Abstract
Mannans are hemicellulosic polysaccharides in the plant primary cell wall (CW). Mature seeds, specially their endosperm cells, have CWs rich in mannan-based polymers that confer a strong mechanical resistance for the radicle protrusion upon germination. The rupture of the seed coat and endosperm are two sequential events during the germination of Arabidopsis thaliana. Endo-β-mannanases (MAN; EC. 3.2.1.78) are hydrolytic enzymes that catalyze cleavage of β1 → 4 bonds in the mannan-polymer. In the genome of Arabidopsis, the endo-β-mannanase (MAN) family is represented by eight members. The expression of these eight MAN genes has been systematically explored in different organs of this plant and only four of them (AtMAN7, AtMAN6, AtMAN2 and AtMAN5) are expressed in the germinating seeds. Moreover, in situ hybridization analysis shows that their transcript accumulation is restricted to the micropylar endosperm and to the radicle and this expression disappears soon after radicle emergence. T-DNA insertion mutants in these genes (K.O. MAN7, K.O. MAN6, K.O. MAN5), except that corresponding to AtMAN2 (K.O. MAN2), germinate later than the wild type (Wt). K.O. MAN6 is the most affected in the germination time course with a t 50 almost double than that of the Wt. These data suggest that AtMAN7, AtMAN5 and specially AtMAN6 are important for the germination of A. thaliana seeds by facilitating the hydrolysis of the mannan-rich endosperm cell walls.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Zygotic embryogenesis is a critical developmental step in the life cycle of higher plants that can influence seed viability and germination potential when the hormone and environmental conditions are suitable (reviewed in Finch-Savage and Leubner-Metzger 2006; Finkelstein et al. 2008; Holdsworth et al. 2008; Matilla and Matilla-Vázquez 2008; Rodríguez-Gacio et al. 2009). Although, during seed development angiosperm seeds are all endospermic, the endosperm is obliterated at maturity in one-third of these species and the reduction of this tissue is accompanied by the use of cotyledons as reserve organs, as occurs in the Brassicaceae where the endosperm of mature seeds consists in many species of a single cell layer that completely encloses the embryo (Debeaujon et al. 2007; Linkies et al. 2010). In these seeds, the endosperm that remains alive at seed maturity is tightly associated with the external seed coat that undergoes a programmed cell death (Haughn and Chaudhury 2005; Debeaujon et al. 2007). The key role of these embryo-surrounding tissues in controlling germination time is coming into focus (Penfield et al. 2006; Bethke et al. 2007). Arabidopsis thaliana, as well as, other closely related species of the Brassicaceae such are Sisymbrium officinale and Lepidium sativum (Müller et al. 2006, 2009; Linkies et al. 2009; Iglesias-Fernández and Matilla 2010) germinates in two sequential steps: (1) initially, the seed-coat layer is ruptured and the radicle still remains covered by the endosperm; and (2) subsequently, after a lag of several hours, the rupture of the micropylar endosperm takes place and the embryo radicle emerges. While abscisic acid (ABA) does not affect the kinetics of the testa rupture, it specifically inhibits endosperm weakening; gibberellins (GA) act as ABA antagonists in a complex network integrating environmental signals (Toorop et al. 2000; Koornneef et al. 2002; Kucera et al. 2005; Matilla and Matilla-Vázquez 2008). Recently, Linkies et al. (2009) showed that ethylene (ET), via the CTR1-pathway, promotes endosperm weakening, counteracting ABA-inhibition through interference with ABA signaling without altering the ABA content.
Since the endosperm weakening is an important factor for radicle emergence in endospermic seeds, identification of endosperm-expressed genes, particularly those encoding cell wall (CW) hydrolytic enzymes is a first step for understanding the role of the endosperm in the control of germination (Dubreucq et al. 2003; Liu et al. 2005; Nonogaki et al. 2007). The enzymatic activity and the gene expression of several members of the endo-β-mannanases (MAN) gene family have been described in several plant species but it is in tomato seeds whose endosperm cell walls are particularly rich in mannans (60% of CW dry weight) where this family has been more deeply studied (Nonogaki et al. 2000; Gong and Bewley 2007; Schröder et al. 2009). One of these, LeMAN2, has been found to be expressed specifically in the micropylar endosperm prior to radicle emergence and MAN activity increases markedly in the remaining lateral endosperm following radicle protrusion (Nonogaki et al. 2000; Belotserkovsky et al. 2007).
To gain insight into the MAN function in germination, the expression of the eight members of the MAN family in A. thaliana is described and the four MAN genes which are expressed upon germination have been more systematically investigated. In order to localize the expression of these MAN transcripts, in situ hybridization analyses are performed in germinating seeds. These results indicate that the MAN transcripts are expressed in the radicle and in the micropylar endosperm before radicle protrusion, its expression not being detected after the radicle has emerged. Moreover, the analysis of T-DNA insertion mutants show that knock-out of three of these MAN genes present a later germination rate than the wild type (Wt) seeds. These data indicate that MAN expression is important for the germination process, probably by diminishing the mechanical resistance of the micropylar endosperm thus facilitating the emergence of the radicle.
Materials and methods
Plant materials
To propagate stocks of A. thaliana ecotype Columbia (Col-0; NASC, Nottingham University, UK) and T-DNA insertion mutants in this ecotype, we proceed as follows: vapor-phase (bleach 37% HCl, 15:1, v/v) sterilization of Arabidopsis seeds was done for 90 min before germination in Petri dishes containing 0.5× MS medium supplemented with vitamins and solidified with 0.8% agar. Immediately after plating, seeds were stratified for 3 days at 4°C and then incubated at 23°C under long-day conditions (16 h/8 h; light/darkness; light intensity 55 mol photons m−2 s−1) during 2 weeks and then transferred to pots in the greenhouse. Mature seeds for Wt and homozygous for T-DNA insertion mutants were stored at 21°C and 30% relative humidity, until used for germination assays.
