopsis and identified a total of 57 GRAS genes from rice. Recently, Tong et al. (2009) reported the cloning of DWARF AND LOW-TILLERING (DLT), which is shown to play positive roles in BR signaling in rice. The dlt mutant is characterized by the production of dwarf and low-tillering phenotypes and insensitivity or less response to BR application (Tong et al. 2009). It is also shown that the transcription of BR biosynthetic genes and BR response genes is altered in dlt mutant (Tong et al. 2009). The DLT gene is mapped within the 1,310-kb region of chromosome 6 and confirmed to be the locus OsGRAS-32, which encodes a new member of the GRAS family (Tian et al. 2004; Tong et al. 2009). In the present study, positional cloning of D62 gene revealed that the D62 was the same locus as DLT/OsGRAS-32 and the d62 was allelic to the dlt mutant reported by Tong et al. (2009). However, the characterization of d62 mutant indicated that the D62 (DLT/OsGRAS-32) was related to the GA pathway. In the d62 mutant, reduced activity of α-amylase was observed in endosperm induction by GA application and decreased level of endogenous bioactive GA1 was also measured in leaves. Furthermore, the expression levels of GA biosynthetic genes increased in the d62 mutant. These observations suggested that the D62 influenced GA metabolism besides BR signaling. In fact, recent molecular studies have provided much evidence for interactions of GA and BR. Microarray analysis of cDNA library in rice treated with GA and BR has demonstrated some specific genes coordinately regulated by GA and BR (Yang et al. 2004). Recently, Wang et al. (2009) reported that OsGSR1, a member of the GAST (GA-stimulated transcript) gene family, plays important roles in both BR and GA pathways, and also mediates an interaction between the two signaling pathways in rice. The OsGSR1 is induced by GA and repressed by BR (Wang et al. 2009). The OsGSR1 RNAi transgenic rice exhibits a reduced sensitivity to GA treatment, an increased expression of the GA biosynthetic gene OsGA20ox2, and an elevated level of endogenous GA (Wang et al. 2009). Furthermore, the OsGSR1 RNAi transgenic plants also show a reduced level of endogenous BR and dwarf phenotypes similar to BR-deficient plants (Wang et al. 2009). Altered GA metabolism also exists in some BR-related mutants. For example, the expression levels of GA biosynthetic genes including the OsKAO, OsGA20ox1, OsGA20ox2/SD1 and OsGA3ox2/D18 increased in the rice BR-deficient brd1, and the high expression levels of the GA biosynthetic genes were suppressed by BR treatment (Mori et al. 2002; Komorisono et al. 2005). Komorisono et al. (2005) also characterized a dwarf mutant, dwarf and gladius leaf 1 (dgl1), which exhibits increased expression of the GA biosynthetic genes and only minimal response to GA or BR treatment. Bouquin et al. (2001) reported that, in Arabidopsis, the bri1-201 (BR-insensitive mutant) and cpd (BR-deficient mutant) displayed decreased expression levels of GA5 (GA20ox1). RNA-blot analysis revealed that BR and GA antagonistically regulate the accumulation of mRNAs of the GA-responsive GASA1 gene and the GA-repressible GA5 gene (Bouquin et al. 2001). In other words, BR may act as a positive regulator of GA 20-oxidation, a key step in GA biosynthesis. These results suggest that cross talk exists between these two important hormones, and that GA and BR modulate the expression of GA5 in Arabidopsis (Bouquin et al. 2001). In contrast, this may not be the case in pea as the GA20 levels are elevated in BR-deficient lkb mutant and application of BR to lkb plants reduced GA20 levels (Jager et al. 2005). These indicate that BR actually negatively regulates GA20 levels in pea (Jager et al. 2005). Even though a clear interaction exists between BR levels and the level of GA20, it is suggested that this interaction is not biologically significant in pea (Jager et al. 2005). Based on the evidence reported by Tong et al. (2009), it indicated that the d62 was a BR-insensitive dwarf mutant. However, we found that the expression levels of several GA biosynthetic genes, including OsGA20ox2/SD1, were elevated in the d62 mutant. This demonstrated that the BR-responsive D62 (DLT/OsGRAS-32) gene also influenced GA metabolism. In this respect, we propose that the D62 may mediate a cross talk between GA and BR in rice.
The inductions of α-amylase and shoot elongation by GA, both of which are GA-mediated control of physiological processes, are classical model systems for studying how GA acts (Lanahan et al. 1992; Matsukura et al. 1998). Although the d62 mutant exhibited reduced activity of α-amylase in endosperm induction by GA3 application, it was normally responsive to GA3 treatment for shoot elongation. These observations suggested that the dwarf phenotype of d62 mutant was caused by some factors independent of GA signaling. It was possible that the reduced α-amylase activity resulted from low background levels of endogenous GA in the d62 mutant. This conclusion was consistent with the result that d62 mutant had low level of endogenous bioactive GA1. Furthermore, the expression level of OsGA2ox3 was up-regulation in the d62 mutant. In rice, the OsGA2ox3 encodes GA2ox protein which functions in the control of GA levels by inactivating the bioactive GA, such as GA1 (Sakamoto et al. 2004). Thus, it was possible that the decreased level of GA1 was caused by the increased expression of OsGA2ox3 in the d62 mutant.