The T-DNA insertion lines, obtained from NASC (Nottingham University, UK), were identified from the GABI-KAT (http://www.gabi-kat.de/) and from the Salk collection (Alonso et al. 2003), and they were: GABI_747H02 (K.O. for AtMAN7), Salk_129977 (K.O. for AtMAN6), Salk_016450 (K.O. for AtMAN2) and GABI_707G06 (K.O. for AtMAN5). The T-DNA mutants used were from GABI-Kat FST population (Rosso et al. 2003) and were gently provided by Bernd Weisshaar (GABI-Kat, MPI for PBR, Köln, Germany). Homozygous plants for the T-DNA insertion were selected by PCR using a gene specific primer and a primer derived from the left border (LBP) of the T-DNA (http://signal.salk.edu/tdnaprimers.2.html). For this purpose, a leaf of each line was collected and processed as described (Iglesias-Fernández and Matilla 2009). The sequences of the primers used for the PCR analysis are shown in Suppl. Table S1, and the amplified fragments were sequenced for confirmation of the location of the T-DNA.
Germination assays
Three replicates of 50 seeds were germinated in 90 mm Petri dishes on two layers of filter paper (Whatman No. 1) moistened with 3 ml of sterile water. Germination was done at 23°C in growth chambers under long-day conditions (16 h/8 h of light/darkness) and with light supplied by cool-white fluorescent lamps (light intensity 55 mol photons m−2 s−1). After-ripened seeds were neither surface-sterilized nor stratified at 4°C in order to avoid influencing their dormancy status, nevertheless fungal infections were not detected by light microscopy. Seeds were scored as germinated when the radicle emergence through the seed coat was visible under a magnifying lens. Germination tests were performed in three biological samples using three technical replicates.
Endo-β-mannanase (EC 3.2.1.78) activity assays
Triplicate lots of 0, 10, 20, 24, 30, 36, 42 and 48 h germinating non-stratified seeds were ground under liquid nitrogen and then extracted with 200 μl in 1 M sodium acetate buffer, pH 4.7 (Sigma-Aldrich Química, Madrid, Spain). After centrifugation at 20,000g at 4°C for 45 min, the supernatants were assayed in duplicate for MAN activity. For enzymatic determination, 100 μl of 0.25% (w/v) AZCL-galactomannan (Megazyme International Ireland Ltd, Wicklow, Ireland) in 100 mM sodium acetate buffer, pH 4.7 were mixed with 80 μl of supernatant and incubated at 37°C for 24 h, with constant agitation in an orbital shaker. Dye release from AZCL-galactomannan was determined spectrophotometrically by measuring the absorbance at 590 nm in supernatant samples of the reaction mixture. One unit of MAN activity was defined as the amount of enzyme that releases one nanomole of reducing sugar equivalent to d-mannose per second under the above conditions.
Genes and bioinformatic tools
The complete deduced amino acid sequences of the eight MAN genes in the A. thaliana genome were used to construct a phylogenetic dendrogram. The alignment of these sequences was carried out by means of the CLUSTAL W program (Thompson et al. 1994) prior to the phylogenetic analysis, that was done by the neighbor-joining method with the MEGA software, version 4.0 (Tamura et al. 2007), using a bootstrap analysis with 1,000 replicates, complete deletion and the Jones–Taylor–Thornton matrix, as settings.
The conserved motives within the deduced protein sequences of the eight MAN were established by means of the MEME program (http://www.meme.sdsc.edu/meme/meme.html), as described by Bailey et al. (2006). Default parameters were used with the following exceptions: the maximum number of motives to find was set to 50 and the minimum width of each motif was set to 6 amino acid residues. Consensus sequences follow the criteria of Joshi et al. (1997): a single capital letter is given if the relative frequency of a single residue at a certain position is greater than 50% and greater than twice that of the second most frequent residue. When no single residue satisfies these criteria, a pair of residues is assigned as capital letters in brackets if the sum of their relative frequencies exceeds 75%. If neither these criteria is fulfilled, a lower-case letter is given if the relative frequency of a residue is greater than 40%. Otherwise, x is given.
Signal peptide cleavage sites were predicted by using SignalP 3.0 available at the website http://www.cbs.dtu.dk/services/SignalP (Emmanuelson et al. 2007) and both pI and Mw were calculated using Compute pI/Mw tool available at the website http://www.expasy.ch/tools/pi_tool.html (Gasteiger et al. 2003).
The accession numbers for the cDNAs and corresponding predicted proteins of the Arabidopsis MAN genes described here are: AtMAN1 (At1g02310; NM_100112; NP_171733), AtMAN2 (At2g20680; NM_127632; NP_179660), AtMAN3 (At3g10890; NM_187700; NP_187700), AtMAN4 (At3g10900, NM_194561; NP_187701), AtMAN5 (At4g28320, NM_118972; NP_194561), AtMAN6 (At5g01930, NM_120271; NP_195813), AtMAN7 (At5g66460, NM_126044; NP_201447) and AtMANP (At3g30540, NM_113955).