In higher plants, the final stage of bioactive GA synthesis, from GA53/GA12 to GA1/GA4, is catalyzed by GA20-oxidase (GA20ox) and GA3-oxidase (GA3ox) (Hedden and Phillips 2000). For the GA biosynthetic genes, OsGA20ox2/SD1 and OsGA3ox2/D18, the expression of which was negatively regulated by the levels of GA in a feedback manner (Itoh et al. 2001; Sasaki et al. 2003). As expected, the expression levels of the two genes were decreased by GA3 treatment. It was noteworthy that the expression level of OsGA20ox2/SD1 was significantly increased in d62 mutant. Although semi-quantitative RT-PCR analysis showed that the expression level of OsGA3ox2/D18 was not significantly altered in d62 mutant (Suppl. Fig. 6), the results of real-time qPCR analysis indicated that the expression level of OsGA3ox2/D18 in d62 mutant was slightly higher than those in wild-type plants (Suppl. Fig. 7). It was possible that the slight elevation of OsGA3ox2/D18 expression was caused by feedback regulation of low GA1 level in d62 mutant. Furthermore, the GA biosynthetic genes, OsCPS1, OsKS1, OsKO1 and OsKAO, the expression of which was independent of the levels of GA, were also significantly increased in d62 mutant. These observations indicated that D62 affected the expression of the GA biosynthetic genes. Based on these results, it was suggested that the D62 influenced GA metabolism in rice. The effect could be considerably down-stream of D62 (DLT/OsGRAS-32) and may be related to developmental changes.
In addition, RT-PCR analysis and promoter activity analysis showed that the D62 gene expressed in all tested tissues including roots, stems, leaves and panicles of rice plant. Consistent with the expression patterns of D62 gene, the phenotypic analysis also showed that the d62 mutant had many morphological disorders, such as dwarfism, abnormal leaves and roots, smaller panicles, reduced tiller numbers, late and asynchronous heading, partial male sterility, etc. These results indicated that the D62 gene was related to multiple processes of morphological development in rice.
Abbreviations
- bp:
-
Base pair
- BR:
-
Brassinosteroid
- GA:
-
Gibberellin
- GRAS:
-
GAI-RGA-SCR
- GUS:
-
β-Glucuronidase
- InDel:
-
Insertion/Deletion
- ORF:
-
Open reading frame
- SSR:
-
Simple sequence repeat
- UTR:
-
Untranslated region
References
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127:450–458
Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131:1055–1064
Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldman KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86:423–433
Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159:777–785
Dolan L (1997) SCARECROW: specifying asymmetric cell divisions throughout development. Trends Plant Sci 2:1–2
Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54:137–164
Gomi K, Matsuoka M (2003) Gibberellin signalling pathway. Curr Opin Plant Biol 6:489–493
Grennan AK (2006) Gibberellin metabolism enzymes in rice. Plant Physiol 141:524–526
Hedden P (2003) The genes of the Green Revolution. Trends Genet 19:5–9
Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5:523–530
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13:999–1010
Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M (2001) Cloning and functional analysis of gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA 98:8909–8914
Itoh H, Shimada A, Ueguchi-Tanaka M, Kamiya N, Hasegawa Y, Ashikari M, Matsuoka M (2005) Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J 44:669–679
Itoh H, Ueguchi-Tanaka M, Matsuoka M (2008) Molecular biology of gibberellins signaling in higher plants. Int Rev Cell Mol Biol 268:191–221
Jager CE, Symons GM, Ross JJ, Smith JJ, Reid JB (2005) The brassinosteroid growth response in pea is not mediated by changes in gibberellin content. Planta 221:141–148
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: betaglucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Kamiya N, Itoh J, Morikami A, Nagato Y, Matsuoka M (2003) The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J 36:45–54
King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159:767–776
Komorisono M, Ueguchi-Tanaka M, Aichi I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M, Sazuka T (2005) Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin-mediated microtubule organization causes up-regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiol 138:1982–1993
Lanahan MB, Ho TDH (1988) Slender barley: a constitutive gibberellin-response mutant. Planta 175:107–114
Lanahan MB, Ho TDH, Rogers SW, Rogers JC (1992) A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell 4:203–211
Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16:646–658
Lim J, Helariutta Y, Specht CD, Jung J, Sims L, Bruce WB, Diehn S, Benfey PN (2000) Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems. Plant Cell 12:1307–1318
Matsukura C, Itoh S, Nemoto K, Tanimoto E, Yamaguchi J (1998) Promotion of leaf sheath growth by gibberellic acid in a dwarf mutant of rice. Planta 205:145–152
Mitsunaga S, Tashiro T, Yamaguchi J (1994) Identification and characterization of gibberellin-insensitive mutants selected from among dwarf mutants of rice. Theor Appl Genet 87:705–712
Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, Hirochika H, Kikuchi S (2002) Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol 130:1152–1161
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
Olszewski N, Sun T, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14:S61–S80
Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194–3205