Real-time quantitative PCR assays
RNA was purified from Arabidopsis siliques, flowers, rosette leaves and roots from adult plants (5 weeks), and from germinating seeds. Seeds were imbibed in sterile water for 0, 10, 20, 24, 30, 36, 42 and 48 h; three replicates of 90 seeds for each time point were collected in 2-ml tubes and immediately frozen in liquid N2 and stored at −80°C until RNA extraction. A grinding ball (stainless steel 0.7 mm) was added to the tubes, and material was finely ground in liquid N2 using a Mikro-Dismembrator-S (Sartorius, Göttingen, Germany) for 2 min at a shaking frequency of 1,500 min−1. Total RNA was isolated using the SV Total RNA isolation System (Promega, Madison, WI, USA) and its concentration was estimated by A 260 measurement. The cDNA was synthesised from 0.1 μg of total RNA using the First-Strand Synthesis kit for RT-PCR (Roche Diagnostics, Mannheim, Germany), using oligo-T as a primer and following manufacturer’s instructions. Samples were stored at −20°C until processed.
The specific primers used in the RT-qPCR analysis appear in supplementary Table S2 and 18S-RNA was used to normalize the data (ΔCT). The RT-qPCR was performed in an iCycler iQ™ Real-time Detection System (Bio-Rad Laboratories, Hercules, CA, USA). For each 25-μl reaction, 1 μl cDNA sample was mixed with 12.5 μl of IQTM SYBR® Green Supermix (Bio-Rad), 0.5 μl of each primer (12 μM, final concentration 240 nM), plus sterile water up to final volume. Samples were subjected to thermal-cycling conditions of 95°C for 4 min and 40 cycles (45 s at 95°C, 45 s at 60°C, 45 s at 72°C and 45 s at 80°C). This analysis was performed with three different biological samples for each time-point and each one was made by triplicate. The amplicon was analyzed by electrophoresis and sequenced for confirmation. Expression levels were determined as the number of cycles needed for the amplification to reach a threshold fixed in the exponential phase of the PCR reaction (CT) (Pfaffl 2001).
In situ hybridization analyses
The protocol described here is a modification of that described in Ferrandiz et al. (2000). Arabidopsis seeds were germinated on filter paper soaked with distilled water at 23°C and collected at 24 h. Fixation was in FAE solution (formaldehyde: acetic acid: ethanol: water, 3.5:5:50:41.5, by vol.) for 45 min under mild vacuum, and left for 2 days with gentle shaking at room temperature. The samples were dehydrated, through a graded ethanol series, and embedded in paraffin, sectioned to 8 μm and de-waxed. Previous to the hybridization protocol, some sections were stained with 2% (w/v) Toluidine Blue (Merck, Darmstadt, Germany) to verify tissue integrity (Fig. S1). A pre-hybridization treatment was performed, by incubating the sections in 0.2 M HCl, neutralizing, and digesting with 1 mg/ml proteinase-K. Then, tissue sections were again dehydrated in an ethanol dilution series, before applying the hybridization solution (100 mg/ml tRNA, 6× SSC; 3% formamide) containing approximately 100 ng/ml antisense or sense digoxigenin (DIG)-labeled RNA probes, corresponding to DNA fragments (200–300 pb) derived from the 3′-non coding regions of the Arabidopsis MAN genes (see Table S3), synthesized with the DIG RNA labeling mix, according to the manufacturer’s specifications (Roche Diagnostics). Hybridization was performed overnight at 52°C followed by two washes in 2× SSC and 50% formamide for 90 min at the same temperature. Antibody incubation and color detection was carried out according to the manufacturer’s instructions.
All observation were made on a Zeiss Axiophot fluorescence microscope and the images were captured with a Leica DFC 300FX CCD color camera under the Leica Application Suite 2.8.1 build 1554 acquisition software.
Results
The Arabidopsis endo-β-mannanase gene family
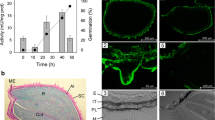
Our previous experience on the germination of S. officinale seeds had demonstrated a peak of MAN activity at 20 h, just prior to radicle protrusion, that decreased afterward (our data not shown). This result led us to explore whether a correlation would exist between MAN activity (EC. 3.2.1.78) and the time course of germination of the A. thaliana seeds. As shown in Fig. 1, dried seeds of A. thaliana have practically no detectable MAN activity but this enzymatic activity progressively increases to ~0.8 × 10−2 nKat/mg prot at 20–24 h. Then, this activity decreases at 30 h to increase again at 48 h when germination is almost completed, as evaluated by a visible protrusion of the radicle through the endosperm and seed coat.
Endo-β-mannanase activity (bars) of Arabidopsis thaliana seeds during and following germination. 50 mg of seeds were used for each time point analysis. Enzymatic activity in nKat was referred to milligrams of protein in the sample. Percentage of germinated seeds is indicated as black circle. Data are means ± standard error (SE) of three independent experiments
In order to get a deeper insight into the function of MAN during and after germination, it was decided to characterize at the molecular level the whole MAN family of A. thaliana. A non-redundant and complete compilation of the Arabidopsis MAN protein sequences was obtained from the TAIR database. A total of eight MAN, with calculated molecular weights between 45 and 51 KDa and all of them with predicted signal peptides were found (Table S4). These MAN proteins were used to construct a phylogenetic unrooted tree (Fig. 2a). Pair-wise amino acid similarities were higher than 50%, a threshold conventionally used to classify a group of genes as a gene family. Consistent with the un-rooted tree obtained by the neighbor-joining algorithm (Fig. 2a), two MAN classes were clearly defined (A and B), supported by bootstrapping values higher than 85% and the occurrence of common protein motives (see MEME analysis). AtMAN4, AtMANP*, AtMAN3, AtMAN7 and AtMAN6 integrate clade A and another group is formed by AtMAN2 and AtMAN5 (clade B). AtMAN1 cannot be included clearly in any of the two clades described above.
Deduced sequence analysis of the Arabidopsis thaliana endo-β-mannanase family. a Phylogenetic tree; bootstrapping values are indicated in the branches. b Schematic distribution of conserved motives among the deduced protein sequences in a that were identified by means of the MEME software. Asterisks indicate a putative pseudogene (AtMANP*: At3g30540) with a stop codon after motif 6 and a gene with expression levels in the detection limit (AtMAN1*: At1g02310)
The search for conserved amino acid motives was done using the MEME software (Fig. 2b, Table 1). The eight MAN putative proteins share motives 2, 6, 3, 1, 5, and 4 with the exception of AtMANP that probably is a pseudogene, due to an early stop codon found in position 255 (G → A) and consequently lacks the motives downstream from motif 6 (Fig. 2b). MAN proteins in clade A also share motif 8 and those in clade B share motives 9, 11 and 10; this last motif is also present in AtMAN6 that has been included in clade A. Finally, AtMAN1 has only the motives shared by all of the A. thaliana MAN proteins, suggesting that this enzyme could be the most primitive or ancestor of all of them. Conserved amino acids predicted as critic for mannanase activity (Yuan et al. 2007) are present in motives 6, 1, 5 and 4 (Table 1).
The expression pattern of the eight MAN genes was investigated by RT-qPCR in different Arabidopsis organs: dry seeds, siliques, flowers, rosette leaves and roots (Fig. 3). Data were normalized to the expression of the 18S-RNA gene. It was found that in siliques expression of AtMAN3, 7, 6, 2 and 5 was quite similar (~15–20 × 10−3/18S-RNA), while AtMAN4 was only barely detected, as occurs with the AtMAN3 transcript accumulation in the other tissues analyzed. The AtMAN7 gene was highly expressed in flowers and in roots and, less so, in leaves, being almost undetectable in dry seeds. AtMAN2 transcript accumulation was similar in leaves, roots and flowers and very scarce in dry seeds. AtMAN5 was the most abundant MAN transcript in dry seeds. Finally, AtMAN1* transcripts were barely detected in all tissues analyzed and AtMANP* was corroborated to be a pseudogene (our data not shown), as previously suggested by Yuan et al. (2007).
RT-qPCR global expression analysis of the endo-β-mannanase genes in different plant organs normalized to the expression of the 18S-RNA gene. Bars represent the standard error of three independent experiments. AtMAN4: At3g10900; AtMAN3: At3g10890; AtMAN7: At5g66460; AtMAN6: At5g01930; AtMAN2: At2g20680 and AtMAN5: At4g28320
Expression of endo-β-mannanase genes during seed germination
The six MAN genes that are ubiquitously expressed (Fig. 4) were studied in greater detail by RT-qPCR analysis during Arabidopsis seed germination which was completed at 48 h under our experimental conditions (see “Materials and methods”). Samples were taken out at 10, 20, 24, 30, 36, 42 and 48 h. Only four of these AtMAN genes are expressed upon seed imbibition: AtMAN7, AtMAN6, AtMAN2 and AtMAN5 (Fig. 4). The transcript accumulation of the four genes reach a peak between 20 and 30 h; AtMAN7, and AtMAN5 transcripts accumulation sharply decreases after 24 h, being practically undetectable at 48 h. A bimodal type of curve is observed for the expression of AtMAN6 upon germination with two peaks of maximum expression at 24 h and 42 h. However, AtMAN2 expression reaches a maximum at 30 h, decreasing slowly thereafter.
Transcript analysis by RT-qPCR of the four mannanase genes expressed during the germination of A. thaliana seeds (AtMAN7, AtMAN6, AtMAN2 and AtMAN5) at different hours (h). Error bars indicate standard deviations of three independent experiments. Arabidopsis seeds reach 100% germination (radicle protrusion) at 42 h in the conditions of the assay: 23°C and a photoperiod of 16 h light/8 h dark
AtMAN7, AtMAN6, AtMAN2 and AtMAN5 are expressed in the micropylar endosperm and in the radicle of A. thaliana seeds during seed germination
To determine the spatial expression of the MAN genes within the Arabidopsis germinating seeds, mRNA in situ hybridization experiments were done (Fig. 5). Samples, at 24 h, including seeds before and after radicle emergence, were hybridized to specific antisense probes for AtMAN7, AtMAN6, AtMAN2 and AtMAN5. In the longitudinal and transversal sections of the water imbibed seeds before radicle protrusion (Germinating), as shown in Fig. 5a–c, f–h, k–m, p–r, a strong signal is detected in the micropylar and lateral endosperm and in the incipient root vascular elements of the radicle, while it is barely detected in the cotyledons. Interestingly, this transcript accumulation in seeds disappears immediately after radicle protrusion (Germinated) (Fig. 5d, i, n, s). The strongest hybridization signal is detected with the antisense probe for AtMAN7 and AtMAN6, followed by that for AtMAN5, and the weakest is for that of AtMAN2. Since these transcripts are not detectable by the in situ mRNA hybridization in the radicle or endosperm after radicle protrusion, the expression of these genes in post-germination seedlings detected by RT-qPCR (Fig. 4), probably account for MAN expression in other tissues, such are the expanded cotyledon leaves, the hypocotyl or the root proper. As expected, no signal is detected, neither during nor after germination, on seed sections hybridized with the corresponding sense probes (Fig. 5e, j, o, t).