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F et al (1999) ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261
Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18:111–119
Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134:1642–1653
Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS et al (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416:701–702
Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299:1896–1898
Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10:155–169
Sun T, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55:197–223
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Tian C, Wan P, Sun S, Li J, Chen M (2004) Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol 54:519–532
Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C (2009) Dwarf and low-tillering, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58:803–816
Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y (2000) Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97:11639–11643
Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I et al (2005) Gibberellin insensitive dwarf1 encodes a soluble receptor for gibberellin. Nature 437:693–698
Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58:183–198
Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, Xu Z, Wang Z, Chong K (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57:498–510
Wen C, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14:87–100
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251
Yang Y, Xu C, Wang B, Jia J (2001) Effects of plant growth regulators on secondary wall thickening of cotton fibres. Plant Growth Regul 35:233–237
Yang GX, Jan A, Shen SH, Yazaki J, Ishikawa M, Shimatani Z, Kishimoto N, Kikuchi S, Matsumoto H, Komatsu S (2004) Microarray analysis of brassinosteroids- and gibberellin-regulated gene expression in rice seedlings. Mol Genet Genomics 271:468–478
Zhang Z, Deng Y, Tan J, Hu S, Yu J, Xue Q (2007) A genome-wide microsatellite polymorphism database for the indica and japonica rice. DNA Res 14:37–45
Zhu S, Gao F, Cao X, Chen M, Ye G, Wei C, Li Y (2005) The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol 139:1935–1945
Acknowledgments
This work was supported by National Natural Science Foundation of China (no. 31071394), Zhejiang Provincial Natural Science Foundation of China (no. Z3100089), National Key Project of GMO Breeding of China (no. 2008ZX08001-006), the Science and Technology Office of Zhejiang Province (no. 2007C12902), and the 151 Foundation for the Talents of Zhejiang Province. We thank Wanli Guo for providing the binary vector pCAMBIA1301. We also appreciate the efforts taken by Alfred Quampah and Mahmood Ul Hassan in revising the English of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
W. Li and J. Wu contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Fig. 1
cDNA and deduced amino acid sequence of D62 from cultivar 93-11 (wild type). The full-length cDNA consisted of 3,084 nucleotides containing an ORF of 1854-bp. The 5′-UTR was 770-bp long, and the 3′-UTR was 460-bp long. The ORF was predicted to encode a polypeptide of 617 amino acids. The 2-bp GC deletion of d62 allele is indicated 199 × 196 mm (300 × 300 DPI) (JPEG 2.05 MB)
Suppl. Fig. 2
Identification of transgenic plants by GUS reporter gene. The constructs consisted of GUS reporter gene driven by the CaMV 35S promoter. GUS staining of leaf blades from regenerated plants containing the empty vector pCAMBIA1301 (+), untransformed d62 mutant plants (−) and independent transgenic lines of d62 mutant plants containing pCT6.1 vector (1–6) are shown 199 × 89 mm (300 × 300 DPI) (JPEG 1.34 MB)
Suppl. Fig. 3
Alignment of D62 and rice DELLA-like proteins. Full-length amino acid sequences of D62, SLRL1 and SLRL2 from rice were aligned using Clustal X program. Black and gray boxes indicate identical and similar amino acids, respectively. The lines above the alignment indicate the locations of the conserved regions in the GRAS proteins as defined by Pysh et al. (1999) 199 × 103 mm (300 × 300 DPI) (JPEG 1.76 MB)
Suppl. Fig. 4
Alignment of D62 and SCR proteins. Full-length amino acid sequences of D62, OsSCR, ZmSCR and AtSCR were aligned using Clustal X program. Black and gray boxes indicate identical and similar amino acids, respectively. The lines above the alignment indicate the locations of the conserved regions in the GRAS proteins as defined by Pysh et al. (1999) 199 × 143 mm (300 × 300 DPI) (JPEG 2.91 MB)
Suppl. Fig. 5
Elongation of the second leaf sheath (a) and the seedling height (b) in response to GA3 treatment. The length of the second leaf sheaths and the seedling height were measured after 7 days when the wild-type and d62 seeds were germinated on 1/2 MS medium containing various concentrations of GA3. mean ± SD, n = 12, 150 × 71 mm (300 × 300 DPI) (JPEG 362 kb)
Suppl. Fig. 6
Expression analysis of the GA biosynthetic genes by RT-PCR. Total RNA was isolated from wild-type (WT) and d62 mutant plants treated with 10−4 M GA3 solution (+) or control solution (−). RT-PCR was performed using primers specific for each gene. The rice Actin1 gene was used as a control 99 × 103 mm (300 × 300 DPI) (JPEG 383 kb)
Suppl. Fig. 7
Expression analysis of the OsGA3ox2/D18 by real-time qPCR159 × 83 mm (200 × 200 DPI) (JPEG 152 kb)
Rights and permissions
About this article
Cite this article
Li, W., Wu, J., Weng, S. et al. Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta 232, 1383–1396 (2010). https://doi.org/10.1007/s00425-010-1263-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1263-1