In situ mRNA hybridization analysis of AtMAN7, AtMAN6, AtMAN2 and AtMAN5 in 24 h seeds. AtMAN7 (a–e), AtMAN6 (f–j), AtMAN2 (k–o) and AtMAN5 (p–t): a, f, k, p longitudinal sections of germinating seeds; b, g, l, q close-up of the radicle tip and the micropylar zone in a, f, k, p. Cross-sections of germinating seeds (c, h, m, r) at the radicle tip zone and (d, i, n, s) longitudinal sections of germinated seeds (after radicle protrusion). e, j, o, t Control sense probes of germinating seeds. AL aleurone layer, ChE chalazal endosperm, Cot cotyledon, ME micropylar endosperm, SC seed coat, R radicle, VE vascular elements
Characterization and germination time course of knock-out mutants for AtMAN7, AtMAN6, AtMAN2 and AtMAN5
To address the question of whether the MAN genes expressed in the radicle and endosperm of water imbibed seeds, had an important functional role during the germination progress of Arabidopsis seeds, homozygous T-DNA insertion mutant lines, described in Fig. 6a, were selected: GABI_747H02 (K.O. for AtMAN7) that has a T-DNA insertion in the 5′UTR, 10 base pairs upstream from the ATG; Salk_129977 (K.O. for AtMAN6) that has a T-DNA insertion in the second exon (598 base pairs down-stream from the ATG); Salk_016450 (K.O. for AtMAN2) that has a T-DNA insertion in the fifth exon (1,856 base pairs down-stream from the ATG); and GABI_707G06 (K.O. for AtMAN5) that has a T-DNA insertion in the third exon (753 base pairs from the ATG). These lines have the T-DNA insertions upstream of one or more of the motives described previously as important for MAN activity (motives 6, 1, 5 and 4).
a Intron–exon schematic representation of the four endo-β-mannanase genes expressed in the germinating seeds. Position of the T-DNA insertion in each mutant line is shown with a triangle and its distance to the ATG initiation codon is indicated as the number of base pairs. LB left border, RB right border. Horizontal arrows represent the localization of the primers used for the RT-qPCR analysis. b In situ hybridization analyses of seeds at 24 h of T-DNA knock-out lines: AtMAN7 (GABI_747H02); AtMAN6 (Salk_129977); AtMAN2 (Salk_016450); AtMAN5 (GABI_707G06). AL aleurone layer, ChE chalazal endosperm, Cot cotyledon, ME micropylar endosperm, SC seed coat, R radicle. c Transcript analysis by RT-qPCR of the four MAN genes expressed in Wt and K.O. mutants in 24 h seeds normalized to the 18S-RNA gene
To check if these T-DNA insertion mutants are affected in the transcription of their corresponding genes, both RT-qPCR and in situ hybridization analyses were done. Transcripts are barely detectable by the in situ analyses (Fig. 6b) in germinating seeds of the K.O. lines for AtMAN7, AtMAN6, AtMAN2 and AtMAN5; and these data were corroborated by RT-qPCR expression analysis at 24 h in each of the four mutants (Fig. 6c).
To explore whether down-expression of the AtMAN7, AtMAN6, AtMAN2 and AtMAN5 affects the germination time course, germination assays were carried out. As it appears in Fig. 7: (1) no significant difference exists in the germination time course between the Wt line and the K.O. MAN2 line (Salk_016450); (2) K.O. MAN 7 (GABI_747H02) and K.O. MAN5 (GABI_707G06) germination is inhibited by 15–20%; and (3) the most affected mutant line in germination rate is the K.O. MAN6 (Salk_129977) reaching only 50% of germinated seeds at 48 h when compared with the control which reached 100% at this time point. Besides, the t 50 (time to reach 50% of germination) for different mutants was 25 h (Wt), 28 h (K.O. MAN2), 34 h (K.O. MAN7), 37 h (K.O. MAN5) and 48 h (K.O. MAN6), respectively (Fig. 7, inset).
Germination time course of Arabidopsis thaliana Wt and T-DNA insertion mutant seeds. Wt closed circles, K.O. MAN7 (GABI_747H02) opened triangles, K.O. MAN6 (Salk_122701) closed triangles, K.O. MAN2 (Salk_016450) closed squares, K.O. MAN5 (GABI_707G06) opened circles. Data are means ± standard error (SE) of three independent experiments. In the inset, the time necessary for 50% germination (t 50) is indicated for the Wt and the four mutants analyzed
The expression of the four MAN genes in 24 h imbibed seeds (Fig. 6c) showed that AtMAN7 transcript accumulation is one order of magnitude higher than that of the AtMAN6, 2 and 5 gene but the effect upon germination time of its corresponding knock-out line is of the same order of the knock-out line for AtMAN5 (t 50 for K.O. AtMAN7 = 34 h; t 50 for K.O. AtMAN5 = 37 h). AtMAN6 with an expression level of the same order than those of AtMAN2 and AtMAN5 seems to be the most important MAN gene for germination. Probably, this effect is not only to be attributed to its mRNA expression level but other functional aspects, such are its enzymatic characteristics, must be also involved.
Discussion
The data presented in this work support the idea that MAN expressed in the micropylar endosperm (ME) and in the radicle of germinating Arabidopsis seeds, are important for their germination. In this physiological process, ME weakening as well as expansion of the apical and sub-apical zones of the radicle are indispensable requisites for the radicle to protrude.
Two major forces play antagonistic roles in germination: the growth potential of the radicle and the mechanical resistance of the covering layers (seed coat or testa and endosperm); in order to complete germination, the growth potential of the radicle must overcome the ME resistance (Nonogaki et al. 2007; Rodríguez-Gacio et al. 2009). It has been suggested that the weakening of the endosperm CW, probably involving several types of hydrolytic enzymes, is required for radicle emergence (Sánchez and de Miguel 1992; Bewley 1997; Kucera et al. 2005; Finch-Savage and Leubner-Metzger 2006; Nonogaki et al. 2007; Holdsworth et al. 2008). Recently, it has been demonstrated that the elongation of embryo cells that provide the expansion force to complete germination in A. thaliana does not occur within the radicle itself, but rather in a discrete region that is immediately proximal to the radicle, spanning the lower hypocotyl and the hypocotyl–radicle transition zone (Sliwinska et al. 2009).
In the classical ‘hatching hypothesis’ of Ikuma and Thimann (1963) it was proposed that “… the final step in the germination control process is the production of an enzyme whose action enables the tip of the radicle to penetrate through the coat”. In looking for this ‘hatching enzyme’, evidences have been uncovered for the contribution of various CW-modifying enzymes, including MAN, endo-β-1,3-glucanases, chitinases, peroxidases, expansins and other proteins and their genes detected through the use of proteomics or transcriptomics (Bewley 1997; Leubner-Metzger and Meins 2000; Wu et al. 2001; Koornneef et al. 2002; Li et al. 2003; Bailly 2004; Kucera et al. 2005; Penfield et al. 2006; Linkies et al. 2009; Müller et al. 2010).
However, there are some questions to be elucidated in relation to the radicle emergence in endospermic seeds: (1) can the embryo itself generate enough expansion force to break the ME without the enzymatic degradation of its CW? If so, has the ME a predisposed structure to permit radicle protrusion? (2) are hydrolytic enzymes acting on CW of ME required for radicle protrusion? (3) is it necessary a cooperation between the expanding embryo and a weakened ME for the radicle protrusion? The CW of A. thaliana endosperm has a rigid structure with a considerable mechanical strength, due to their composition rich in hemicelluloses, such as mannans (galactomannans, glucomannans), xyloglucans and (1,3;1,4)-β-glucans. Degradation of galactomannans, abundant in seeds, is initiated by mannanases (Moreira and Filho 2008; Nonogaki et al. 2010).
In endospermic seeds of S. officcinale, it was demonstrated that the MAN activity of dry seeds was positively affected by dry after-ripening (i.e. ~12-fold more than in non-after-ripened seeds) and that MAN activity was considerably stronger prior to radicle emergence (Iglesias-Fernández and Matilla 2009). In contrast, MAN activity in A. thaliana seeds has a maximum at 20–25 h (Fig. 1). The contribution of the different seed tissues (endosperm, radicle, cotyledons, etc.) was difficult to study in both S. officinale and A. thaliana due to the small size of their seeds. Other authors detected MAN activity in tomato germinating seeds and this activity increased in the ME prior to radicle emergence (Nonogaki et al. 2000) and different MAN isoforms were reported to be expressed in the ME and in the embryo (Toorop et al. 1996). In lettuce seeds, there is no evidence of transcription or increase in enzymatic activity of these CW-degrading enzymes prior to radicle emergence, and degradation of the CW in the ME was not observed (Nonogaki and Morohashi 1999). A possibility would be that the ME in these seeds exerts less physical constraint for the radicle to protrude than in Arabidopsis and the MAN activity in lettuce endosperm could be associated with reserve mobilization (Wang et al. 2004).
In hard seeds, morphological differences seem to exist between the CW of the micropyle cells (thin) and those of the lateral endosperm (thick) cells (da Silva et al. 2004; Gong et al. 2005). In these seeds, the ME presents a lower physical constraint to the completion of germination than the lateral endosperm, and hence its structure is predisposed to permit radicle protrusion; the activity of MAN that increases only after radicle emergence, will have a role in hydrolyzing mannans to provide energy for the growing seedling. These observations are in contrast with the results in Arabidopsis seeds, where the endosperm is the constraining structure for radicle emergence (Finch-Savage and Leubner-Metzger 2006; Nonogaki et al. 2007).
In the genome of Arabidopsis, the MAN family is represented by eight members among which AtMAN7, AtMAN6, AtMAN2 and AtMAN5 are expressed in the dry seeds and are induced upon germination (Figs. 3, 4). These MAN genes expressed upon germination are only detected in the micropylar endosperm and in the radicle, before radicle protrusion, as shown by in situ hybridization experiments, the signal disappearing soon after germination was completed (Fig. 5). In tomato seeds two MAN genes have been characterized: LeMAN1 and LeMAN2. While LeMAN2 transcripts were detectable at 12–18 h (Nonogaki et al. 2000; Belotserkovsky et al. 2007), LeMAN1 was not expressed until germination was completed at about 48 h, these latter transcripts appeared in the lateral endosperm cells surrounding the cotyledons (Bewley et al. 1997). In short, the temporal and spatial expression pattern of LeMAN1 and LeMAN2 genes is strictly regulated between the germinative and post-germinative stages that are clearly separated by the occurrence of radicle protrusion. Recently, four putative MAN genes have been described in rice (OsMAN1, OsMAN2, OsMAN6 and OsMANP) and these were expressed during and following germination (Ren et al. 2008). As in the present work, DcMAN1 transcripts were also detected in both embryo and endosperm of carrot seeds, but the amounts were much higher in the endosperm than in the embryo, this being consistent with results from the activity assay (Homrichhausen et al. 2003).
To get new insight about the role of MAN enzymes during germination of A. thaliana seeds, the T-DNA insertion mutants of AtMAN7, AtMAN6, AtMAN2 and AtMAN5 have been analyzed: (1) the MAN gene expression by RT-qPCR in each of these mutants was almost undetectable, and this was corroborated by the in situ hybridization experiments, and (2) the germination time course was negatively affected in the K.O. MAN7 and K.O. MAN5 mutant seeds and strongly inhibited in those of K.O. MAN6, and these phenotypic differences were easily appreciated when the kinetics of germination were expressed as t 50 (Fig. 7).
Taken together, these results point to a possible cooperation between embryo and endosperm MAN activities, prior to radicle emergence in A. thaliana seeds. This assumption is made on the basis that there are not known physiological reasons nor contrasting results for the MAN activity to take place in the embryo because as in all growing organs, their CWs are not mannan rich. However, MAN expression is high in the vascular elements (Fig. 5) of the radicle, probably indicating a role in emptying the nascent conducting vessels that will ultimately develop into the vascular systems of xylem and phloem. An important point to be considered is that all the annotated MAN genes in Arabidopsis have predicted signal peptides, an indication that their corresponding proteins may be exported through the secretory pathway into the apoplastic space, thus reaching the CW of the ME, even if synthesized in the radicle. Thus, radicle and endosperm MAN together could contribute to the weakening of the ME. This assumption regarding the collaboration radicle–endosperm is not a new fact since it is well known the role of the embryo in the synthesis and export of gibberellins, and perhaps also cytokinins, to the endosperm in germinating seeds (Vicente-Carbajosa and Carbonero 2005; Arana et al. 2006). The transport of α-amylase induced by GAs from the aleurone layer towards the endosperm of cereals through the secretory pathway is a clear example of cooperation between different seed tissues. On the other hand, based on a recent view on MAN in plants (Schröder et al. 2009 and references therein), another way to explain the presence of AtMAN transcripts in the radicle could be that AtMAN has a dual enzymatic activity (e.g. hydrolase and transglycosylase). The transglycosylase activity would be involved in the expansion process of the cells as occurs in the radicle before protrusion, while the hydrolase activity would be more important in the loosening of the endosperm CWs. Fruit LeMAN4a was the first (and currently the only) enzyme identified that exhibits both MAN and mannan transglycosylase activity. However, its physiological role has not been yet clarified (Schröder et al. 2006). The dual characteristic of AtMAN is currently under study.
Abbreviations
- CW:
-
Cell-wall
- ECWE:
-
Enhancing cell wall extensibility
- MAN:
-
Endo-β-mannanase
- ME:
-
Micropylar endosperm
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Arana MV, de Miguel LC, Sánchez RA (2006) A phytochrome dependent embryonic factor modulates gibberellin responses in the embryo and micropylar endosperm of Datura ferox seeds. Planta 223:847–857
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acid Res 34:369–373
Bailly C (2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14:93–107
Belotserkovsky H, Berger Y, Shahar R, Wolf S (2007) Specific role of LeMAN2 in the control of seed germination exposed by overexpression of the LeMAN3 gene in tomato plants. Planta 227:199–209
Bethke PC, Libourel IGL, Aoyama N, Chung YY, Still DW, Jones RL (2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143:1173–1188
Bewley JD (1997) Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci 2:464–469
Bewley JD, Burton RA, Morohashi Y, Fincher GB (1997) Molecular cloning of a cDNA encoding a (1 → 4)-β-mannan endo-hydrolase from the seeds of germinated tomato (Lycopersicon esculentum). Planta 203:454–459
da Silva EAA, Toorop PE, van Aelst AC, Hilhorst HWM (2004) Abscisic acid controls embryo growth potential and endosperm cap weakening during coffee (Coffea arabica cv. Rubi) seed germination. Planta 220:251–261
Debeaujon I, Lepiniec L, Pourcel L, Routaboul J-M (2007) Seed development, dormancy and germination. In: Bradford K, Nonogaki H (eds) Seed coat development and dormancy. Blackwell, Oxford, pp 25–49
Dubreucq B, Berger N, Vincent E, Boisson M, Pelletier G, Caboche M, Lepiniec L (2003) The Arabidopsis AtEPR1 extensin-like gene is specifically expressed in endosperm during seed germination. Plant J 23:643–652
Emmanuelson O, Brunnak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971
Ferrandiz C, Liljegren S, Yanofsky M (2000) FRUITFULL negatively regulates the SHATTERPROOF genes during Arabidopsis fruit development. Science 289:436–438
Finch-Savage W, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171:501–523
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415
Gasteiger E, Gattiker A, Googland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acid Res 31:3784–3788
Gong XM, Bewley JD (2007) Sorting out the LeMANs: endo-β-mannanase genes and their encoded proteins in tomato. Seed Sci Res 17:143–154
Gong XM, Bassel GW, Wang A, Greenwood JS, Bewley JD (2005) The emergence of embryos from hard seeds is related to the structure of the cell walls of the micropylar endosperm, and not to endo-β-mannanase activity. Ann Bot 96:1–9
Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10:472–477
Holdsworth MJ, Soppe WJJ, Bentsink L (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179:33–54
Homrichhausen TM, Hewitt JR, Nonogaki H (2003) Endo-β-mannanase activity is associated with the completion of embryogenesis in imbibed carrot (Daucus carota L.) seeds. Seed Sci Res 13:219–227
Iglesias-Fernández R, Matilla AJ (2009) After-ripening alters the gene expression pattern of oxidases involved in the ethylene and gibberellins pathways during early imbibitions of Sisymbrium officinale L. seeds. J Exp Bot 6:1645–1661
Iglesias-Fernández R, Matilla AJ (2010) Genes involved in ethylene and gibberellins metabolism are required for endosperm-limited germination of Sisymbrium officinale L. seeds. Planta 231:653–664
Ikuma H, Thimann KV (1963) The role of the seed-coats in germination of photosensitive lettuce seeds. Plant Cell Physiol 4:169–185
Joshi CP, Zhou H, Huang XQ, Chiang VL (1997) Context sequences of translation initiation codon in plants. Plant Mol Biol 35:993–1001
Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5:33–36
Kucera B, Cohn MA, Leubner-Metzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15:281–307
Leubner-Metzger G, Meins F Jr (2000) Sense transformation reveals a novel role for class I β-1,3-glucanases in tobacco seed germination. Plant J 23:215–221
Li Y, Jones J, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6:603–610
Linkies A, Müller K, Morris K, Turecková V, Wenk M, Cadman CSC, Corbineau F, Strnad M, Lynn JR, Finch-Savage WE, Leubner-Metzger G (2009) Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21:3803–3822
Linkies A, Graeber K, Knight C, Leubner-Metzger G (2010) The evolution of seeds. New Phytol 186:817–831
Liu P-P, Koizuka N, Homrichhausen TM, Hewitt JR, Martin RC, Nonogaki H (2005) Large-scale screening of Arabidopsis enhancer-trap lines for seed germination-associated genes. Plant J 41:936–944
Matilla AJ, Matilla-Vázquez MA (2008) Involvement of ethylene in seed physiology. Plant Sci 175:87–97
Moreira LRS, Filho EXF (2008) An overview of mannan structure and mannan-degrading enzyme systems. Appl Microbiol Biotechnol 79:165–178
Müller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47:864–977
Müller K, Linkies A, Vreeburg RAM, Fry SC, Krieger-Liszkay A, Leubner-Metzger G (2009) In vivo cell wall loosening by hydroxyl radicals during cress (Lepidium sativum L.) seed germination and elongation growth. Plant Physiol 150:1855–1865
Müller K, Job C, Belghazi M, Job D, Leubner-Metzger G (2010) Proteomics reveal tissue-specific features of the cress (Lepidium sativum L.) endosperm cap proteome and its hormone-induced changes during seed germination. Proteomics 10:406–416
Nonogaki H, Morohashi Y (1999) Temporal and spatial pattern of the development of endo-β-mannanase activity in germinating and germinated seeds. J Exp Bot 50:1307–1313
Nonogaki H, Gee OH, Bradford KJ (2000) A germination-specific endo-beta-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol 123:1235–1245
Nonogaki H, Chen F, Bradford KJ (2007) Mechanisms and genes involved in germination “senso stricto”. In: Bradford K, Nonogaki H (eds) Seed development, dormancy and germination. Blackwell, Oxford, pp 264–304
Nonogaki H, Bassel GW, Bewley JD (2010) Germination—still a mystery. Plant Sci. doi:10.1016/j.plantsci.2010.02.010
Penfield S, Li Y, Gilday A, Graham S, Graham I (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination in the endosperm. Plant Cell 18:1887–1899
Pfaffl MW (2001) A new mathematical model for relative quantification in real time RT-PCR. Nucleic Acid Res 29:2002–2007
Ren Y, Bewley JD, Wang X (2008) Protein and gene expression patterns of endo-β-mannanase following germination of rice. Seed Sci Res 18:139–149
Rodríguez-Gacio MC, Matilla-Vázquez MA, Matilla AJ (2009) Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal Behav 4:1035–1048
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53:247–259
Sánchez RA, de Miguel L (1992) Phytochrome promotion of mannan-degrading enzyme activities in the micropylar endosperm of Datura ferox seeds requires the presence of embryo and gibberellin synthesis. Seed Sci Res 7:27–33
Schröder R, Wegrzyn TF, Sharma NN, Atkinson RG (2006) LeMAN4 endo-β-mannanase from ripe tomato fruit has dual enzyme activity and can act as a mannan transglycosylase and hydrolase. Planta 224:1091–1102
Schröder R, Atkinson RG, Redgwell RJ (2009) Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann Bot 104:197–204
Sliwinska E, Bassel GW, Bewley JD (2009) Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocoty. J Exp Bot 60:3587–3594
Tamura K, Dudley J, Nei M, Kumar C (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Toorop PE, Bewley JD, Hilhorst HWM (1996) Endo-β-mannanase isoforms are present in the endosperm and embryo of tomato seeds, but are not essentially linked to the completion of germination. Planta 200:153–158
Toorop PE, van Aelst AC, Hilhorst HWM (2000) The second step of the biphasic endosperm cap weakening that mediates tomato (Lycopersicon esculentum) seed germination is under control of ABA. J Exp Bot 51:1371–1379
Vicente-Carbajosa J, Carbonero P (2005) Seed maturation: developing and intrusive phase to accomplish a quiescent state. Int J Dev Biol 49:645–651
Wang AX, Li JR, Bewley JD (2004) Molecular cloning and characterization of an endo-β-mannanase gene expressed in the lettuce endosperm following radicle emergence. Seed Sci Res 14:267–276
Wu CT, Leubner-Metzger G, Meins F Jr, Bradford KJ (2001) Class I β-1,3-glucanases and chitinases are expressed in the micropylar endosperm of tomato seeds prior to radicle emergence. Plant Physiol 126:1299–1313
Yuan JS, Yang X, Lai J, Lin H, Cheng Z-M, Nonogaki H, Chen F (2007) The endo-β mannanase gene families in Arabidopsis, rice, and poplar. Funct Integr Genom 7:1–16
Acknowledgments
This work was financially supported by grants from Ministerio de Ciencia e Innovación (MICINN, Spain) (CGL2004-01996; CGL2009-11425 and Consolider CSD2007-00057). R I–F and C B–S are supported by post-doctoral contracts from CSD2007-00057 and JdC-UPM (BFU2006-07258), respectively. We thank B. Lueiro for expert technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Iglesias-Fernández, R., Rodríguez-Gacio, M.C., Barrero-Sicilia, C. et al. Three endo-β-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233, 25–36 (2011). https://doi.org/10.1007/s00425-010-1257-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1257-z